Abstract
Base excision repair (BER) can protect a cell after endogenous or exogenous genotoxic stress, and a deficiency in BER can render a cell hypersensitive to stress-induced apoptotic and necrotic cell death, mutagenesis, and chromosomal rearrangements. However, understanding of the mammalian BER system is not yet complete as it is extraordinarily complex and has many back-up processes that complement a deficiency in any one step. Due of this lack of information, we are unable to make accurate predictions on therapeutic approaches targeting BER. A deeper understanding of BER will eventually allow us to conduct more meaningful clinical interventions. In this review, we will cover historical and recent information on mammalian BER and DNA polymerase β and discuss approaches toward development and use of small molecule inhibitors to manipulate BER. With apologies to others, we will emphasize results obtained in our laboratory and those of our collaborators.
Keywords: Base excision repair, DNA polymerase β, PARP-1, Small molecule inhibitors, PARP inhibitors, Structural biology, Mouse models, DNA repair deficiency
Overview on historical aspects of mammalian BER
In every part of nature, cells contain multiple and overlapping DNA repair pathways that are essential for maintaining the integrity of genomic DNA. The DNA repair pathway known as base excision repair (BER) purifies genomic DNA of damaged nucleotides, abasic sites, and single-strand breaks arising from a variety of exogenous and endogenous sources. The BER pathway is found in organisms throughout nature, and the pathway in various organisms has similar features and enzymatic reactions, yet there are subtle, but absolutely key, differences from organism to organism. Both spontaneous and enzymatic removal of bases, through N-glycosidic bond cleavage, lead to the abasic site “lesion” that is repaired by BER. Deaminations of cytosine and adenine and oxidation of bases produce lesions that are repaired by the BER pathway. Base alkylations from endogenous alkylating agents (e.g., S-adenosylmethionine) and from exposure of cells to alkylating agents [e.g., methyl methanesulfonate (MMS)] are also sources of lesions and abasic sites that are repaired by BER. Perturbations of normal nucleotide metabolism can lead to incorporation of unnatural bases into DNA. Such DNA bases are often processed to the abasic site in prokaryotic and mammalian cells and eventually repaired by BER. BER is generally thought to accurately repair thousands of lesions per day in a mammalian cell.
BER was described initially in Escherichia coli as a pathway of sequential enzymatic steps initiated by spontaneous or enzymatic N-glycosidic bond cleavage (i.e., by DNA glycosylases) creating an abasic site in double-stranded DNA, termed the apurinic/apyrimidinic (AP) site [1–3]. In operational terms, the BER pathway has been distinguished from other DNA repair pathways by the relatively short excision gap generated in double-stranded DNA after removal of the lesion, and also by the fact that DNA lesions repaired by BER are generally limited to single nucleotide modifications that are similar in size and shape to the normal bases. The core steps of BER are base removal, strand cleavage at the AP site, processing of the incised strand providing a suitable substrate for the next steps, gap filling DNA synthesis, and DNA ligation. Since the DNA intermediates in BER are themselves cytotoxic, or lead to genomic instability via recombination, a sequential handoff from one step to the next during BER is likely. Reasonable models for explaining such step-to-step coordination in BER suggests action by multi-protein complexes carrying the necessary enzymatic machinery and accessory proteins or intimate step-to-step coordination so that the intermediates are sequestered [4]. However, evidence for such models is limited. Although nucleotide excision repair (NER) functions on bulky adducts, such as UV-induced dimers and benzo[a]pyrene adducts, NER can overlap with BER in repairing AP sites and a variety of single-base DNA modifications [5]. Similarly, mismatch repair and recombinational repair can overlap with BER and provide robust protection against base pair mismatches and strand breaks [5–7]. As yet, there is little understanding of potential mechanisms coordinating these three repair pathways when they are functioning simultaneously in the same cell.
For each step in BER, mammalian enzymes performing functions similar to the E. coli counterpart have been identified, cloned, and over-expressed as recombinant proteins. Many mammalian DNA glycosylases have been studied in this way, including the well-known proteins 3-methyladenine-DNA glycosylase (AAG), 8-oxoguanine-DNA glycosylase (OGG), uracil-DNA glycosylase (UDG) and endonuclease III-like DNA glycosylases. These DNA glycosylases recognize and remove altered bases, creating an intact or cleaved AP site in double-stranded DNA. For the monofunctional DNA glycosylases, the DNA product is the AP site itself. In many cases, however, DNA glycosylases are bifunctional, catalyzing lyase cleavage of the AP site and δ-elimination, liberating the deoxyribose group from the AP site. In addition, several mammalian “base mismatch” DNA glycosylases have been identified, purified, and characterized, including two uracil-DNA glycosylases (UDGs) and the GT mismatch-DNA glycosylase [8–10].
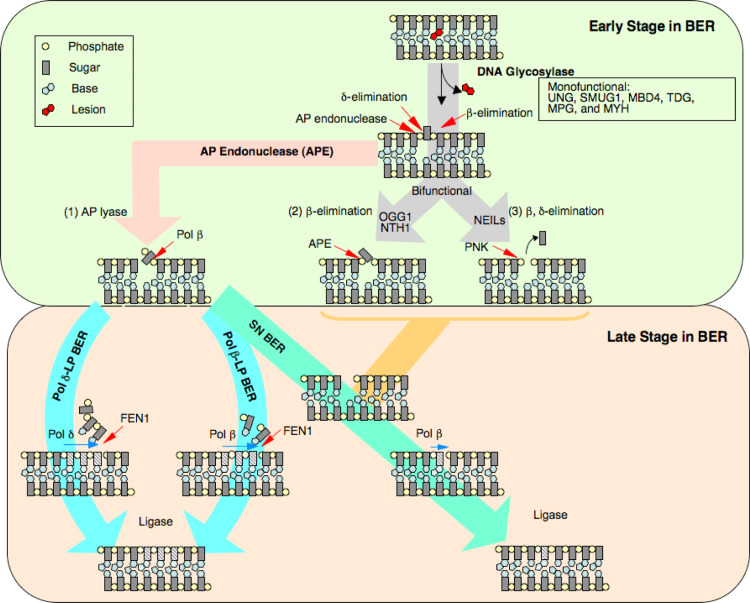
Even though the steps in the BER pathway are consistent with the activities of the cloned enzymes, there are many uncertainties concerning enzymatic specificity and step-to-step coordination in BER. Features of enzyme specificity and coordination of enzyme function that appear to lead to alternate sub-pathways of BER are illustrated in Fig. 1. For example, if the AP site is processed by a bifunctional DNA glycosylase with AP lyase activity followed by δ-elimination, the product will be a single-nucleotide gap with 3′-O-phosphate and 5′-phosphate at the margins of the gap. This gap will represent a dead-end product of BER if the 3′-O-phosphate is not further processed to liberate the 3′-OH necessary for DNA synthesis. When δ-elimination does not occur, as with OGG1 activity, the 3′-OH will be blocked by a phosphodeoxyribose or phosphoglycolate group. These dead-end intermediates of BER will accumulate unless the 3′-OH blocking groups are removed. Similarly, if the 3′-OH product of AP endonuclease (APE) or another enzyme is extended by a DNA polymerase and the 5′-deoxyribose phosphate (dRP) has not been removed, the possibility arises that the dRP group will be processed in a mechanistically different manner (e.g., leading to long-patch BER, recombination or cell death). As noted above, an interesting question is the extent to which coupling of the DNA glycosylase, APE, dRP lyase, DNA synthesis, and ligation steps could facilitate the overall BER pathway. Such coupling has been demonstrated for the APE and DNA polymerase steps [11, 12], and in addition, several BER protein complexes have been observed (e.g., DNA ligase I and Pol β, and Pol β and XRCC1/DNA ligase III) [13–15]. A further uncertainty is the extent to which various cellular isoforms and polymorphic variants of the BER enzymes can influence the capacity of the cell for conducting BER. Other uncertainties are also apparent, including the influence of chromatin/nucleosomal structure on BER efficiency and substrate specificities. Finally, recent studies have shown that BER occurs by distinct sub-pathways [16, 17] (Fig. 1). These are generally grouped into single-nucleotide (SN) BER and long-patch (LP) BER. Both of these sub-pathways are considered to have branch points that depend upon the initiating lesion and the expression level of individual BER proteins. Kinetic characterization of the BER sub-pathways in vivo is somewhat lacking, but is a prerequisite to understanding the roles of the sub-pathways in the cellular responses to genotoxic stress.
Fig. 1.
Working model illustrating early and late stages of SN and LP BER sub-pathways for the repair of uracil-DNA. Both sub-pathways operate in parallel, yet SN BER is considered to be more active. Several DNA polymerases are involved in each sub-pathway; this is not illustrated for SN BER. Regulation of DNA ligase activity is critical, as it can reverse the pathway after AP endonuclease strand incision, and ligase cannot function on a nick with a 5′-dRP group. In the presence of Pol β, removal of the dRP group is much faster than gap filling. From Ref. [16]
Overview on historical aspects of mammalian DNA polymerase β
Our research focuses on understanding BER in mammalian cells, and on an atomic level understanding of DNA polymerase β (Pol β) structure–activity relationships. Pol β is found in all vertebrate species as a ~39-kDa DNA polymerase lacking intrinsic nuclease activities such as 3′ or 5′ exonuclease and endonuclease, but containing 5′-dRP lyase activity [18–22]. In light of its size, this enzyme is considered the simplest naturally occurring cellular DNA polymerase, and thus provides an ideal model for studies of the DNA synthetic nucleotidyl transferase and dRP lyase mechanisms [21, 23]. Based upon the high level of sequence conservation of Pol β among mammalian species, it seems highly likely that Pol β is conducting a role that is essential for animal survival [24]. The discovery of a Pol β homologous sequence in yeast and our finding that a protein over-expressed in E. coli, from the open reading frame of the yeast Pol β-homologous gene, has Pol β-like enzymatic activity suggested that Pol β homologues occur throughout eukaryotes [25]. The yeast enzyme is referred to as Pol IV and is now known to be homologous to another X-family DNA polymerase identified in higher eukaryotic cells, Pol λ. Additional studies of yeast Pol IV were conducted in parallel by the Sugino group in Japan [26]. Earlier inhibitor-based studies implicated Pol β in some types of mammalian DNA repair, such as the repair of bleomycin damage. Other studies of HeLa cell nuclear extracts also implicated Pol β in gap-filling DNA synthesis during BER of monofunctional alkylation damage and in the repair of UV-damaged DNA, while studies using Xenopus laevis oocyte extract implicated Pol β in the repair of abasic site DNA lesions [27–29].
Although the inhibitor studies noted above implicated Pol β in BER and in other types of short gap-filling DNA repair [30, 31], the role of Pol β in the cell was widely considered as undocumented. Approaches toward such in vivo documentation were conducted in our laboratory and others and are still ongoing. These projects included knock-out experiments deleting the mouse Pol β gene [32] and the yeast homologue [33], anti-sense RNA experiments using a zinc-inducible stably transformed anti-sense gene in 3T3 cells [34, 35], and comparisons of DNA repair in cells expressing different levels of Pol β. Our laboratory also developed an in vitro BER assay system for correction of the G-U mismatch [36]. By virtue of spontaneous and enzymatic deamination of cytosine, this mismatch is a common form of genomic DNA damage repaired by BER and is mutagenic if not corrected. We found that a crude nuclear extract from bovine testis was efficient at conducting BER of the G-U lesion by insertion of dCTP opposite G. This reaction involved a single-nucleotide gap-filling DNA synthesis event and was catalyzed by Pol β in the extract. The extract was nearly devoid of gap-filling DNA synthesis when a neutralizing Pol β antibody was added, and we demonstrated that the replicative cellular DNA polymerases were relatively inefficient in single-nucleotide gap-filling synthesis [36]. Overall, the results of these various studies indicated a role for Pol β in mammalian BER, at least to the limits of the methodology at the time [22].
Mammalian Pol β was cloned and overexpressed in E. coli [37–40]. The recombinant proteins from human and rodent sources were fully active and well behaved in solution and possessed substrate specificity and catalytic properties similar to those of the natural enzymes [41, 42]. These recombinant proteins are monomeric (like their natural analogues), and the enzyme was found to have a slightly elongated structure in solution [43]. We performed detailed studies of Pol β domains by proteolytic and chemical cleavage of the enzyme under controlled conditions [44, 45]. Use of the purified domain fragments provided important information on the relationship between structural elements of the enzyme and various activities, such as single-stranded (ss) and double-stranded (ds) nucleic acid (NA) binding, nucleoside triphosphate (dNTP) binding, and the dRP lyase and nucleotidyl transferase catalytic activities [44, 46, 47]. The association of these properties with defined portions of the protein is consistent with the existence of tightly folded domains and subdomains linked by short solvent-exposed regions. Thus, the full-length enzyme consists of an amino-terminal lyase domain (8 kDa) connected by a short protease-sensitive segment to a carboxyl-terminal polymerase domain (31 kDa) (Fig. 2). NA-binding studies have shown that the purified 8-kDa domain binds ssNA with an affinity almost the same as that of the intact enzyme. However, unlike the full-length protein, the 8-kDa domain has little, if any, dsNA-binding activity. By contrast, the 31-kDa C-terminal domain has little or no affinity for single-stranded lattices, but binds dsDNA, such as a template-primer. The 31-kDa domain has nucleotidyl transferase activity, whereas the 8-kDa domain has dRP lyase activity [48, 49].
Fig. 2.
Domain and subdomain organization of DNA polymerase β. Pol β is composed of a polymerase (colored) and an amino-terminal lyase domain (grey). The polymerase domain is composed of three subdomains: D (purple), C (gold), and N (green) subdomains. These subdomains are involved in duplex DNA binding, nucleotidyl transfer, and binding of the nascent base pair, respectively. The lyase domain provides the enzymatic activity required to remove a 5′-dRP intermediate during BER. The panel on the right includes gapped DNA. The template strand (red) bends 90° as it exits the nascent base pair binding pocket. The templating (coding) nucleotide is shown with the incoming nucleotide (blue surface). The remaining nucleotides are omitted for clarity. From Ref. [21]
Crystallographic structures of Pol β in a variety of liganded states have been solved. In many instances, these structures identified molecular interactions that are involved in DNA-binding and/or binding of the correct dNTP [20, 50–59]. The structures of Pol β indicate that the enzyme has a modular domain organization (Fig. 2). The polymerase domain is typically composed of three functionally distinct subdomains. The catalytic subdomain coordinates two divalent metal cations that assist the nucleotidyl transferase reaction. The other subdomains have principal roles in binding duplex DNA and nascent base pair (dNTP and templating nucleotide) and are spatially situated on opposite sides of the catalytic subdomain. These subdomains are referred to as C– (Catalytic), D– (Duplex DNA binding), and N– subdomains (Nascent base pair binding) to highlight their intrinsic function [50]. These would correspond to the palm, thumb, and fingers sub-domains, respectively, according to the nomenclature that utilizes the architectural analogy to a right hand. In addition to the polymerase domain, DNA polymerases often have an accessory domain that contributes a complementary enzymatic activity necessary for the polymerase to fulfill its biological task. For example, the amino-terminal lyase domain of Pol β contributes a lyase activity required during BER in vivo [60].
In summary, Pol β is a multi-domain protein whose domains contribute distinct biochemical functions. Many crystal structures of Pol β have been solved in different liganded forms. Solution studies of Pol β and its domain fragments by NMR spectroscopy have provided a picture of the multiple interactions and conformations of Pol β with various ligands. Thus, we use both crystallographic and NMR approaches to determine the enzyme’s structure [54–58, 61–63]. This structural information is the basis for understanding the mechanism of Pol β at the atomic level and facilitates the understanding of DNA polymerase function in general. The information also facilitates design of small molecule inhibitors, as discussed below.
Insight on DNA polymerase mechanism
DNA polymerase fidelity or specificity represents the ability of a polymerase to select a correct dNTP from a pool of structurally similar molecules. Fidelity is quantified from the ratio of catalytic efficiencies for alternate substrates (i.e., correct and incorrect dNTPs). An analysis of the efficiency of dNTP (correct and incorrect) insertion for a low-fidelity mutant of Pol β (R283A) and published work from exonuclease-deficient DNA polymerases from five families derived from a variety of biological sources was conducted [50]. The analysis revealed that a strong correlation exists between the ability to synthesize DNA and the probability that the polymerase will make a mistake (i.e., base substitution error) (Fig. 3). Unexpectedly, this analysis indicates that the difference between low- and high-fidelity DNA polymerases is related to the efficiency of correct, but not incorrect, nucleotide insertion. Thus, low-fidelity DNA polymerases lack the ability to efficiently insert the correct nucleotide. Inefficient enzymes (i.e., poor correct nucleotide insertion efficiency) exhibit low fidelity, whereas efficient polymerases display high fidelity. Consequently, low-fidelity naturally occurring polymerases generally insert wrong nucleotides with an efficiency that is similar to, or lower than, a polymerase with higher fidelity. The observation that low- and high-fidelity polymerases insert incorrect nucleotides with similar efficiencies suggests that the ‘structure’ of the transition state for incorrect insertions may be similar for all DNA polymerases. In contrast, since fidelity is modulated by the efficiency of correct nucleotide insertion, the molecular interactions that contribute to efficient DNA synthesis (i.e., formation of the transition state) are dependent on the specific polymerase. More importantly, an understanding of fidelity at the molecular level requires structural insight into the polymerase-dependent attributes that contribute to correct insertion efficiency. A comparison of crystallographic ternary substrate complexes of DNA polymerases from five families exhibiting a range of nucleotide insertion rates reveals that the geometry of the nascent base pair-binding pockets are very similar and is consistent with the similar binding affinities reported for the correct nucleotide for these DNA polymerases [64]. Thus, the loss in catalytic efficiency for correct insertion by low-fidelity polymerases is due to a decreased rate of nucleotide insertion.
Fig. 3.
Relative misinsertion efficiency for formation of the dG–dTTP mispair as a function of catalytic efficiency for nucleotide insertion. The reported catalytic efficiencies for dCTP or dTTP insertion opposite guanine were analyzed for exonuclease-deficient polymerases that span five polymerase families: A-family filled circle; B-family filled square; RT-family filled triangle; X-family open circle; Y-family open square. The DNA polymerases included are T7 Pol (T7), Klenow fragment (Kf), Pol ζ (ζ), RB69 Pol (RB69), Pol X (X), rat and human Pol β (rβ and hβ, respectively) on non-gapped or gapped (g) DNA, R283A mutant of Pol β (βR283A), Pol η (η), Pol ι (ι), and Pol κ (κ). For some of these polymerases, several reported catalytic efficiencies are given. These represent alternate determinations from independent laboratories. The dotted line (f ins = 0.6) represents the calculated free energy difference determined for matched and mismatched terminal base pairs (ΔΔG ~ 0.3 kcal/mol at 37°C. The efficiencies for correct nucleotide insertion (k cat/K m,dCTP) span five orders of magnitude for the DNA polymerases surveyed with a corresponding decrease in the relative misinsertion efficiencies (f ins). From Ref. [64]
In light of the analysis in Fig. 3, the question of the molecular mechanisms surrounding control of DNA polymerase catalytic efficiency for a matched incoming nucleotide becomes paramount. The discovery of the structure of a Pol β ternary complex with the complete active site poised for chemistry was a step forward [65]. The structure was obtained using a non-hydrolyzable incoming nucleotide. This structure facilitated computational experiments that refined a working model for the nucleotidyl transferase reaction with matched incoming nucleotide [66, 67]. Additional structures were obtained using a similar approach, including a structure with a mismatched incoming nucleotide [68]. This structure revealed that the spatial arrangement of substrate and enzyme atoms in the active site was somewhat different than the arrangement in the structure with the matched incoming nucleotide. These results along with NMR experiments conducted in collaboration with R. London and associates (NIEHS, NIH) indicated that conformational changes necessary to assemble the complete active site observed with a matched incoming nucleotide failed to occur with the mismatched incoming nucleotide [68].
Adaptive response in BER
Even though mammalian cells maintain a constitutive level of “defense systems” against genotoxicants and endogenous genotoxic stress, cells are able to respond to such stress by altering gene expression and to enhance the defensive processes. In the DNA repair field, this type of response is known as the “adaptive response” from early work in both prokaryotic and eukaryotic systems. Several components of the BER system in mammalian cells have been recognized as being subject to an adaptive response. Thus, a picture emerged that mammalian BER is under dynamic regulation in the cell and that this may influence the relative efficiency of one BER sub-pathway versus another, or the rate of repair of an individual type of DNA lesion versus another. Such a picture of dynamic regulation of BER protein expression may explain a range of observations indicating significant differences in BER capacity across tissues and from cell type to cell type [5]. Our group initially observed up-regulation of Pol β gene expression in mammalian cells after exposure to a DNA alkylating agent or to an oxidative stress agent [69–71]. Such up-regulation of Pol β expression was later shown to protect cells against toxicant-induced cytotoxicity [69]. Similarly, deletion of the Pol β gene in the mouse is embryonic lethal, and mouse fibroblasts with a Pol β null genotype, although viable, are hypersensitive to alkylating and oxidative stress agents (that induces DNA damage repaired primarily by BER) [32, 72]. Transfection of Pol β expression vectors reversed the hypersensitivity phenotype of Pol β null cells such that the complemented cells were as resistant to stress as wild-type cells.
Coordination of steps in BER
Taken together, the results summarized thus far indicate the importance of BER and of Pol β in cellular protection against base lesion stress. Yet, there are many uncertainties about BER that confound our ability to predict the cellular response to genotoxic stress. For example, the extent to which the sub-pathways of BER can cross-complement each other is unknown, as is the biological consequence of such complementation. Similarly, other DNA repair pathways are able to complement a deficiency in BER, but the extent to which this occurs and the attendant biological implications (e.g., mutagenesis and cell death) are unknown. Recent progress included better definition of the two sub-pathways of BER that appear to operate on the same lesions in parallel [73]. The role of Pol β in both sub-pathways appears to have been established [74], especially regarding recent information on the LP sub-pathway [75]. In addition, several other DNA polymerases were found to play a role in BER, either in back-up functions (i.e., Pol λ) or in specialized capacities. The special features of oxidized base BER were revealed regarding tailoring of the gapped intermediate, such as that of polynucleotide kinase (PNK), now well established as a BER co-factor [76].
Using purified human Pol β and FEN1, the repair patch synthesis process for LP BER was shown to occur in a stepwise fashion of successive single-nucleotide gap forming steps by FEN1 and gap-filling steps by Pol β; we named this process the “hit and run” mechanism. The “hit and run” mechanism challenged the traditional paradigm for LP BER, and has important implications. This is because the alternate model involving strand-displacement of a long single-stranded molecule makes way for strand slippage, hairpin formation and frameshift mutagenesis. One form of such mutagenesis is trinucleotide repeat (TNR) expansion, where the “hit and run” mechanism for LP BER is circumvented by strand slippage and hairpin formation and eventual ligation of the slipped strand [77].
A highlight has been the observation that PARP-1 has the capacity to bind to the AP site-containing BER intermediate and also to facilitate enzymatic steps in BER, identifying PARP-1 as a critical component of the BER machinery, as illustrated here. This scheme is intended to emphasize the important notion that the abundant PARP-1 protein is involved in early stages once BER is initiated.
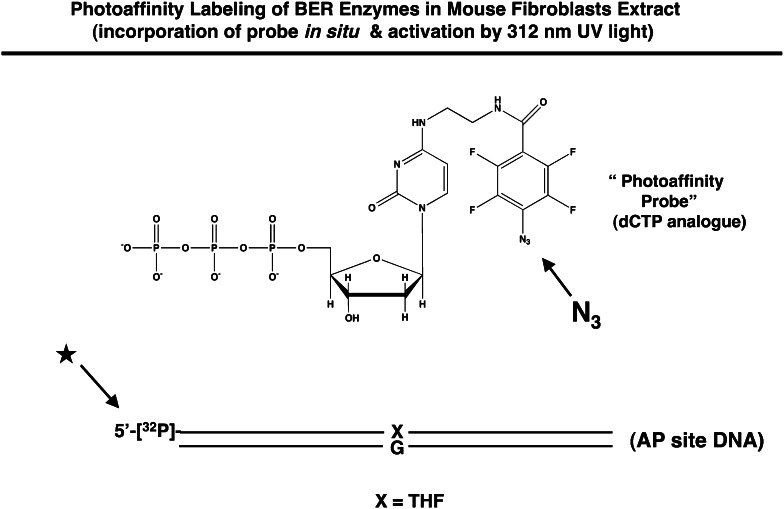
To establish this concept, we evaluated BER factors in the cell extract. We used gentle photoaffinity cross-linking and a labeled oligonucleotide representing a BER intermediate. This cross-linking probe is illustrated below. The symbol X represents the abasic site analog tetrahydrofuran, and after APE incision the photoaffinity probe is incorporated in situ into the BER intermediate. The extract is then exposed to low-energy UV light, activating the azido (N3) cross-linking group. The approach was developed by O. Lavrik and associates (Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia), and our experiments have been conducted in collaboration with this group.
Using extract from Pol β null mouse fibroblasts expressing flag-tagged Pol β, we found that PARP-1, FEN-1, APE, HMGB1 and Pol β were labeled by this highly sensitive photo-probe as illustrated here [78]. The protein bands were identified by immunological and peptide chemical techniques.
Our interpretation of the results was that these proteins in the crude extract bind to the BER intermediate, are in proximity to the affinity probe, and hence are cross-linked. Initially, we were surprised and impressed to observe that PARP-1 was by far the most abundantly labeled product. Interestingly, the robust labeling of PARP-1 depended on the presence of the 5′-sugar phosphate group in the BER intermediate, as illustrated below [79]. Only a minor amount of PARP-1 was cross-linked in the absence of the 5′-dRP group, and in addition, APE labeling strictly depended on the presence of the 5′-dRP group.
Based upon this lead concerning the importance of the 5′-dRP group, we investigated the possibility that PARP-1, under certain circumstances, could remain bound to the 5′-dRP-containing intermediate and promote LP BER in combination with other LP BER factors. Indeed, in the presence of FEN-1, PARP-1 is capable of stimulating LP BER DNA synthesis, and this stimulation depended on the presence of the 5′dRP group in the BER intermediate. The current working hypothesis is that APE and PARP-1 are able to bind to and incise the AP site. Thus, after cutting the DNA backbone 5′ to the AP site, APE and PARP-1 remain bound at the 5′dRP group until Pol β removes it or until LP BER is initiated [79]. These findings place PARP-1 at the center of events as the AP site is processed. This information has stimulated research on the importance of PARP-1 in BER and on the possibility that PARP-1 could serve as a regulatory molecule in the DNA damage response system.
In collaboration with A. Yasui and associates (Tohoku University), we made use of novel microscopic imaging techniques for definition of dynamic aspects of the recruitment of BER factors at sites of oxidative DNA damage in living cells. The approach revealed rapid recruitment of oxidized base DNA glycosylases and the core BER factors Pol β, XRCC1, and PARP-1 [80]. A partnership between Pol β and XRCC1 in the recruitment also was revealed. In addition, we recently made use of an alternate system for microscopic imaging, but in this case the damage and BER corresponded exclusively to LP BER [75]. We demonstrated that Pol β was involved in the synthesis step in this LP BER system and that Pol β and FEN1 were immediately recruited to focal sites of DNA damage and LP BER [75].
HMGB1 is a recently described BER co-factor. This well-known DNA-binding and non-histone chromosomal protein was found to be a modulator of BER steps: it stimulates both APE activity and FEN1 activity, probably through its known DNA bending effect on the respective BER intermediates. Steps in the LP BER sub-pathway that are modulated by HMGB1 were revealed using several approaches [77]. Knowledge of the interaction of HMGB1 with the BER intermediates will be an important consideration in future research.
Efficiency of and step-to-step coordination of BER
In recent unpublished experiments, it was determined that the three steps in the “late stage” of SN BER (dRP lyase, gap-filling synthesis, and ligation) are conducted in a processive fashion by purified Pol β and DNA ligase. In other words, the product of one reaction was passed to the next reaction without cycling into the solution compartment of the reaction mixture. This was the first direct evidence for the “passing the baton” model in BER that was proposed several years ago [4]. Direct transfer of the product of an enzymatic step to the next enzyme where it serves as the substrate is referred to as “substrate channeling.” This phenomenon of substrate channeling appears to explain how BER is conducted while sequestering the toxic intermediates of BER from cell signaling and recombination processes. Thus, we have recently shown that SN BER can proceed from an intact AP site to a ligated product through channeling (unpublished observations).
In collaboration with M. Smerdon and associates (Washington State University), we have developed a program to evaluate the influence of nucleosomes on BER. Chromatin structure obviously presents a challenge to BER enzymes, yet this topic is an open question in the BER field and has not received much attention to date. Nevertheless, the topic represents an important consideration in understanding hot spots for genetic instability. We found that Pol β activity in in vitro BER of uracil-DNA is completely inhibited in nucleosomal substrates, whereas both uracil-DNA glycosylase and APE activities are functional on nucleosomes, albeit at a lower rate than on naked DNA substrates [64]. These results suggested that the first or “early steps” in BER of uracil-DNA may be stimulated by nucleosomal remodeling activity, but the gap-filling synthesis step by Pol β absolutely requires remodeling [81]. Nucleosomal remodeling, therefore may be an important regulatory event for BER efficiency and for accumulation of BER intermediates. This is especially interesting, in view of the well-known ability of PARP-1 to poly(ADP-ribosyl)ate nucleosomal histones, triggering nucleosomal remodeling, as pointed out in a recent review by Rouleau et al. [82]. It is also interesting that well-known PIKK kinases, such as ATR, phosphorylate histone H1A after DNA damage, and this is associated with remodeling.
Studies of the enzymatic activities of BER enzymes have indicated that functionally significant partnerships may occur through interactions in multi-protein complexes. Changes in enzyme activity as a function of complexes could have significance for the overall rate of BER and also for the accuracy of the repair patch synthesis. Thus, stimulation of the APE step by HMGB1 could increase the rate of BER. In a similar fashion, stimulation of APE by Pol β/DNA ligase I could have functional significance in cases where the activity of APE is weak, such as with the blocked 3′-O group generated by bifunctional DNA glycosylases, or by agents such as bleomycin and ionizing radiation since products of such incision, 3′-O-phosphate and 3′-O-phosphoglycolate are poor substrates for APE. Such stimulation of APE could be sufficient to change it from a rate-limiting activity in BER on these substrates to a non-rate-limiting activity. Earlier, a bovine testis nuclear extract [15] had been examined for BER-proficient macromolecular complexes using affinity columns, and physicochemical techniques such as velocity sedimentation. We found that the extract contained a complex or “repairosome” that was proficient in SN BER of uracil-DNA [15]. In current experiments, we used immuno-affinity chromatography to isolate Pol β-associated BER proteins in extracts from mammalian cells in culture. Using mass spectrometric analysis, the Pol β-associated fraction or “interactome” was found to contain a set of core components that are active in the late stage of single-nucleotide BER: Pol β, XRCC1, DNA ligase III, PNK, PARP-1, and Ku70/80 (unpublished observations). Interestingly, the interactome does not contain APE1, HMGB1, FEN1, or any of the DNA glycosylases.
We have been impressed that XRCC1 deficiency in mouse fibroblasts is associated with greater cellular hypersensitivity to the monofunctional DNA alkylating agent MMS than the deficiency in Pol β [83]. This marked hypersensitivity points to a deficiency in BER, yet the role of XRCC1 in BER is not at all apparent, as XRCC1 has no enzymatic activity and has had only a modest, if any, influence on BER enzymes in BER reactions in vitro in our hands. Biochemical experiments demonstrating protein–protein interaction between XRCC1 and Pol β have been conducted by others and by our group [84, 85]. The domains in XRCC1 have been determined, and characterization of the domain–domain interaction in the XRCC1–Pol β complex has provided information on the specificity and mechanism of binding [84]. Residues in Pol β that interface with XRCC1 in the binary complex have been determined by NMR. Since XRCC1 is known to interact with other BER proteins, it could, therefore recruit several factors to the BER reaction. Experiments to understand the role of XRCC1 as an accessory factor in BER are underway.
DNA damage signaling
Experiments are underway to explore the hypothesis that cellular differences in BER can influence susceptibility to environmental and endogenous genotoxic agents and hence have public health significance. Even though cells maintain a constitutive level of BER “defense proteins” against environmental and endogenous genotoxicants, they are able to respond to such agents by altering their pattern of gene expression so as to adapt to stresses. Interestingly, such adaptation in expression of BER components must be coordinated across all of the components participating in the BER pathway, because it appears that imbalanced expression of one component, like Pol β, can impose a dominant negative effect on the entire pathway. For example, we used this concept to explain the negative effects of gross Pol β over-expression in the mouse cataract model developed earlier in our laboratory [86]. This idea is consistent with the many shared protein–protein interactions for BER components: where there is strong over-expression of one factor, it can easily be envisioned that dead-end complexes are being formed with other factors and with BER intermediates.
Another example of an adverse effect of a BER imbalance or lack of coordination is the extreme cell killing effect of PARP inhibition during MMS-induced BER (Fig. 4). In cells exposed to a DNA-damaging agent such as MMS, PARP is rapidly activated by virtue of binding to strand break intermediates of BER and the auto-(ribosyl)ated PARP is believed to recruit other DNA repair proteins to the site of damage. In the presence of an inhibitor of PARP activity, PARP is expected to remain bound to DNA and BER will be blocked.
Fig. 4.
Extreme hypersensitivity of both wild-type and Pol β null cells to the DNA methylating agent MMS (1 h exposure) when treated for 24 h in the presence of the PARP inhibitor 4-amino-1,8-naphthalimide (4-AN). From Ref. [83]
Both wild-type and Pol β null cells are extremely sensitized to MMS and other DNA methylating agents, such as the chemotherapeutic triazene derivative temozolomide, in the presence of the potent PARP inhibitor 4-AN (e.g., see Fig. 4). In wild-type cells, a non-toxic concentration of MMS (0.25 mM for 1 h) combined with a non-toxic concentration of 4-AN (10 μM for 24 h) results in >99% cell killing by apoptosis [87, 88]. We observed that this sensitivity to the PARP inhibitor requires its exposure during the S-phase of the cell cycle. We have chosen to investigate the mechanism of this cell killing effect to learn more about linkages between stalled BER and cell cycle arrest and apoptosis, and also because of the importance of therapeutic approaches involving PARP inhibitors. Using human fibroblasts capable of expressing a kinase-dead form of ATR, treatment with a combination of MMS and 4-AN was shown to result in an ATR-dependent S-phase checkpoint [89]. The accumulation of S-phase cells was accompanied by phosphorylation of the effector kinase Chk1. In the absence of ATR-mediated signaling, MMS plus 4-AN resulted in a G2/M arrest. We also found ATR to be a substrate for poly(ADP-ribosyl)ation by PARP-1 [90]. A more recent study demonstrated the involvement of a second PIKK kinase, ATM. Loss of ATM, or of its downstream effector kinase Chk2, limited duration of the MMS plus 4-AN S-phase delay [91]. Chk2 phosphorylation that was still observed in the absence of ATM appeared to be dependent on both ATR and DNA-PK. The working model for research in this area is as follows: (1) stalled BER intermediates containing a strand break and dRP group are cytotoxic to cells in culture, (2) the stalled BER intermediates bind activity-inhibited PARP protein at the strand break, (3) extreme toxicity occurs by virtue of DNA replication-dependent DSB formation and attendant cell signaling, as illustrated in this scheme.

PARP-1 −/− mouse fibroblasts and isogenic wild-type cells also have been examined. We were interested to compare results in the PARP-1 −/− cells with those obtained in wild-type cells with the PARP inhibitor 4-AN. PARP-1 −/− cells were only moderately sensitive to MMS compared with the PARP-1 +/+ cells, and this was a quite different picture from the extreme sensitivity to MMS in the presence PARP-1 inhibitor. These data indicated that the inhibited PARP protein plays an active role in cell killing, or in other words, the absence of PARP-1 is not equivalent to the presence of inhibited PARP-1 [87]. Interestingly, in the absence of PARP-1, cells treated with MMS plus 4-AN bypass the S-phase delay and arrest directly in G2/M suggesting that the presence of inhibited PARP is a critical component of this model. Further, DSBs could be readily measured in the wild-type cells treated with MMS plus 4-AN, but not in the PARP-1 −/− cells [92].
We also have found that Pol β null cells are hypersensitive to the thymidine analog hmdUrd. This agent is incorporated into cellular DNA and elicits cytotoxicity only when removed by glycosylase-initiated BER. Sensitization to this agent was seen in both wild-type and Pol β null cells in the presence of 4-AN, further substantiating the model that the hypersensitivity to PARP inhibitor reflects accumulation of cytotoxic BER intermediates following removal of base lesions from DNA. In addition, and consistent with the proposal that there is a common repair intermediate resulting in cytotoxicity following treatment with MMS or hmdUrd, cell death initiated by both agents is preceded by a rapid suppression of DNA synthesis (S-phase checkpoint) and a later cell cycle arrest in G2M phase of the cell cycle [93]. For the methylated base and hmdUrd DNA, monofunctional glycosylases result in formation of the 5′-dRP-containing intermediate, that we believe to be cytotoxic and a binding site for the abundant PARP-1 protein.
Unpublished data indicate that a separate branch of the S-phase checkpoint pathway, distinct from the ATR/ATM/Chk pathway involves phosphorylation of SMC1 by ATM and ATR. This is facilitated by NBS1 and would explain the S-phase checkpoint defect we have observed in NBS1-deficient cells treated with MMS plus 4-AN. A manuscript describing a study of the ATR- or ATM-dependence of NBS1 and SMC1 activation in MMS plus 4-AN-treated cells is in preparation.
BER roles of Pol β and PARP-1 as revealed in cellular models
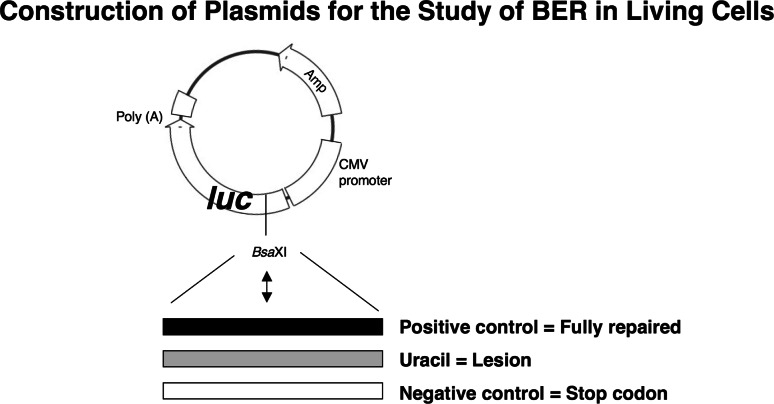
We developed a novel plasmid-based assay for measurement of BER capacity in vivo [74], as illustrated here. The assay involves real-time measurement of expression of a luciferase reporter gene product and is routinely conducted with positive control, negative control and a lesion-containing plasmid, as illustrated. Repair of the lesion enables expression of luciferase whereas there is no expression in the absence of repair due to a stop codon in the transcript.
This assay was used to verify a role for Pol β in both SN and LP BER. Typical results for SN BER assays obtained with three mouse fibroblasts cell lines as illustrated here. The reason(s) for the residual repair in the absence of Pol β (as can be seen in the middle panel) is unknown.
The assay also was used to evaluate the effect of PARP inhibition as a function of Pol β expression. PARP inhibition with 4-AN blocked Pol β-dependent BER measured in both sub-pathways. An illustration of this effect with the SN BER plasmid (uracil-DNA) is shown below (adapted from Masaoka et al.). This result suggested that Pol β and PARP function in the same pathway and represent a demonstration that PARP activity is actually required for BER in vivo.
Small molecule inhibitors of Pol β and translational research in BER
In recent years, there has been considerable effort toward isolating and identifying natural product inhibitors of Pol β. One aim for use of such compounds is inhibiting repair of DNA adducts formed after treatment with DNA-damaging anticancer agents, and thus potentiating chemotherapeutic effects. The discovery of specific Pol β inhibitors also would enable use of these agents as probes to dissect Pol β functions in the various sub-pathways of BER and the cross-talk between repair pathways in vivo.
In collaboration with G. Mullen and associates, the binding site for a known natural product Pol β inhibitor, koetjapic acid, was localized by NMR chemical shift mapping to the Pol β 8-kDa domain [94]. The interaction at this region of the enzyme inhibited both DNA synthesis activity with a single-nucleotide gapped substrate and dRP lyase activity with a similar substrate structure. The NMR-binding study was then extended to include structurally similar synthetic terpenoids and finally other synthetic compounds containing aromatic or other hydrophobicity in combination with two carboxylate groups. Dissociation constants for binding were determined for these compounds and the binding sites on the Pol β 8-kDa domain were mapped. Since a predominant phenotype of Pol β null cells is hypersensitivity to monofunctional alkylating agents such as MMS, an appropriate way to assay for a block of intracellular Pol β activity was to look for an increase in cellular sensitivity to MMS. The ability of nine synthetic compounds to potentiate MMS-induced cytotoxicity in wild-type and Pol β null mouse fibroblasts was ascertained. Treatment of cells with the most active and specific compound identified, pamoic acid (PA), resulted in sizeable sensitization of wild-type cells to MMS with only a negligible effect in Pol β null cells. PA is now in use as a Pol β inhibitor, even though it binds to Pol β with only relatively low affinity. PA inhibits the DNA polymerase activity of Pol β.
To develop stronger and more specific Pol β inhibitors, we are collaborating with several groups. First, in collaboration with A. Simeonov and associates (NCGC, NHGRI, NIH), we recently reported development of a robotics-based assay system for screening very large chemical libraries for potent and specific Pol β inhibitors [95], and further experiments with this approach are underway. Next, use of specific chemical modifications of the nucleotide substrates and products of DNA synthesis is under evaluation. This approach for DNA polymerase inhibition has been widely successful in therapeutic applications for AIDS and cancer.
dNTP analogs
An alternate approach toward small molecule inhibitor design has been use of targeted analogues of the natural DNA synthesis dNTP substrates. Progress in this approach includes enhanced understanding of the Pol β nucleotide insertion mechanism, and this has been through use of various dNTP analogs with substitutions in the bridging oxygens of the triphosphate moiety, as illustrated below. Some of the analogs are commercially available and others have been obtained through collaborations with the synthetic organic chemists C. McKenna (USC), S. Prakash (USC) and B. Cho (University of Rhode Island) and their associates. Highlights of the approach have included determination of the structure of the complete active site for a DNA polymerase [65] and the use of analogs to examine the influence of differences in protonation of the transfer reaction leaving group (pyrophosphate) on features of the overall insertion reaction [96, 97]. The latter experiments also have been in collaboration with A. Warshel and M. Goodman and their associates (USC).
Additionally, kinetic analyses of Pol β insertion efficiency with incoming nucleotides with modified sugars indicated that Pol β can insert dideoxy-nucleotides and the clinically useful analog araCTP with high efficiency [98]. Of course, the enzyme discriminates against rNTP insertion, but nevertheless inserts ribonucleotides with surprisingly high efficiency, given the predominance of rNTPs in the cellular nucleotide pool. Once a ribonucleotide is inserted by Pol β, the question of further extension from the ribonucleotide primer in LP BER or ligation in SN BER are important considerations. In kinetic experiments with a ribonucleoside at the primer terminus, we found that Pol β was able to efficiently conduct extension [98].
Strategy for a combinatorial small molecule approach toward blocking BER and enhancing cell killing
A strategy for cell killing through use of a combination of agents is under evaluation. These agents are: (1) a BER initiating chemical, such as MMS or temozolomide; (2) a PARP inhibitor for blocking repair and promoting DBS formation; and (3) a Pol β inhibitor for blocking BER. The impressive cytotoxic effect of PARP inhibition in the presence of BER initiated with a low dose of MMS is illustrated here. Cell survival at the non-toxic MMS dose of 0.25 mM is reduced by two orders of magnitude by the presence of the PARP inhibitor; the PARP inhibitor alone is not cytotoxic.
Yet, even though PARP inhibition in a cell undergoing BER causes extreme cytotoxicity, Pol β elimination causes additional toxicity beyond that from PARP inhibition. These results indicate that the combination of both Pol β inhibition and PARP inhibition has the potential to cause even more profound cytotoxicity, as a function of triggering BER, than PARP inhibition alone [89].
Conclusions
In addition to the PARP inhibitor effects described involving blocking of BER and possibly other repair pathways (i.e., DSB repair), opportunities to impose even stronger cytotoxicity seem to be at hand, through targeting the cell signaling system that detects blocked repair intermediates. However, results from such experiments are not yet available. An additional feature for consideration in the future is targeting the cell-killing effect of combination therapy to the most appropriate cell types. The requirement for DNA replication in the PARP inhibitor effect represents an intrinsic feature of the consequence of PARP inhibition that targets the effect to dividing cells. Nevertheless, more specific targeting to individual cell types would seen to be advantageous.
Acknowledgments
The authors thank Bonnie E. Mesmer for editorial assistance. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES050158 & Z01-ES050159).
References
- 1.Clarke ND, Kvaal M, Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197:368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 4.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 5.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 6.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae . Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 7.Russo MT, De Luca G, Degan P, Bignami M. Different DNA repair strategies to combat the threat from 8-oxoguanine. Mutat Res. 2007;614:69–76. doi: 10.1016/j.mrfmmm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 10.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu YA, Prasad R, Beard WA, Kedar PS, Hou EW, Shock DD, Wilson SH. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J Biol Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 16.Asagoshi K, Wilson SH. Base excision repair. In: Wafik E-D, Penning TM, editors. Current cancer research. Berlin Heidelberg New York: Springer; 2009. [Google Scholar]
- 17.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 18.Beard WA, Prasad R, Wilson SH (2006) Activities and mechanism of DNA polymerase beta. In: Campbell JL, Modrich P (eds) Methods in enzymology, DNA repair, Part A. Academic Press, San Diego [DOI] [PubMed]
- 19.Beard WA, Shock DD, Wilson SH. Influence of DNA structure on DNA polymerase β active site function: extension of mutagenic DNA intermediates. J Biol Chem. 2004;279:31921–31929. doi: 10.1074/jbc.M404016200. [DOI] [PubMed] [Google Scholar]
- 20.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat Res. 2000;460:231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 21.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase beta. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 22.Wilson SH. Mammalian base excision repair and DNA polymerase β. Mutat Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 23.Beard WA, Wilson SH. Structural insights into DNA polymerase β fidelity: hold tight if you want it right. Chem Biol. 1998;5:R7–R13. doi: 10.1016/s1074-5521(98)90081-3. [DOI] [PubMed] [Google Scholar]
- 24.Wilson SH, Abbotts J, Widen S. Progress towards molecular biology of DNA polymerase β. Biochim Biophys Acta. 1988;949:149–157. doi: 10.1016/0167-4781(88)90078-4. [DOI] [PubMed] [Google Scholar]
- 25.Prasad R, Widen SG, Singhal RK, Watkins J, Prakash L, Wilson SH. Yeast open reading frame YCR14C encodes a DNA β-polymerase-like enzyme. Nucleic Acids Res. 1993;21:5301–5307. doi: 10.1093/nar/21.23.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu K, Santocanale C, Ropp PA, Longhese MP, Plevani P, Lucchini G, Sugino A. Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae. A probable homolog of mammalian DNA polymerase beta. J Biol Chem. 1993;268:27148–27153. [PubMed] [Google Scholar]
- 27.Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto Y, Bogenhagen DF. Repair of a synthetic abasic site involves concerted reactions of DNA synthesis followed by excision and ligation. Mol Cell Biol. 1991;11:4441–4447. doi: 10.1128/mcb.11.9.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiebauer K, Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci USA. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase β is processive. J Biol Chem. 1993;268:15906–15911. [PubMed] [Google Scholar]
- 31.Wilson SH, Singhal R, Kumar A. Structural and functional studies of mammalian DNA polymerase β. In: Bohr VA, Wassermann K, Kraemer KH, editors. Alfred Benzon symposium 35: DNA repair mechanisms. Copenhagen: Munksgaard; 1993. pp. 343–360. [Google Scholar]
- 32.Sobol RW, Horton JK, Kühn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase β in base excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 33.Prasad R, Kumar A, Widen SG, Casa-Finet JR, Wilson SH. Identification of residues in the single-stranded DNA-binding site of the 8-kDa domain of the rat DNA polymerase β by UV cross-linking. J Biol Chem. 1993;268:22746–22755. [PubMed] [Google Scholar]
- 34.Horton JK, Srivastava DK, Zmudzka BZ, Wilson SH. Strategic down-regulation of DNA polymerase β by antisense RNA sensitizes mammalian cells to specific DNA damaging agents. Nucleic Acids Res. 1995;23:3810–3815. doi: 10.1093/nar/23.19.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zmudzka B, Wilson SH. Deregulation of DNA polymerase β by sense and antisense RNA expression in mouse 3T3 cells alters cell growth. Somat Cell Mol Genet. 1990;16:311–320. doi: 10.1007/BF01232459. [DOI] [PubMed] [Google Scholar]
- 36.Singhal RK, Prasad R, Wilson SH. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 37.Abbotts J, SenGupta DN, Zon G, Wilson SH. Studies on the mechanism of Escherichia coli DNA polymerase I large fragment: effect of template sequence and substrate variation on termination of synthesis. J Biol Chem. 1988;263:15094–15103. [PubMed] [Google Scholar]
- 38.Kumar A, Casas-finet JR, Luneau CJ, Karpel RL, Merrill BM, Williams KR, Wilson SH. Mammalian heterogeneous nuclear ribonucleoprotein A1: nucleic acid binding properties of the COOH-terminal domain. J Biol Chem. 1990;265:17094–17100. [PubMed] [Google Scholar]
- 39.SenGupta DN, Zmudzka BZ, Kumar P, Cobianchi F, Skowronski J, Wilson SH. Sequence of human DNA polymerase β mRNA obtained through cDNA cloning. Biochem Biophys Res Commun. 1986;136:341–347. doi: 10.1016/0006-291x(86)90916-2. [DOI] [PubMed] [Google Scholar]
- 40.Zmudzka BZ, SenGupta D, Matsukage A, Cobianchi F, Kumar P, Wilson SH. Structure of rat DNA polymerase β revealed by partial amino acid sequencing and cDNA cloning. Proc Natl Acad Sci USA. 1986;83:5106–5110. doi: 10.1073/pnas.83.14.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbotts J, SenGupta DN, Zmudzka B, Widen SG, Notario V, Wilson SH. Expression of human DNA polymerase β in Escherichia coli and characterization of the recombinant enzyme. Biochemistry. 1988;27:901–909. doi: 10.1021/bi00403a010. [DOI] [PubMed] [Google Scholar]
- 42.Patterson TA, Little W, Cheng X, Widen SG, Kumar A, Beard WA, Wilson SH. Molecular cloning and high-level expression of human polymerase β cDNA and comparison of the purified recombinant human and rat enzymes. Protein Expr Purif. 2000;18:100–110. doi: 10.1006/prep.1999.1167. [DOI] [PubMed] [Google Scholar]
- 43.Kim SJ, Lewis MS, Knutson JR, Porter DK, Kumar A, Wilson SH. Characterization of the tryptophan fluorescence and hydrodynamic properties of rat DNA polymerase β. J Mol Biol. 1994;244:224–235. doi: 10.1006/jmbi.1994.1720. [DOI] [PubMed] [Google Scholar]
- 44.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A, Widen SG, Williams KR, Kedar P, Karpel RL, Wilson SH. Studies of the domain structure of mammalian DNA polymerase β: identification of a discrete template binding domain. J Biol Chem. 1990;265:2124–2131. [PubMed] [Google Scholar]
- 46.Casas-Finet JR, Kumar A, Karpel RL, Wilson SH. Mammalian DNA polymerase β: characterization of a 16-kDa transdomain fragment containing the nucleic acid-binding activities of the native enzyme. Biochemistry. 1992;31:10272–10280. doi: 10.1021/bi00157a014. [DOI] [PubMed] [Google Scholar]
- 47.Casas-Finet JR, Kumar A, Morris G, Wilson SH. Spectroscopic studies of the structural domains of mammalian DNA β-polymerase. J Biol Chem. 1991;29:19618–19625. [PubMed] [Google Scholar]
- 48.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 49.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase β as revealed by site-directed mutagenesis: DNA binding and 5′-deoxyribose phosphate lyase activities. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 50.Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J Biol Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 51.Beard WA, Wilson SH. DNA lesion bypass polymerases open up. Structure. 2001;9:759–764. doi: 10.1016/s0969-2126(01)00646-3. [DOI] [PubMed] [Google Scholar]
- 52.Beard WA, Wilson SH. DNA polymerases lose their grip. Nat Struct Biol. 2001;8:915–917. doi: 10.1038/nsb1101-915. [DOI] [PubMed] [Google Scholar]
- 53.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 54.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 55.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase β complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 56.Pelletier H, Sawaya MR, Wolfle W, Wilson SJ, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 57.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 58.Sawaya MR, Prasad P, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 59.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 61.Liu DJ, Derose EF, Prasad R, Wilson SH, Mullen GP. Assignments of 1H, 15N, and 13C resonances for the backbone and side chains of the N-terminal domain of DNA polymerase β. Determination of the secondary structure and tertiary contacts. Biochemistry. 1994;33:9537–9545. doi: 10.1021/bi00198a020. [DOI] [PubMed] [Google Scholar]
- 62.Liu DJ, Prasad R, Wilson SH, Derose EF, Mullen GP. Three-dimensional solution structure of the N-terminal domain of DNA polymerase β and mapping of the ssDNA interaction interface. Biochemistry. 1996;35:6188–6200. doi: 10.1021/bi952656o. [DOI] [PubMed] [Google Scholar]
- 63.Mullen GP, Antuch W, Maciejewski MW, Prasad R, Wilson SH. Insights into the mechanism of the β-elimination catalyzed by the N-terminal domain of DNA polymerase β. Tetrahedron. 1997;53:12057–12066. [Google Scholar]
- 64.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 65.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymerase beta. Proc Natl Acad Sci USA. 2006;103:13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Radhakrishnan R, Arora K, Wang Y, Beard WA, Wilson SH, Schlick T. Regulation of DNA repair fidelity by molecular checkpoints: “gates” in DNA polymerase beta’s substrate selection. Biochemistry. 2006;45:15142–15156. doi: 10.1021/bi061353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen K-H, Yakes FM, Srivastava DK, Singhal RK, Sobol RW, Horton JK, Van Houten B, Wilson SH. Up-regulation of base excision repair correlates with enhanced protection against a DNA damaging agent in mouse cell lines. Nucleic Acids Res. 1998;26:2001–2007. doi: 10.1093/nar/26.8.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fornace J, Albert J, Zmudzka B, Hollander MC, Wilson SH. Induction of β-polymerase mRNA by DNA-damaging agents in Chinese hamster ovary cells. Mol Cell Biol. 1989;9:851–853. doi: 10.1128/mcb.9.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase β expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 72.Horton JK, Baker A, Vande Berg BJ, Sobol RW, Wilson SH. Involvement of DNA polymerase β in protection against the cytotoxicity of oxidative DNA damage. DNA Repair (Amst) 2002;1:317–333. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 73.Hou EW, Prasad R, Asagoshi K, Masaoka A, Wilson SH. Comparative assessment of plasmid and oligonucleotide DNA substrates in measurement of in vitro base excision repair activity. Nucleic Acids Res. 2007;35(17):e112. doi: 10.1093/nar/gkm639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst) 2009;8:1290–1299. doi: 10.1016/j.dnarep.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asagoshi K, Liu Y, Masaoka A, Lan L, Prasad R, Horton JK, Brown AR, Wang XH, Bdour HM, Sobol RW, Taylor JS, Yasui A, Wilson SH. DNA polymerase β-dependent long patch base excision repair in living cells. DNA Repair (Amst) 2009;9:109–119. doi: 10.1016/j.dnarep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between polymerase β and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate: evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 79.Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair (Amst) 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 80.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson SH. Gene regulation and structure-function studies of mammalian DNA polymerase β. In: Strauss PR, Wilson SH, editors. The eukaryotic nucleus: molecular biochemistry and macromolecular assemblies. Caldwell: Telford Press, Inc.; 1990. pp. 199–233. [Google Scholar]
- 82.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gryk MR, Marintchev A, Maciejewski MW, Robertson A, Wilson SH, Mullen GP. Mapping of the interaction interface of DNA polymerase β with XRCC1. Structure. 2002;10:1709–1720. doi: 10.1016/s0969-2126(02)00908-5. [DOI] [PubMed] [Google Scholar]
- 85.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. Domain specific interaction in the XRCC1-DNA polymerase β complex. Nucleic Acids Res. 2000;28:2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sobol RW, Foley JF, Nyska A, Davidson MG, Wilson SH. Regulated over-expression of DNA polymerase β mediates early onset cataract in mice. DNA Repair (Amst) 2003;2:609–622. doi: 10.1016/s1568-7864(03)00026-0. [DOI] [PubMed] [Google Scholar]
- 87.Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest after DNA methylating agent exposure. J Biol Chem. 2005;280:15773–15785. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- 88.Horton JK, Stefanick DF, Wilson SH. Involvement of poly(ADP-ribose) polymerase activity in regulating Chk1-dependent apoptotic cell death. DNA Repair (Amst) 2005;4:1111–1120. doi: 10.1016/j.dnarep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 89.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kedar PS, Stefanick DF, Horton JK, Wilson SH. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Repair (Amst) 2008;7:1787–1798. doi: 10.1016/j.dnarep.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carrozza MJ, Stefanick DF, Horton JK, Kedar PS, Wilson SH. PARP inhibition during alkylation-induced genotoxic stress signals a cell cycle checkpoint response mediated by ATM. DNA Repair (Amst) 2009;8:1264–1272. doi: 10.1016/j.dnarep.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heacock M, Stefanick DF, Horton JK, Wilson SH. Alkylation damage combined with PARP inhibition in cells results in S-phase dependent double-strand breaks. DNA Repair (Amst) 2010;9:929–936. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase β null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 94.Hu HY, Horton JK, Gryk MR, Prasad R, Naron JM, Sun DA, Hecht SM, Wilson SH, Mullen GP. Identification of small molecule synthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. J Biol Chem. 2004;279:39736–39744. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- 95.Dorjsuren D, Wilson DM, Beard WA, McDonald JP, Austin CP, Woodgate R, Wilson SH, Simeonov A. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sucato CA, Upton TG, Kashemirov BA, Batra VK, Martinek V, Xiang Y, Beard WA, Pedersen LC, Wilson SH, McKenna CE, Florian J, Warshel A, Goodman MF. Modifying the beta, gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 97.Sucato CA, Upton TG, Kashemirov BA, Osuna J, Oertell K, Beard WA, Wilson SH, Florian J, Warshel A, McKenna CE, Goodman MF. DNA polymerase beta fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry. 2008;47:870–879. doi: 10.1021/bi7014162. [DOI] [PubMed] [Google Scholar]
- 98.Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J Biol Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]