Abstract
Background
CYP3A5 genotyping might be useful to guide tacrolimus and sirolimus dosing. The aim of this study was to assess the influence of CYP3A5 polymorphism on everolimus metabolism and pharmacokinetics.
Methods
We investigated the effect of CYP3A5 A6986G polymorphism (CYP3A5*1/*3 alleles) on the pharmacokinetics of everolimus in 28 renal transplant patients as well as on its in vitro hepatic metabolism using a bank of genotyped human liver microsomes (n=49). We further evaluated in vitro the contribution of CYP3A4, CYP3A5 and CYP2C8 to everolimus hepatic metabolism using recombinant enzymes.
Results
We found no association between CYP3A5 polymorphism and everolimus pharmacokinetics in renal transplant patients. On the other hand, no effect of CYP3A5 polymorphism was observed on the CLint of everolimus by human liver microsomes, while that of tacrolimus (positive control) was 1.5-fold higher in microsomes carrying the CYP3A5*1 allele than in non-carriers. In vitro data showed that CYP3A4 is a better catalyst of everolimus metabolism than CYP3A5, while the opposite was observed for tacrolimus.
Conclusions
This study provides direct and indirect evidence that CYP3A5 genotyping cannot help improve everolimus therapy.
Keywords: Adult; Aged; Cytochrome P-450 CYP3A; genetics; Female; Genotype; Humans; Immunosuppressive Agents; metabolism; Kidney Transplantation; Male; Middle Aged; Polymorphism, Single Nucleotide; Sirolimus; analogs & derivatives; metabolism
Keywords: everolimus, pharmacogenetics, pharmacokinetics, drug metabolism
Introduction
Everolimus (Certican®, 40-O-(2-hydroxyethyl)—rapamycin) is a potent immunosuppressive drug used for the prevention of acute and chronic rejection after solid organ transplantation. It is a derivative of sirolimus (rapamycin). Because of the introduction of the 40-O-(2-hydroxyethyl) group, everolimus is more hydrophilic, has a shorter half-life and a relatively higher bioavailability compared to sirolimus (1). Everolimus and sirolimus are metabolized in the liver and in the gut wall primarily by the cytochrome P450 (P450) 3A enzymes and to a minor extent by CYP2C8 (2). CYP3A5 is highly polymorphic. The CYP3A5*3 allele results in a splicing defect, leading to a truncated protein with no enzyme activity; CYP3A5*1 is the wild-type allele, associated with enzyme activity. A large number of studies have investigated the influence of CYP3A5 polymorphism on the dose requirement and clearance of the immunosuppressant substrates of CYP3A. The CYP3A5*3 genotype appears to have a sufficiently large influence on tacrolimus metabolism to be used to guide drug dosing (3, 4). A similar association between the CYP3A5*3 genotype and dose requirement has been found for sirolimus, but not in all studies and only in patients not treated with a calcineurin inhibitor (5–7). The influence of this polymorphism on everolimus pharmacokinetics remains however unclear, as only one such study has been reported, showing no association between CYP3A5 genotype and everolimus blood levels or dose requirement in 30 adult cardiac transplants (8). In the present study, we investigated the impact of the CYP3A5*3 polymorphism on everolimus pharmacokinetics in 28 renal transplant patients receiving everolimus with no associated calcineurin inhibitor. In order to provide confirmation data, we studied the respective roles of CYP3A4 and CYP3A5 and the effect of the CYP3A5*3 polymorphism on everolimus hepatic metabolism in vitro.
Results
Clinical pharmacogenetics of everolimus in renal transplant patients
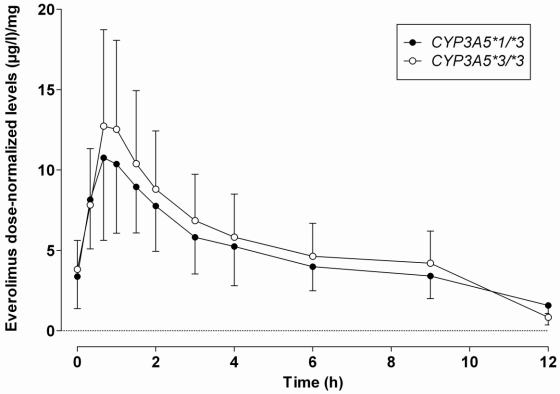
None of the patients were homozygous carriers of the CYP3A5*1 allele. Seven were heterozygous carriers of this allele and 21 non carriers. Patients carriers of the CYP3A5*1 active allele (i.e. expressers) and non carriers (i.e. non expressers) received similar doses of everolimus (1.6±0.9 versus 2.2±1.1 mg; p=0.2959) and had similar everolimus blood levels at all sampling times (Figure 1). The two groups had no significantly different dose-normalized exposure indices (Table 1). Corticosteroid doses and patients’ renal function at the time of blood sampling were similar in CYP3A5*1 and CYP3A5*3 allele carriers (Table 2).
Figure 1.
Mean ± SD of dose-normalized concentration-time profiles of everolimus as a function of the CYP3A5 genotype (CYP3A5*1/*3: n=7; CYP3A5*3/*3: n= 21).
Table 1.
Patients exposure to everolimus according to the CYP3A5*3 genotype.
| CYP3A5 Genotypes (n) | dn-C0 (μg/l)m/mg | dn-Cmax (μg/l)/mg | Tmax (h) | dn-AUC0–12h (μg.h/l)/mg |
|---|---|---|---|---|
| CYP3A5*1/*3 (n=7) | 3.4±2.0 | 11.6±4.6 | 1.0±0.4 | 61.1±20.3 |
| CYP3A5*3/*3 (n=21) | 3.8±1.8 | 13.7±5.8 | 0.7±0.2 | 69.6±29.2 |
| p values | 0.4417 | 0.4417 | 0.2125 | 0.5418 |
Table 2.
Characteristics of the study population according to CYP3A5 genotype.
| CYP3A5*1/*3 (n=7) | CYP3A5*3/*3 (n=21) | P values | |
|---|---|---|---|
| Age of recipient (years) | 53±12 | 51±12 | 0.5926 |
| Male sex (n; %) | 3 (43%) | 15 (71%) | 0.2075 |
| Weight (kg) | 64±11 | 66±19 | 0.5211 |
| Years since transplantation (range) | 10 (0.6–19) | 7 (1.1–18) | 0.7908 |
| Mycophenolate mofetil cotreatment (n; %) | 5 (71%) | 18 (85%) | 0.4318 |
| Corticosteroid dose (range) | 2.1 mg (0–5) | 2.5 mg (0–10) | 0.2057 |
| Serum creatinine levels | 130.7 μmol/l | 131.2 μmol/l | 0.6863 |
In vitro metabolism of everolimus by P450
Initial experiments with recombinant CYP3A4, CYP3A5 and CYP2C8 confirmed previous findings (2) that the three P450 isoforms are able to metabolize everolimus. The relative contribution of CYP3A and CYP2C8 to everolimus hepatic metabolism estimated using the Relative Activity Factor (RAF) approach was 93% and 7%, respectively. Chemical inhibition of CYP3A4 and CYP2C8 confirmed these results: troleandomycin and trimethoprim resulted in 87.7±10.7% and 4.2±3.8% inhibition of everolimus metabolic depletion by human liver microsomes (HLM).
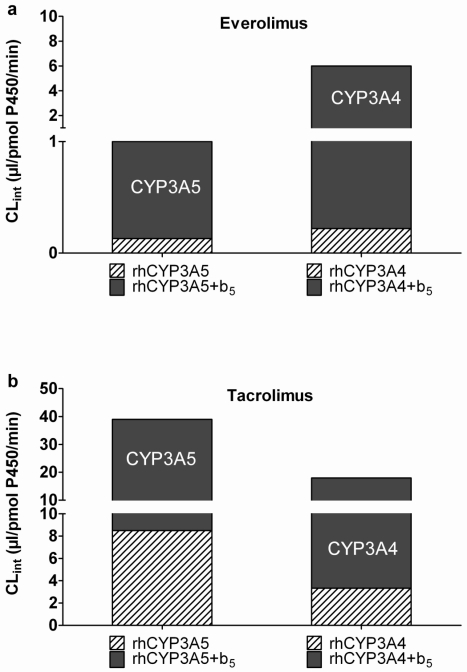
The metabolic intrinsic clearance (CLint) of everolimus by CYP3A4 and CYP3A5 was estimated from its in vitro half-life, using tacrolimus as a positive control. The CLint of everolimus was 1.7-fold lower with recombinant CYP3A5 than CYP3A4, while it was 2.7-fold higher for tacrolimus (Figure 2). The activity of both P450s was greatly enhanced when they were co-expressed with cytochrome b5. However, CYP3A5+b5 still had a lower CLint than CYP3A4+b5 towards everolimus and a higher CLint towards tacrolimus (Figure 2).
Figure 2.
Estimated in vitro intrinsic clearance (CLint) of everolimus (a) and tacrolimus (b) metabolism by human recombinant cytochrome P450 (rhCYP) 3A4 and 3A5. Grayed box represents the activity of rhCYP co-expressed with cytochrome b5 and hatched box the activity of rhCYP without cytochrome b5. The CLint were estimated from tacrolimus and everolimus in vitro metabolic half-lives as described in the materials and methods section. Results are derived from duplicate experiments.
Metabolism of everolimus by genotyped microsomes
Among the 49 hepatic microsomal preparations, 40 (81.6%) came from homozygous carriers of CYP3A5*3 and 9 (18.4%) from heterozygotes. Pools of CYP3A5 expressing and non expressing hepatic microsomes, prepared based on the genotype, showed similar CLint for everolimus (632 [CI95% 515–748] versus 723 [CI95% 358–1088] μl/mg protein/min, respectively; p = 0.1107), while the former had a 1.75-fold higher CLint for tacrolimus, the positive control (2717 [CI95% 2401–3034] versus 1547 [CI95% 1176–1919] μl/mg protein/min; p = 0.0280).
Discussion
Immunosuppressants are narrow therapeutic index drugs and many of them are substrates for CYP3A enzymes. This exploratory study showed that the CYP3A5*3 polymorphism did not influence everolimus dose requirement, exposure or dose-normalized exposure in 28 stable renal transplants. Further experimental studies demonstrated that CYP3A4 is more active than CYP3A5 in everolimus metabolism and confirmed, using genotyped HLM, that the CYP3A5*3 polymorphism has no significant influence on the metabolism of everolimus.
Only one former clinical study in cardiac transplants investigated the effect of the CYP3A5*3 genotype on everolimus pharmacokinetics, and no association was found either (8). The number of subjects in this previous study was quite small (n=30), as in the present one, but sample size calculation showed that it was sufficient to detect an effect similar to that observed for the parent drug sirolimus (6), with a power of 80%. In both studies, patients were not cotreated with calcineurin inhibitors which ensures that enzyme inhibition cannot have interacted with the pharmacogenetic association. A limitation of these two studies is that none of the patients studied were homozygous carriers of the CYP3A5*1 active allele, a rare genotype in patients of European ancestry. We cannot totally exclude a significantly higher metabolic clearance of everolimus in patients of this particular genotype. This should be explored in patients of African ancestry, known to carry the CYP3A5*1 allele at much higher frequency (9).
We provide here experimental evidence that the CYP3A5*1/*3 genotype does not influence the hepatic metabolism of everolimus. In contrast, the metabolism of tacrolimus, used as positive control, was substantially higher in microsomes derived from CYP3A5 expressers, a genotype effect consistent with numerous clinical reports (3, 10–12).
We found that CYP3A4 is a better catalyst of everolimus than CYP3A5, while the opposite was observed for tacrolimus. This provides a first explanation to the fact that CYP3A5 expression does not significantly influence everolimus metabolism. CYP2C8 is the only other P450 isoform reported to be involved in the metabolism of everolimus (2). In the present study, we found that it accounts for less than 7% of everolimus overall in vitro metabolism. This excludes that this isoform can substantially contribute to everolimus metabolism and thus to its pharmacokinetic variability.
It is also noteworthy that everolimus overall metabolic clearance is 2-to-4 fold lower than that of tacrolimus, suggesting that the hepatic first-pass effect is less critical for everolimus than tacrolimus. The hepatic microsomal CLint of everolimus observed here is also 2-fold lower than what we previously reported for sirolimus (677 versus 1271 μl/mg protein/min) (13). This is consistent with the knowledge that the 40-O-(2-hydroxyethyl) group on everolimus results in a 3-fold lower total CLint of P450-dependent metabolism than sirolimus by inhibiting demethylation at position 39 (the major metabolism of sirolimus) and hydroxylation in the C10 to C14 region (the major hydroxylation region of sirolimus) (2). This probably explains the difference in oral bioavailability between the two drugs and suggests that everolimus would be less subject to interindividual variability in first-pass metabolism.
CYP3A5 expression does not appear to contribute substantially to everolimus pharmacokinetic variability. A potential explanation for the observed variability is the contribution of CYP3A4 and P-gp in the small intestine and the liver. It is well established that there is considerable inter-individual variation in CYP3A4 activity, but no polymorphism that would result in absence of activity has been described. SNPs giving rise to alterations in catalytic activity are seen at very low population frequencies, precluding their inclusion in the present study (14). We did not consider P-gp polymorphisms. The literature remains unclear on the precise consequences of P-gp genetic variations on drug pharmacokinetics and most studies investigating the parent drug sirolimus reported no association (5, 7, 15). Everolimus pharmacokinetic variability is likely to be explained by a combination of factors including genetics, epigenetics, gender, diets, diseases, and comedications.
In conclusion, this study provides direct and indirect lines of evidence strongly suggesting that CYP3A5 genotyping is of no clinical relevance in either kidney graft recipients on everolimus, or in candidates to receive it.
Materials and Methods
Patients
28 kidney graft recipients followed as outpatients at Toulouse Rangueil hospital were enrolled in the study. The median time from transplant to the study was 7.2 years (range: 7 months to 18.5 years). All patients received induction therapy with basiliximab or antithymocyte globulin and a standard tapered corticosteroid regimen. The patients had been administered everolimus for 3 to 44 months (median: 24 months) prior to the study. The drug was administered twice daily, targeting a whole blood trough concentration between 5 and 15 μg/l. There was no dose modification for at least 2 months before the study. Twenty-three of the twenty-eight patients received mycophenolate mofetil given at an initial fixed dose of 1g twice daily, reduced if necessary based on clinical criteria. Blood samples were collected in each patient at 0, 20, 40, 60, 90, 120, 180, 240, 360, 540 and 720 min after the morning dose of everolimus. No drugs interacting with CYP3A or P-glycoprotein (P-gp) were allowed to be used in this study.
The study was approved by the ethics committee of Rangueil hospital and informed consent was obtained from each patient.
Drug determination
Everolimus whole blood determination was performed using turbulent flow chromatography-tandem mass spectrometry, as previously described for sirolimus (13). Two MS/MS transitions were monitored for everolimus (m/z 975.5→908.5; m/z 975.5→858.5) and for the internal standard desmethoxysirolimus (m/z 901.3→816.4; m/z 901.3→834.2). The method was fully validated for everolimus determination in whole blood and adapted to in vitro incubation supernatant, as previously described for sirolimus (13). Briefly, sample preparation consisted of mixing for 15 sec 100 μl of samples with 150 μl of a mixture of methanol/aqueous zinc sulfate (70:30; v/v) and 50 μl of internal standard (100 μg/l in acetonitrile). The limit of quantitation was 0.5 μg/l and calibration curves obtained using quadratic regression from the limit of quantitation up to 100 μg/l yielded r2 > 0.99. Tacrolimus was determined by liquid chromatography-tandem mass spectrometry as previously described (16).
Everolimus area under the concentration curve (AUC0–12h) was estimated using the trapezoidal rule.
Chemicals
Everolimus was obtained from Novartis Pharma AG (Basel, Switzerland) and tacrolimus from Fujisawa Pharmaceuticals (Osaka, Japan). All other chemicals were purchased from Sigma-Aldrich (St-Louis, MO).
Enzymes and human microsomes
Recombinant CYP3A4 and 3A5 (each co-expressed with or without cytochrome b5) and CYP2C8 (with cytochrome b5) Supersomes® were purchased from BD Biosciences (Bedford, MA).
Human liver (n=49) tissue samples derived from surgical specimens were obtained from Biopredic International (Rennes, France). All samples were collected after donors had given their informed consent, in accordance with the French bioethics laws. Each sample was examined by a pathologist and only histologically nontumorous tissue was used. Genomic DNA was extracted from each tissue sample (13) and HLM prepared (17) as described previously.
In vitro metabolic experiments
The metabolism of everolimus was studied by incubating HLM (0.1–0.2 mg protein/ml) or P450 Supersomes® (10–75 pmol/ml) in 0.1 mol/l Tris-HCl, pH 7.4, with 10 mmol/l MgCl2 and 100 μg/l everolimus. Following preincubation for 5 min at 37°C, the reaction was initiated by adding 2 mmol/l NADPH. After 0, 2, 5, 10, 15, 20, 30 minutes, aliquots (50 μl) were removed from the mixture and dispensed into tubes containing 50 μl of ice-cold acetonitrile to stop the reaction. Similar experiments were performed using tacrolimus (100 μg/l) as a substrate.
Drug determination was performed as described above and the half-life of everolimus or tacrolimus metabolic depletion estimated using an exponential decay model in GraphPad Prism 5. The total in vitro CLint was then estimated using the formula described by Obach et al. (18). In order to estimate the specific contribution of CYP3A and CYP2C8 to everolimus metabolism, the activity obtained with the respective recombinant P450s after 30 min of incubation was scaled to HLM using the previously described RAF approach (19). The RAF of CYP3A (0.240) and CYP2C8 (0.145) were calculated using the catalytic activities reported by BD Biosciences for Supersomes® and pooled human microsomes towards selective substrates (i.e., testosterone for CYP3A and paclitaxel for CYP2C8). The relative contribution of each isoform to everolimus metabolism was estimated as the ratio of individual to total P450 scaled activity. Chemical inhibition of everolimus metabolism after 30 minutes incubation with HLM was also studied using troleandomycin as CYP3A inhibitor or trimethoprim as CYP2C8 inhibitor.
Genotyping
Patients DNA was isolated from EDTA-treated blood using the Qiagen blood mini kit (Qiagen, Courtaboeuf, France) and was genotyped for the CYP3A5*3 (A6986G, rs776746) single nucleotide polymorphism using a validated TaqMan allelic discrimination assay on an ABI PRISM 7000 Sequence Detection System (Applied-Biosystems, Courtaboeuf, France).
Statistical analysis
The number of patients required for the clinical study was calculated assuming a ratio between CYP3A5 expressers and non-expressers similar to that observed in our bank of HLM from donors operated on in France (i.e. 1:4), an expected difference in everolimus dose-normalized AUC between expressers and non expressers similar to that reported for sirolimus (6), a one-sided significance of 0.05 and a statistical power of 0.8. A minimum of 5 expressers/20 non-expressers was required. Deviations of genotype frequencies from the Hardy-Weinberg equilibrium were investigated using the Fisher-exact test. Categorical variables were compared using the Fisher’s exact test and continuous variables using the non-parametric Mann-Whitney test in StatView (SAS Institute, Cary, NC, version 5.0).
Acknowledgments
We thank Biopredic International for their support.
Abbreviations
- CLint
intrinsic clearance
- HLM
Human Liver Microsomes
- P450
cytochromes P450
- RAF
relative activity factor
- rhCYP
recombinant human CYP
Footnotes
The authors declare no conflict of interest.
N.P. and K.R-M. contributed equally to this work: N.P. supervised the experimental research and participated in data analysis and writing the manuscript; K.R-M. conducted the experimental research and participated in data analysis and writing the manuscript; N.K. and L.R. participated in clinical research design, patients enrolment and data collection; P.M. participated in research design and performance and critical review of the manuscript.
References
- 1.Formica RN, Jr, Lorber KM, Friedman AL, et al. The evolving experience using everolimus in clinical transplantation. Transplant Proc. 2004;36 (2 Suppl):495S. doi: 10.1016/j.transproceed.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen W, Serkova N, Hausen B, Morris RE, Benet LZ, Christians U. Comparison of the in vitro metabolism of the macrolide immunosuppressants sirolimus and RAD. Transplant Proc. 2001;33 (1–2):514. doi: 10.1016/s0041-1345(00)02116-3. [DOI] [PubMed] [Google Scholar]
- 3.Thervet E, Loriot MA, Barbier S, et al. Optimization of Initial Tacrolimus Dose Using Pharmacogenetic Testing. Clin Pharmacol Ther. 2010;87 (6):721. doi: 10.1038/clpt.2010.17. [DOI] [PubMed] [Google Scholar]
- 4.MacPhee IA, Holt DW. A pharmacogenetic strategy for immunosuppression based on the CYP3A5 genotype. Transplantation. 2008;85 (2):163. doi: 10.1097/TP.0b013e3181609054. [DOI] [PubMed] [Google Scholar]
- 5.Anglicheau D, Le Corre D, Lechaton S, et al. Consequences of genetic polymorphisms for sirolimus requirements after renal transplant in patients on primary sirolimus therapy. Am J Transplant. 2005;5 (3):595. doi: 10.1111/j.1600-6143.2005.00745.x. [DOI] [PubMed] [Google Scholar]
- 6.Le Meur Y, Djebli N, Szelag JC, et al. CYP3A5*3 influences sirolimus oral clearance in de novo and stable renal transplant recipients. Clin Pharmacol Ther. 2006;80 (1):51. doi: 10.1016/j.clpt.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Mourad M, Mourad G, Wallemacq P, et al. Sirolimus and tacrolimus trough concentrations and dose requirements after kidney transplantation in relation to CYP3A5 and MDR1 polymorphisms and steroids. Transplantation. 2005;80 (7):977. doi: 10.1097/01.tp.0000174131.47469.d2. [DOI] [PubMed] [Google Scholar]
- 8.Kniepeiss D, Renner W, Trummer O, et al. The role of CYP3A5 genotypes in dose requirements of tacrolimus and everolimus after heart transplantation. Clin Transplant. 2009 doi: 10.1111/j.1399-0012.2009.01198.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27 (4):383. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 10.Haufroid V, Wallemacq P, VanKerckhove V, et al. CYP3A5 and ABCB1 polymorphisms and tacrolimus pharmacokinetics in renal transplant candidates: guidelines from an experimental study. Am J Transplant. 2006;6 (11):2706. doi: 10.1111/j.1600-6143.2006.01518.x. [DOI] [PubMed] [Google Scholar]
- 11.Macphee IA, Fredericks S, Mohamed M, et al. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79 (4):499. doi: 10.1097/01.tp.0000151766.73249.12. [DOI] [PubMed] [Google Scholar]
- 12.Tsuchiya N, Satoh S, Tada H, et al. Influence of CYP3A5 and MDR1 (ABCB1) polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplantation. 2004;78 (8):1182. doi: 10.1097/01.tp.0000137789.58694.b4. [DOI] [PubMed] [Google Scholar]
- 13.Picard N, Djebli N, Sauvage FL, Marquet P. Metabolism of sirolimus in the presence or absence of cyclosporine by genotyped human liver microsomes and recombinant cytochromes P450 3A4 and 3A5. Drug Metab Dispos. 2007;35 (3):350. doi: 10.1124/dmd.106.012161. [DOI] [PubMed] [Google Scholar]
- 14.Daly AK. Pharmacogenetics and human genetic polymorphisms. Biochem J. 2010;429 (3):435. doi: 10.1042/BJ20100522. [DOI] [PubMed] [Google Scholar]
- 15.Miura M, Kagaya H, Satoh S, et al. Influence of drug transporters and UGT polymorphisms on pharmacokinetics of phenolic glucuronide metabolite of mycophenolic acid in Japanese renal transplant recipients. Ther Drug Monit. 2008;30 (5):559. doi: 10.1097/FTD.0b013e3181838063. [DOI] [PubMed] [Google Scholar]
- 16.Benkali K, Premaud A, Picard N, et al. Tacrolimus population pharmacokinetic-pharmacogenetic analysis and Bayesian estimation in renal transplant recipients. Clin Pharmacokinet. 2009;48 (12):805. doi: 10.2165/11318080-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Picard N, Cresteil T, Premaud A, Marquet P. Characterization of a phase 1 metabolite of mycophenolic acid produced by CYP3A4/5. Ther Drug Monit. 2004;26 (6):600. doi: 10.1097/00007691-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Obach RS. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos. 1999;27 (11):1350. [PubMed] [Google Scholar]
- 19.Venkatakrishnan K, von Moltke LL, Greenblatt DJ. Application of the relative activity factor approach in scaling from heterologously expressed cytochromes p450 to human liver microsomes: studies on amitriptyline as a model substrate. J Pharmacol Exp Ther. 2001;297 (1):326. [PubMed] [Google Scholar]