Abstract
The interactions of a newly synthesized platinum-modified perylene derivative, compound 7 ([{Pt(dien)}2(μ-4-S,S')](NO3)4 (dien = diethylenetriamine, 4 = N,N'-bis(1-(2-aminoethyl)-1,3-dimethylthiourea)-3,4,9,10-perylenetetracarboxylic acid diimide), with the human telomeric repeat were studied using various model oligo(deoxy)ribonucleotides to mimic the polymorphic nature of the telomeric G-quadruplex. UV/visible spectroscopy, CD spectropolarimetry, electrospray mass spectrometry (ES-MS), and isothermal titration calorimetry (ITC) were used to demonstrate that compound 7 selectively recognizes the antiparallel form of the unimolecular telomeric G-quadruplex formed by the sequence d(TTAGGG)4 (dG-24), to which it binds with a 2:1 stoichiometry and nanomolar affinity. Compared with telomeric DNA, the first binding event of compound 7 in titrations with the RNA quadruplex formed by r(UUAGGG)4 (rG-24) is an order of magnitude weaker. Compound 7 does not induce the antiparallel G-quadruplex RNA, which invariably exists in a parallel form and dimerizes in solution. Based on the cumulative experimental data, two distinct mechanisms are proposed for the recognition of G-quadruplex DNA and RNA by compound 7. Potential biomedical and biochemical applications of the platinum–perylene technology are discussed.
Keywords: Platinum–perylene, telomeric G-quadruplex, circular dichroism, calorimetry, ligand binding
INTRODUCTION
Guanine (G)-rich nucleic acid sequences have the propensity to adopt remarkably stable secondary structures known as G-quadruplexes 1. The (transient) formation of the quadruplex motif within gene promoter regions and in untranslated regions (UTR) of gene transcripts has been implicated as a potential modulator of gene expression 2. Several quadruplex-forming sequences have been identified within the promoters of oncogenes, which has sparked an interest in G-quadruplexes as targets for cancer chemotherapy 3,4. G-rich DNA sequences prone to G-quadruplex formation are also found in the telomeres, a multifunctional nucleoprotein complex located at the termini of the chromosomes in most eukaryotic organisms, which protects the chromosome from degradation and rearrangements 5. The shortening of the telomeres with each replication cycle limits the life span of normal cells. By contrast, in the majority of tumors, the telomeres are efficiently restored due to overexpressed telomerase, the reverse transcriptase involved in telomere elongation, rendering cancer cells immortal 3,6. One way to intercept the limitless proliferation of cancer cells, therefore, would be to inhibit telomerase-catalyzed strand elongation with small molecules that stabilize the G-quadruplex form of the single-stranded terminus 3,7.
The core of G-quadruplexes comprises multiple π-stacked G-tetrads that are stabilized by monovalent metal ions (Na+ or K+) 1. The topology of unimolecular quadruplex structures is dominated by complex strand folding, resulting in a diverse array of conformations that differ in their strand polarities, loop orientations, and groove dimensions. As a consequence, G-quadruplexes show a high degree of polymorphism 8. The human telomeric DNA consists of tandem repeats of the sequence d(TTAGGG)/(CCCTAA), ending in a single-stranded 3´ overhang of the G-rich strand. The structures and biophysical properties of G-quadruplexes formed by the sequence TTAGGG are being intensively investigated 8,9. While the existence in live cells and potential biological roles of the human telomeric quadruplex are still being debated, the damage response observed in cancer cells treated with quadruplex-binding/inducing ligands, indeed, is consistent with cell kill mechanisms triggered by G-quadruplex formation 10. Among the most promising agents shown to induce senescence and apoptosis in cancer cells are telomestatin, an oxazoline–thiazoline macrocyclic natural product, and synthetic acridine derivatives 10.
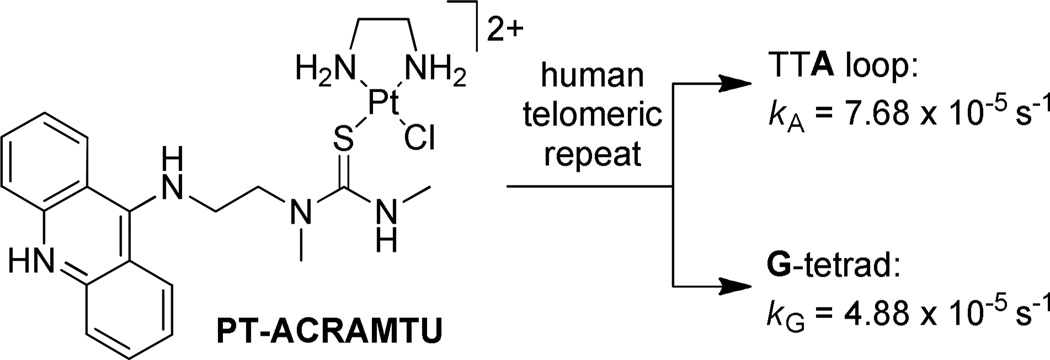
While the vast majority of G-quadruplex-binding molecules are reversible binders containing planar moieties that associate with the G-tetrad via π-stacking interactions, several approaches of targeting the human telomeres with agents that form covalent adducts have also been reported 11–13. We have previously demonstrated that the cytotoxic platinum–acridine hybrid agent [Pt(en)Cl(ACRAMTU)](NO3)2 (“PT-ACRAMTU”; en = ethane-1,2-diamine; ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea) targets adenine (A) in the TTA loops of the telomeric G-quadruplex (Figure 1) 14. The most striking observation in this study was that binding of PT-ACRAMTU to adenine is kinetically favored over binding to guanine, because G-N7, the preferred binding site for platinum in double-stranded DNA, is protected by Hoogsteen hydrogen bonding in the G-tetrad. While targeting the loop adenines in the telomeric quadruplex appears to be a unique approach in telomere-directed chemotherapeutic intervention, PT-ACRAMTU was designed to induce monofunctional–intercalative adducts in double-stranded DNA 15. One possible strategy to target the G-quadruplex structure more efficiently would be to reduce the tendency of classical intercalation into B-form DNA. This could be achieved by replacing the acridine chromophore with a polyaromatic scaffold complementary to the dimensions of a G-tetrad rather than a Watson–Crick base pair. Towards this goal we have begun to investigate a new class of platinum agents containing a perylenediimide moiety, one of the first quadruplex-affinic chromophores studied 16,17. Based on this design rationale, we have synthesized a model platinum–perylene conjugate and have studied here its reversible interactions with nucleic acid sequences mimicking the telomeric DNA and RNA G-quadruplexes, as well as double-stranded DNA, using optical spectroscopies, electrospray mass spectrometry, and isothermal microcalorimetry.
Figure 1.
G-quadruplex interactions of the platinum–acridine agent PT-ACRAMTU.
EXPERIMENTAL SECTION
General Procedures
(2-Aminoethyl)methylcarbamic acid tert-butyl ester was synthesized according to a published procedure 18. [Pt(dien)Cl]Cl (5) and [Pt(dien)(NO3)]NO3 (6) were synthesized following the procedure described by Djuran et al. 19. All other reagents were obtained from common vendors and used as supplied. 1-D 1H NMR and 2-D COSY spectra (for 7, see Supporting Information) were acquired on Bruker Avance 300 MHz and 500 MHz spectrometers. Chemical shifts (δ, ppm) were referenced to residual solvent peaks. UV-visible spectra were recorded on a HP 8453 spectrophotometer. Combustion analyses were carried out by QTI (Whitehouse, NJ).
Preparation of Compounds. N,N´-Bis[2-(tert-butoxycarbonylmethylamino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide (2)
A mixture of 1.97 g (25 mmol) of protected diamine and 4.39 g (5 mmol) of 1 in 20 mL DMF/20 mL 1,4-dioxane was heated under reflux for 20 h. The dark red precipitated 5 was filtered off, washed with diethyl ether, and dried in a vacuum. Yield: 3.16 g (89 %) 1H NMR (300 MHz, CDCl3): δ 1.16 (18 H, s), 2.99 (6 H, s), 3.65 (4 H, s), 4.40 (4 H, s), 8.51 (8 H, d).
N,N´-Bis[2-(methylamino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide (3)
To remove the Boc protecting groups, 1.72 g (2.4 mmol) of 2 were stirred in a mixture of 250 mL of 6 M HCl and 75 mL of methanol for 48 h at 60 °C. The precipitated dihydrochloride salt (3·2HCl) was filtered off and washed with cold ethanol and diethyl ether. To generate the free base, 3·2HCl was stirred in 500 mL of 5 M NH4OH for 48 h. Precipitated 3 was filtered off, washed with cold ethanol and diethyl ether, and dried in a vacuum. Yield: 1.01g (82 %, two steps). 1H NMR (300 MHz, TFA-d): δ 2.96 (6 H, s), 3.72 (4 H, s), 4.75 (4 H, s), 7.45 (2 H, s), 8.74 (8 H, m).
N,N´-Bis[1,3-dimethylthioureaethyl]-3,4,9,10-perylenetetracarboxylic diimide (4)
A mixture of 0.37 g (0.73 mmol) of 3 and 0.12 g (1.6 mmol) of methylisothiocyanate in 80 mL of dry CHCl3 was heated under reflux for 6 h and then placed in the refrigerator overnight. Precipitated 4 was filtered off, washed with CHCl3 and ethanol, and dried in a vacuum at 60 °C. Yield: 0.41g of 4·0.9CHCl3 (74 %). 1H NMR (500 MHz, TFA-d, 65 °C): δ 3.89 (6 H, s), 4.09 (6 H, s), 4.68 (4 H, s), 5.23 (4 H, s), 9.41 (8 H, br s). Anal. Calcd for C34H30N6O4S2·0.9CHCl3: C, 55.28; H, 4.11; N, 11.08. Found: C, 55.21; H, 4.35; N, 11.00.
[{Pt(dien)}2(μ-4-S,S´)](NO3)4 (LPt2(NO3)4, 7)
The complex [Pt(dien)(NO3)]NO3 (6) was generated in situ by reacting 71 mg (0.19 mmol) of [Pt(dien)Cl]Cl (5) with 65 mg (0.38 mmol) of AgNO3 in 5 mL of DMF for 16 h in the dark. Precipitated AgCl was filtered off using a 0.2 µm syringe filter. The filtrate was combined with 61.5 mg (0.082 mmol) of 4·0.9CHCl3 and stirred for 5 h in the dark. DMF was removed in a vacuum and the resulting residue was dissolved in 100 mL of water. Activated carbon was added and the mixture was stirred for 10 min. The mixture was passed through a Celite pad and the solvent was removed by rotary evaporation. The residue was recrystallized from hot methanol. The product was obtained after storing the solution at 4 °C for 24 h. Yield: 50 mg (40 %) of 7·2H2O. 1H NMR (500 MHz, DMSO-d6): δ 2.70 (12 H, m), 2.95 (4 H, s), 3.16 (6 H, d), 3.21 (6 H, s), 4.34 (4 H, s), 4.42 (4 H, s), 5.35 (4 H, s), 5.43 (4 H, s), 7.01 (2 H, s), 8.45 (2 H, d), 8.56 (4 H, d), 8.92 (4 H, d). Anal. Calcd for C42H56N16O16Pt2S2·2H2O: C, 32.94; H, 3.95; N, 14.64. Found: C, 32.85; H, 3.97; N, 14.44. UV-visible spectrum (20 mM potassium phosphate buffer): ε504 = 44,400 M−1 cm−1.
Nucleic Acids and Sample Preparation
The oligo(deoxy)ribonucleotide sequences d(TAGGGTTA) (dG-8), d(TAGGGT)2 (dG-12), d(TTAGGG)4 (dG-24), r(UUAGGG)4 (rG-24), d(AGGG(TTAGGG)3/d(CCCTAA)3CCCT) (ds-22) were synthesized using standard phosphoramidite chemistry and HPLC purified by Integrated DNA Technologies, Inc. (Coralville, IA). Stock solutions of all sequences in their single-stranded form were dissolved in Millipore water obtained from a Milli-Q A10 synthesis water purification system, dialyzed against water, and stored at −20 °C. Concentrations of the oligo(deoxy)ribonucleotides were determined spectrophotometrically using the extinction coefficients provided by the vendor (note: the concentrations of the G-quadruplexes and duplexes used in this study were expressed in µM assembled/folded quadruplex structure or, alternatively, in µM nucleotides (n.t.).). DNA from calf thymus (sodium salt) was from Sigma. Appropriately diluted solutions of the sequences suitable for ligand–DNA/RNA titrations were prepared either in Tris buffer (20 mM, pH 7.2) containing 20 mM KCl (for dG-8 and dG-12), or in potassium phosphate buffer (20 mM KH2PO4, pH 7.2, adjusted with KOH to produce a total K+ concentration of 35 mM) (for dG-24 and rG-24). Samples of dG-24 for electrospray mass spectrometry experiments were prepared in 50 mM ammonium acetate buffer (pH 7.2). The sequences were annealed into their quadruplex or double-stranded forms by heating of the samples to 90 °C and slow cooling to room temperature at a cooling rate of 0.5–0.7 °C/min. Solutions containing dG-8 were stored for an additional 12 h at 4 °C to allow formation of the tertramolecular quadruplex. Stock solutions of compound 7 (0.5 mM) were prepared in the appropriate buffers and stored at −20 °C. Biochemical grade, DNAse- and RNAse-free reagents and HPLC grade solvents were used in all experiments.
UV-Visible Spectroscopy and Circular Dichroism (CD) Spectropolarimetry
UV-visible spectra of compounds 3·2HCl and 7 in complex with dG-24, rG-24, and calf thymus DNA in Tris buffer were recorded on a HP-8453 diode array spectrophotometer. CD spectra were recorded in polystyrene cuvettes at 25 °C on an AVIV Model 215 Circular Dichroism Spectrometer equipped with a thermoelectrically controlled cell holder in the 210–650 nm range with 1 nm resolution and an averaging time of 1 s. A smoothing function was applied to the baseline-corrected spectra. In a typical experiment, solutions of the DNA and RNA sequences (200 µM n.t. for dG-8, equivalent to 12.5 µM assembled quadruplex, 200 µM n.t. for dG-12, equivalent to 8.5 µM assembled quadruplex, and 10 µM (folded) strand for dG-24 and rG-24) were titrated with suitable aliquots (0.2 or 0.25 mole equivalents) of compound 7, and a CD spectrum was acquired after each addition until the ligand-to-DNA/RNA ratio reached 4.0 (dilution effect < 2%). CD intensities were converted from ellipticity (θ) to molar ellipticity, [θ] = θ / c · l (deg · cm2 · dmol−1). Variable-temperature CD spectra were recorded in the range 20–90 °C with a hold time of 30 s and an averaging time of 10 s.
Equilibrium Dialysis Experiments
Dialysis experiments were carried out in 20 mM Tris-HCl buffer (pH 7.2) containing 20 mM KCl in a 28-well microdialyzer (Gibco BRL) using a regenerated cellulose membrane with a 3,500 Da molecular weight cut-off (Fisher). Solutions containing 200 µM (n.t.) DNA, along with DNA-free controls, were equilibrated against a circulating 7.5 mM solution of compound 7 at 25 °C for 72 h. Equal volumes of the samples were withdrawn from the wells and treated with 10% sodium dodecyl sulfate (SDS) solution to a final concentration in SDS of 1%. The amount of DNA-bound drug was determined spectrophotometrically from absorbances at 504 nm. The reported data are averages of three individual determinations.
Electrospray Mass Spectrometry (ES-MS)
Samples of the 1:1 and 2:1 ligand–quadruplex complexes were prepared by combining appropriate volumes of solutions of thoroughly annealed dG-24 and compound 7 in 50 mM ammonium acetate (pH 7.2). Ammonium ions rather than potassium ions were chosen to stabilize the quadruplex structure since ammonium salts are also amenable to ES-MS analysis due to their volatility.20 The final G-quadruplex concentration was 3.4 µM.
ES-MS analyses were performed using an Agilent 1100 model SL LC/MSD ion trap system equipped with an electrospray ionization source. Samples were introduced via direct injection from an in-line LC unit at a flow rate of 0.2 mL/min using 85% water/15% methanol as mobile phase. The injection volume was 10 µL. Desolvation in the source was achieved using N2 drying gas (325 °C, nebulizer pressure 20 psi) at a flow rate of 10 L/min. The mass spectrometer settings were as follows: capillary voltage, 3.5 kV; capillary exit, −200 V; skimmer, 0 V; trap drive, 150. Mass spectra were acquired in negative ion mode using a 13,000 m/z s−1 scan rate over a 200–2,200 m/z range.
Isothermal Titration Calorimetry (ITC)
ITC experiments were carried out at 25 °C using a VP-ITC Microcalorimeter (MicroCal, LLC, Northhampton, MA). Titrations were performed in potassium phosphate buffer (pH 7.2, 35 mM K+) for sequences dG-24 and rG-24 (strand concentration 10 µM). All solutions were thoroughly degassed at 25 °C using the ThermoVac unit. The reference cell was filled with degassed phosphate buffer. A working volume of 1.412 mL of the DNA/RNA solutions was injected into the cell and equilibrated for 30 min. 5-µL aliquots of a 0.5 mM solution of compound 7 were then titrated into the cell with the automated microsyringe at 6-min intervals during which the system was allowed to reach equilibrium. The change in heat was recorded and integrated to provide the heat produced per mole of injected ligand. Titrations of compound 7 into phosphate buffer were performed to correct for the background heat of dilution and deaggregation prior to calculating the binding enthalpy. The integrated heat data were analyzed and fit to the sequential binding site model using the nonlinear curve fitting routine in Origin 7.0, with the exception of the first two binding events in dG-24, which were fit to a non-standard model containing a DNA pre-equilibrium step (see Supporting Information) by self-consistent least-squares optimization using the Simplex minimizer in MATLAB (version R2008b, The MathWorks, Inc., Natick, MA). No constraints were applied to the parameters ΔH° (binding enthalpy, kJ mol−1), Kb (binding constant, M−1), and n (number of sites). The standard Gibbs free energy, ΔG°, and standard entropy, ΔS°, for each binding event were calculated from the equation, − RT lnKb = ΔG° = ΔH° − TΔS°. Titrations for each sequence were performed in triplicate.
Molecular modeling
All structural manipulations and molecular simulations were performed with the Discovery Studio software package (version 2.1, Accelrys, San Diego, CA, 2008). The CHARMm27 force field for nucleic acids was used for all molecular mechanics/molecular dynamics (MM/MD) simulations. Details of the simulations and energy calculations are provided as Supporting Information.
RESULTS
Ligand Design
The design of a G-quadruplex-targeted platinating agent requires linking the metal to a ligand that promotes efficient binding to this DNA secondary structure. A survey of the literature on G-quadruplex interactive molecules suggests that perylene-based ligands should be a suitable carrier system. The interaction of perylenetetracarboxylic diimides with G-quadruplexes is mediated by π-stacking between the ligands’ large hydrophobic polyaromatic scaffold and the G-tetrad 21–28. While perylene diimides have been demonstrated to unwind plasmid DNA relaxed by topoisomerase I, which has been interpreted as a result of intercalative binding 29, circular dichroism spectra strongly suggest that these chromophores associate with the grooves/backbone of the biopolymer nonspecifically in the form of (exciton-coupled) stacked multimers 30. Certain perylene ligands have demonstrated selectivity for the human telomeric G-quadruplex over other G-quadruplexes 31.
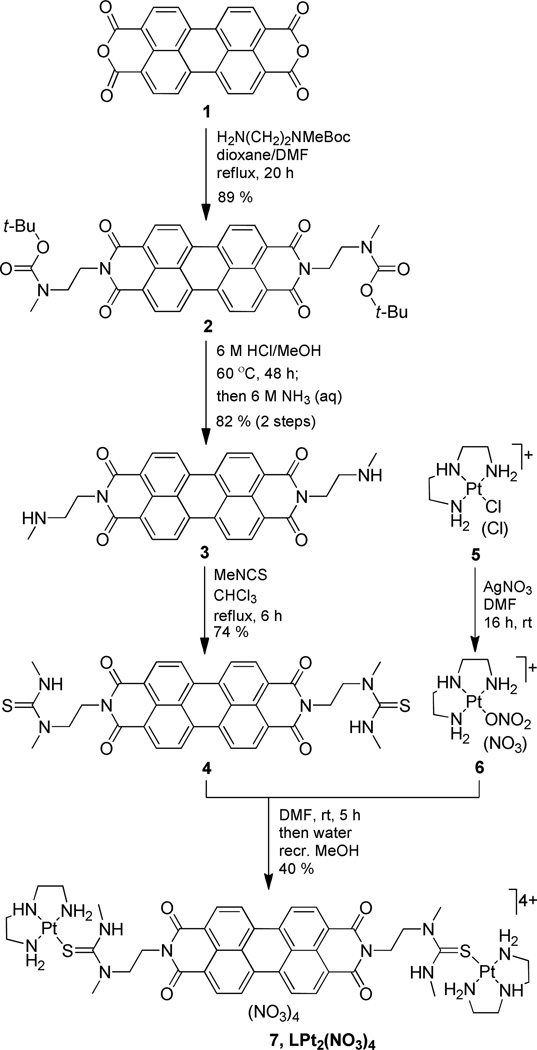
The proposed mechanism of a G-quadruplex-platinating agent involves rapid (large kon) reversible binding 32 and subsequent slow (kPt ≈ 10−4 s−1) formation of adducts with loop adenine 14 by the electrophilic metal. To study the reversible binding event without interference from irreversible reactions of the metal, we designed a model compound containing substitution inert platinum moieties. The carrier ligand, N,N´-bis[1,3-dimethylthioureaethyl]-3,4,9,10-perylenetetracarboxylicdiimide (L, 4), and the corresponding platinum derivative, [{Pt(dien)}2(μ-4-S,S´)](NO3)4 (LPt2(NO3)4, 7) (dien = diethylenetriamine), were generated as follows (Scheme 1): dianhydride 1 was reacted with selectively protected N-methylethylenediamine to give compound 2. Subsequent transformation of the pendant deprotected secondary amino groups in diamine 3 into thiourea yielded carrier ligand 4, which was treated with activated [Pt(dien)] complex 6 in DMF to produce the desired target compound as the water soluble tetranitrate salt, 7. The lack of chloro leaving groups on the platinum centers (both thiourea and the dien chelate are not replaced by DNA nitrogen) renders compound 7 a suitable model for mimicking the reversible binding step. Simple molecular docking experiments and molecular dynamics simulations were performed with the lowest-energy conformation of 7 (C2/syn conformer; dPt⋯Pt ~ 20 Å) to confirm that the geometry of the platinum-containing side chains is compatible with the dimensions of the G-quadruplex (Supporting Information).
Scheme 1.
Synthesis of platinum–perylene agent LPt2(NO3)4 (7).
Absorption spectra recorded of compound 7 in Tris or phosphate buffered solution show features characteristic of concentration- and temperature-dependent self-aggregation, a hallmark of perylene dyes 26,33,34. The isosbestic behavior observed in variable-temperature absorption spectra suggests that 7 exists in solution as a mixture of monomers and dimers at concentrations as low as 10 µM, but does not undergo extensive aggregation beyond the dimer state at concentrations as high as 200 µM. At concentrations below 1 µM and at 25 °C the compound appears to exist entirely in its monomeric form (Supporting Information).
DNA and RNA Binding Studied by UV-visible Spectroscopy and Circular Dichroism (CD) Spectropolarimetry
The interactions of compound 7 with G-quadruplex forming sequences mimicking human telomeric DNA and RNA, along with double-stranded DNA, were studied by UV-visible spectroscopy and CD spectropolarimetry. Short sequences known to assemble into G-quadruplex structures intermolecularly as well as four-repeat sequences that fold into distinct intramolecular G-quadruplex structures were tested in this study for their ability to bind compound 7. Random-sequence native DNA and the duplex form of the telomeric DNA repeat were also studied. The sequences and the conformations adopted by them in K+ solution are summarized in Table 1.
Table 1.
Oligo(deoxy)ribonucleotide sequences studied
| Name | Sequence | Major conformation(s) in K+ solution |
|---|---|---|
| dG-8 | d(TAGGGTTA) | parallel tetramolecular G-quadruplexa |
| dG-12 | d(TAGGGT)2 | mixture of parallel and antiparallel bimolecular G-quadruplexesb |
| dG-24 | d(TTAGGG)4 | mixture of hybrid [3+1] and antiparallel [2+2] unimolecular G-quadruplexesc |
| rG-24 | r(UUAGGG)4 | parallel unimolecular G-quadruplexd |
| ds-22 | d(AGGG(TTAGGG)3)/d(CCCTAA)3CCCT) | Watson–Crick duplex |
UV-visible spectroscopy was used to monitor spectral changes in the absorption spectrum of compound 7 in the presence of unimolecular G-quadruplexes, dG-24 and rG-24, and native double-stranded DNA. The platinum-free perylene precursor 3·2HCl (Scheme 1) was titrated into the same sequences to determine if the platinum moieties modulate the interactions of the perylene chromophore with the secondary structures. In all of the spectra, the characteristic feature in the 400–600 nm region (π−π* transition within the perylene core) showed a pronounced bathochromic shift of 8–10 nm and altered relative band intensities within the vibronic fine structure, suggesting strong association of 3·2HCl and 7 with the nucleic acids (Supporting Information). Both derivatives showed a binding preference for the quadruplex nucleic acids, as judged by the relative hyperchromicities observed in the UV-visible spectra. Interestingly, the relative order of affinities observed for the platinum–perylene 7 was dG-24 > rG-24 > calf thymus DNA, whereas the order for 3·2HCl was rG-24 > dG-24 > calf thymus DNA, suggesting that the platinum-containing side chains increase the binding selectivity of the perylene derivative for the DNA secondary structure. Equilibrium dialysis experiments performed with compound 7 in dG-24 and calf thymus DNA suggest an approximately 2-fold higher binding affinity of the platinum–perylene for the quadruplex DNA (Supporting Information).
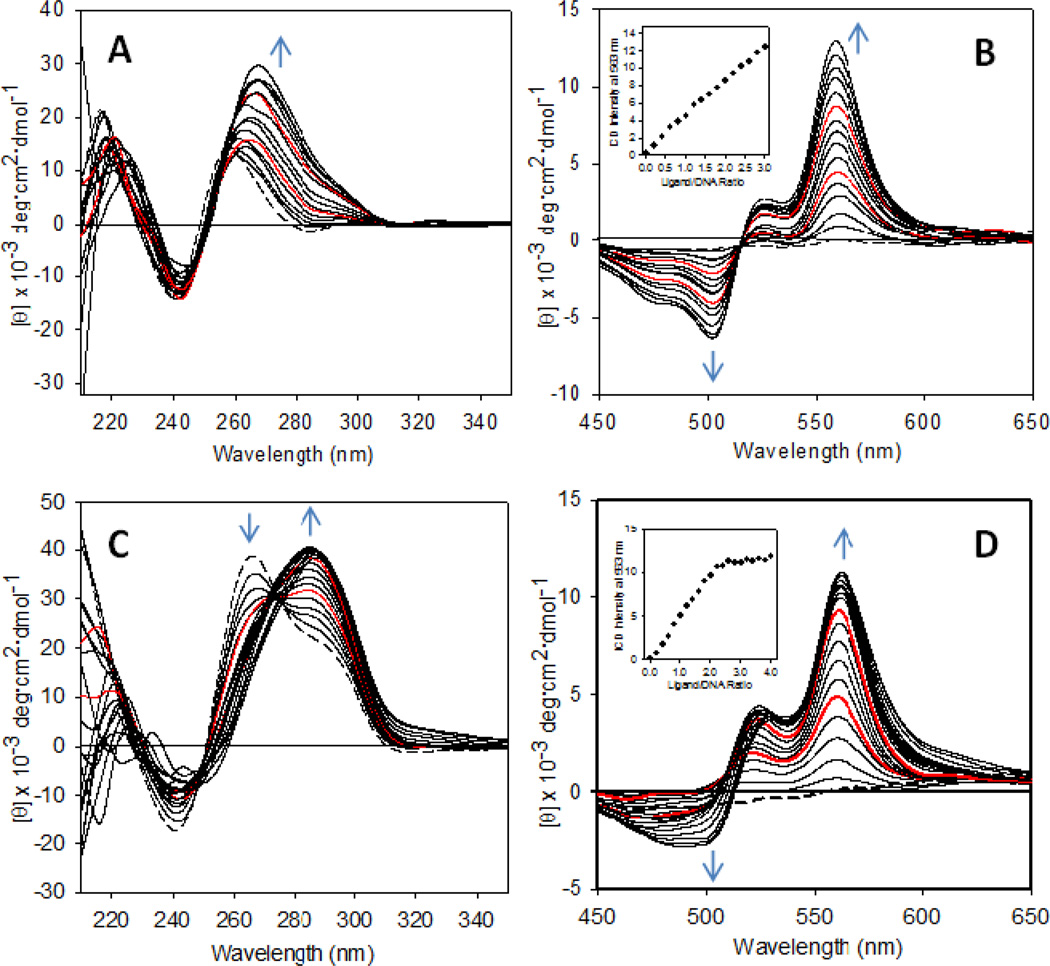
To gain further insight into structural details of the binding geometry, stoichiometry, and selectivity, as well as the conformational changes in the human telomeric G-quadruplex caused by compound 7, titrations of this agent into solutions of selected model sequences were performed and monitored by CD spectropolarimetry. Each structure was titrated with 0.2–0.25 equivalents of 7 up to a four-fold excess of perylene agent. When the sequence dG-8, which forms a parallel tetramolecular G-quadruplex in K+-containing solution 17, was titrated with three equivalents of compound 7, the positive CD band at 260 nm increased in intensity and shifted to 265 nm (Figure 2A). This behavior suggests that the platinum–perylene agent binds to the parallel G-quadruplex and alters its conformation, however, without changing the strand polarity of the structure. Binding of compound 7 to this form of G-quadruplex is supported by the appearance of an induced CD (ICD) signal in the ligand region consisting of a negative band at 502 nm and a positive band at 559 nm, both increasing linearly with the addition of ligand (Figure 2B). The distinct bisignate nature of this signal indicates exciton coupling between multiple bound perylene chromophores 30,35. Previous NMR studies have demonstrated that the perylenediimide agent PIPER forms a 1:1 complex with the parallel G-quadruplex, in which the ligand stacks with the G-tetrad formed at the 5´-GT step 17. The ICD data acquired in this study suggest that additional ligand exceeding this stoichiometry self-aggregates on the G-quadruplex.
Figure 2.
CD spectra recorded for the titration of sequences dG-8 and dG-12 with compound 7 in K+-containing solution. (A) and (C) show changes in the DNA region and (B) and (D) show changes in the ligand (ICD) region during the addition of ligand to dG-8 and dG-12, respectively. Spectral changes with increasing ligand concentration are indicated by blue arrows. The red traces are spectra recorded after addition of 1 and 2 equivalents of ligand, and the dashed traces are spectra of the unmodified DNA. Plots of ICD intensity at 560 nm vs. ligand-to-DNA ratio are shown as insets in (B) and (D).
An entirely different situation is observed when the bimolecular G-quadruplex formed by dG-12 is titrated with compound 7. The DNA in the absence of ligand shows a CD signal consisting of a band of maximum intensity at 265 nm and a shoulder around 290 nm 36,37 (Figure 2C), confirming that this sequence assembles into a parallel and an antiparallel structure, which coexist in a dynamic equilibrium in solution 38. When compound 7 is added to the sequence, the parallel form is efficiently converted into the antiparallel form, as suggested by the disappearance of the band at 265 nm and transformation of the long-wavelength shoulder into a band centered at 287 nm (Figure 2C). Two equivalents of compound 7 are required to effect complete conversion, and additional equivalents do not seem to have a significant effect on the resulting DNA conformation. Likewise, changes in the ICD signal indicate saturation is reached after addition of two equivalents of ligand (Figure 2D). This observation and the fact that the negative band of the ICD signal develops only after addition of additional equivalents of compound 7 suggest that the first two equivalents bind to specific sites to form a 2:1 ligand–DNA complex. To study potential binding modes in this system and to rationalize the observed binding selectivity, we performed molecular dynamics simulations of the 2:1 ligand–DNA complexes and calculated their free energies using the hybrid molecular mechanics–Poisson Boltzmann surface area (MM–PBSA) approach 39,40 (Supporting Information). Contrary to our expectation, the energies extracted from the ensembles of minimized structures did not show a significant difference between the ligand-stabilized parallel and antiparallel forms, which is most likely the result of shortcomings of the force field parameterization 40 (Supporting Information).
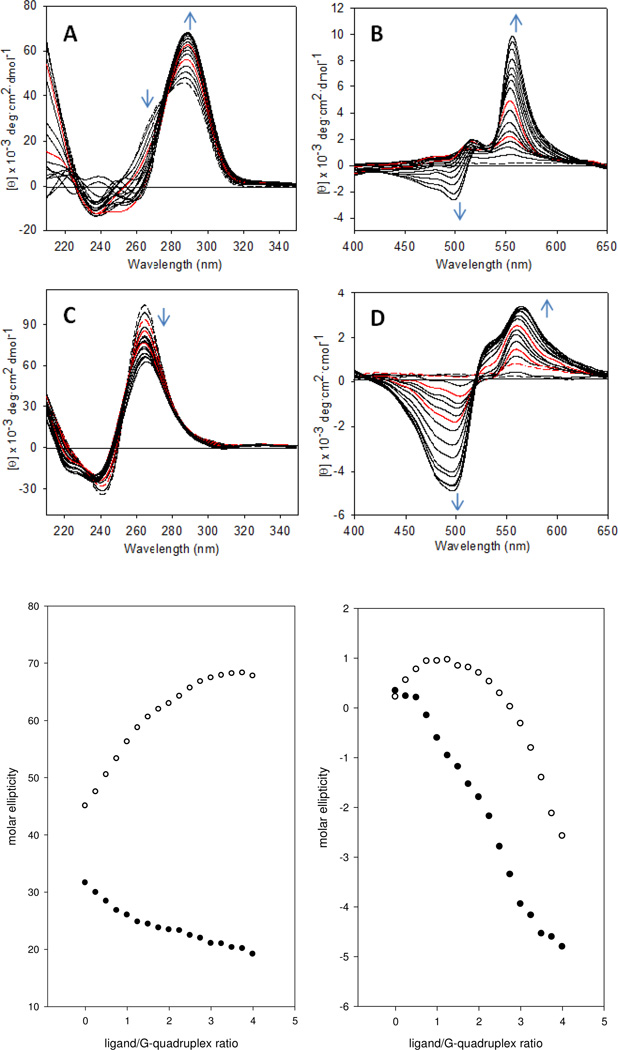
The interactions of compound 7 with the unimolecular G-quadruplexes formed by the sequences dG-24 and rG-24 were also studied. Titrations of these sequences were performed at the same concentration of nucleic acid and in the same buffer used in the isothermal titration calorimetry experiments (see below) to allow for a direct comparison of the spectroscopic with the thermodynamic signatures of both binding processes. The CD signature observed for the DNA sequence in the absence of ligand is consistent with the presence of multiple conformations with pairwise antiparallel ([2+2], chair- or basket-type) and hybrid ([3+1]) strand orientation, respectively 41–45 (Figure 3A). Upon titration with compound 7, the shoulder of the CD band at ~270 nm in the DNA region disappears, and the band develops into a symmetrical signal centered at 290 nm, suggesting the ligand stabilizes one specific conformation (Figure 3A,E). Changes in the ICD signal (Figure 3B,F) observed during the course of the addition of four equivalents of compound 7 to this sequence are consistent with the formation of a 2:1 ligand–DNA complex with additional ligand associating in a non-specific manner, similar to the situation observed for the sequence dG-12. Variable-temperature CD spectra were also recorded to demonstrate that compound 7 increases the thermal stability of the G-quadruplex formed by dG-24 in K+-containing solution and is able to induce this structure from random-coil sequence in the absence of monocation (Supporting Information).
Figure 3.
CD spectra recorded for the titration of sequences dG-24 and rG-24 with compound 7 in K+-containing solution. (A) and (C) show changes in the DNA region and (B) and (D) show changes in the ligand (ICD) region during the addition of ligand to dG-24 and rG-24, respectively. Spectral changes with increasing ligand concentration are indicated by blue arrows. The red traces are spectra recorded after addition of 1 and 2 equivalents (A, B) or 0.5 (dashes), 1, and 2 equivalents (C, D) of ligand, and the dashed black traces are spectra of the unmodified DNA. (E) Plot of nucleic acid-based CD signal intensities vs. ligand concentration for dG-24 (288 nm, open circles) and rG-24 (264 nm, filled circles). (F) Plot of ligand ICD signal intensities vs. ligand concentration for dG-24 (500 nm, open circles) and rG-24 (500 nm, filled circles).
The CD spectra recorded for the titration of the sequence rG-24 are strikingly different from those recorded for dG-24. Unlike human telomeric quadruplex DNA, the corresponding RNA sequence exists in a single conformation in which the four strands adopt a parallel orientation and form propeller-type double-chain reversal loops 8. Folding into this structure appears to be independent of the nuclearity of the G-quadruplex, the flanking sequences, and the nature of the monocation present 46–49. The inability of telomeric RNA to adopt antiparallel or hybrid conformations can be explained with an energetically prohibitive anti→syn conformational switch within the rG residues 50. The CD signature observed for the sequence rG-24 is in agreement with the formation of a parallel G-quadruplex, based on the presence of the positive band at 264 nm and a minimum at 242 nm, which is characteristic of this topology 37 (Figure 3C). When compound 7 is added to this sequence, the positive band at 265 nm decreases in intensity (Figure 3C,E). This decrease in ellipticity in the RNA region of the CD spectrum suggests that compound 7 perturbs the G-quadruplex without causing a transition from one structure to another. The ligand region of the CD spectra provides important clues about both the structure of rG-24 in solution and the binding mode of compound 7. Unlike in titrations of dG-24, the ligand produces a pronounced bisignate ICD signal with an intense negative band at 500 nm (Figure 3D). Interestingly, this band appears at an earlier stage of the titration compared to the situation in dG-24 when the ligand-to-RNA ratio exceeds 0.5:1 (Figure 3F). This observation suggests that the first half equivalent of compound 7 binds to rG-24 in a unique geometry, and additional ligand molecules most likely form stacked ligand–ligand aggregates, giving rise to exciton coupling. This interesting observation can be explained with the fact that telomeric G-quadruplex RNA has a high tendency to aggregate in solution, which has recently been demonstrated using electrospray mass spectrometry (ES–MS) 46. The parallel RNA G-quadruplex has a flattened, propeller-type structure and does not contain diagonal loops that span across the G-quartet 8. This facilitates inter-quadruplex π-stacking between G-quartets leading to the formation of 5´-5´ stacked dimers 48. Thus, the first binding event observed in titrations of rG-24 with compound 7 most likely involves association of a single ligand molecule with a G-quadruplex RNA dimer. Addition of a second equivalent of ligand produces a negative ICD band at 500 nm (Figure 3D), suggesting interactions between the first two bound guest chromophores. The CD data would therefore be in agreement with a binding mode that involves intercalation of two stacked molecules of compound 7 between two RNA G-quadruplexes, which is strongly supported by the microcalorimetry data (see below).
Previous studies have demonstrated that the perylene derivative PIPER is able to induce the G-quadruplex structure from double-stranded DNA in the promoter region of the oncogene c-myc 51. To test if this reactivity exists in telomeric DNA, compound 7 was titrated into a solution containing the duplex ds-22 and CD spectra were recorded immediately after adding ligand and after incubating the samples for 24 h at 37 °C (Supporting Information). The spectra indicate that the CD signature of the duplex ds-22, which shows a positive band at 267 nm and a negative band at 242 nm characteristic of a B-form duplex 36, persists in the presence of compound 7, while there is a noticeable decrease in intensity of the positive CD band. Unlike CD spectra recorded for compound 7 in complex with the G-quadruplex, the spectra recorded for ds-22 in the ligand region show only a weak ICD signal (Supporting Information). The weak negative ICD band at ~580 nm observed for the 2:1 complex can be attributed to the formation of exciton-coupled stacked dimers of compound 7 in the DNA minor groove, which is reminiscent of the non-intercalative binding mode proposed for perylenediimide derivatives 30. Incubation at elevated temperature had no effect on the CD spectrum. Thus, it can be firmly concluded that the induction of telomeric G-quadruplex DNA from double-stranded DNA is unfavorable in this system.
Binding Stoichiometries of Compound 7 in Complex with dG-24 Studied by Electrospray Mass Spectrometry (ES–MS)
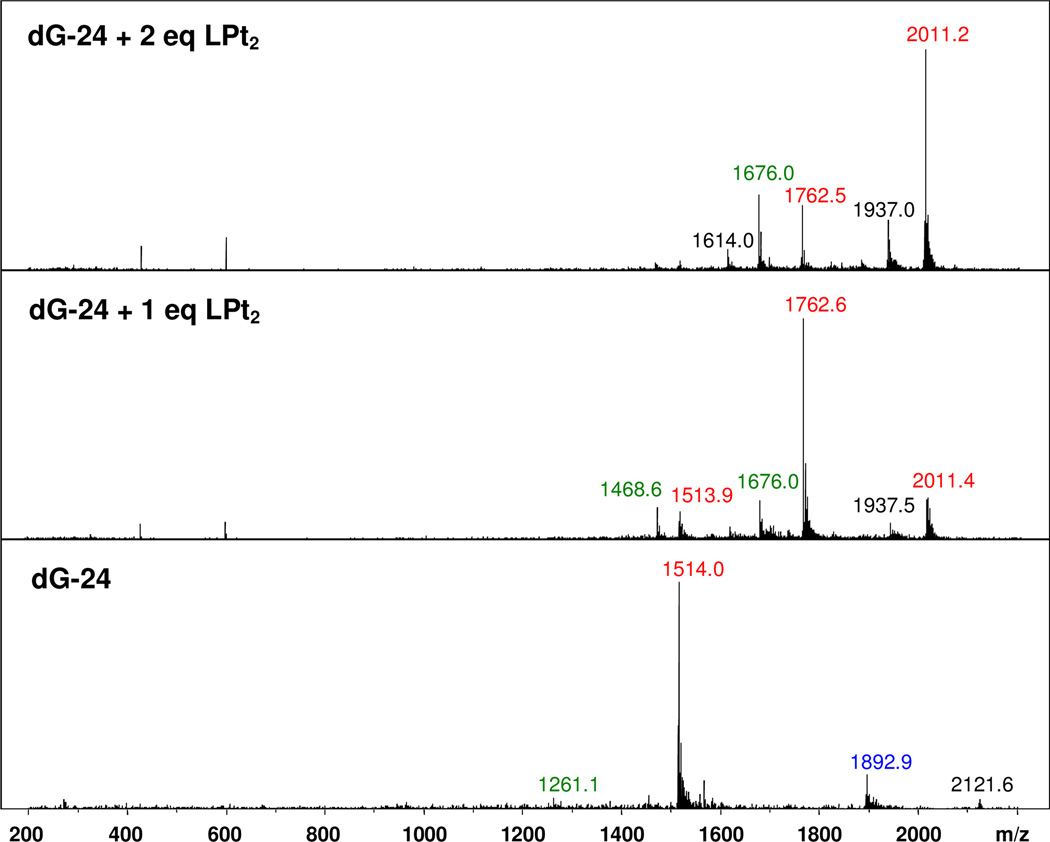
Electrospray mass spectrometry (ES–MS) is a useful analytical tool for the detection of non-covalent complexes formed between small-molecule ligands and nucleic acids52. The method has previously been used to study G-quadruplexes and the complexes of perylene derivatives formed with this type of DNA20,53. Solutions of dG-24 and the 1:1 and 2:1 ligand–DNA complexes were generated in a buffer containing NH4+. The mass spectra recorded in negative–ion mode are shown in Figure 4. A summary of molecular ions is given in Table 2. For the unmodified G-quadruplex DNA, a major peak at m/z 1514 is observed, which can be assigned to the mass of the DNA with z = 5. The two minor peaks at m/z 1893 and m/z 1261 correspond to the DNA in its 4– and 6–charge states, respectively. The mass spectrum of the solution containing one ligand molecule per DNA shows a molecular ion peak at m/z 1762, which can be assigned to the 1:1 ligand–DNA complex carrying a charge of 5–. A minor peak for the 6– ion at m/z 1468 is also observed. At this stoichiometry, a small amount of the 2:1 ligand–DNA complex forms, as evidenced by molecular ion peaks at m/z 2011 (z = 5) and m/z 1676 (z = 6). This, and the observation that free DNA is also present (m/z 1514), suggests a dynamic equilibrium exists between the three species in solution. In the mass spectrum recorded for the 2:1 ligand–DNA mixture, the molecular ion peak at m/z 2011 (z = 5) assigned to the 2:1 complex shows the highest intensity, confirming it is the dominant species in solution. No unbound monomeric or dimerized compound 7 is observed. Likewise, no complexes exceeding the 2:1 stoichiometry are detectable in spectra recorded in the presence of additional ligand. Thus, it can be concluded that compound 7 forms a stable 2:1 complex with the telomeric sequence dG-24 and that additional ligand binds with significantly reduced affinity.
Figure 4.
Negative-ion electrospray mass spectra recorded for solutions containing dG-24, dG-24 + 1 eq. compound 7, and dG-24 + 2 eq. compound 7. Molecular ions with z = 4, 5, and 6 are labeled with blue, red, and green m/z values, respectively. Peaks labeled in black are unassigned species.
Table 2.
Assignment of molecular ions detected in electrospray mass spectraa for titrations of sequence dG-24 with compound 7 (LPt2(NO3)4)
| m/zb | Molecular ion |
|---|---|
| 1261 | (dG-24)6− |
| 1468 | [(dG-24)·LPt2]6− |
| 1514 | (dG-24)5− |
| 1676 | [(dG-24)·(LPt2)2]6− |
| 1762 | [(dG-24)·LPt2]5− |
| 1892 | (dG-24)4− |
| 2011 | [(dG-24)·(LPt2)2]5− |
Recorded in negative-ion mode in 50 mM ammonium acetate buffer.
The molecular weights of dG-24 (neutral form) and LPt2 are 7574.9 g mol−1 and 1247.3 g mol−1, respectively.
Interactions Between Compound 7 and the Sequences dG-24 and rG-24 Studied by Isothermal Titration Calorimetry (ITC)
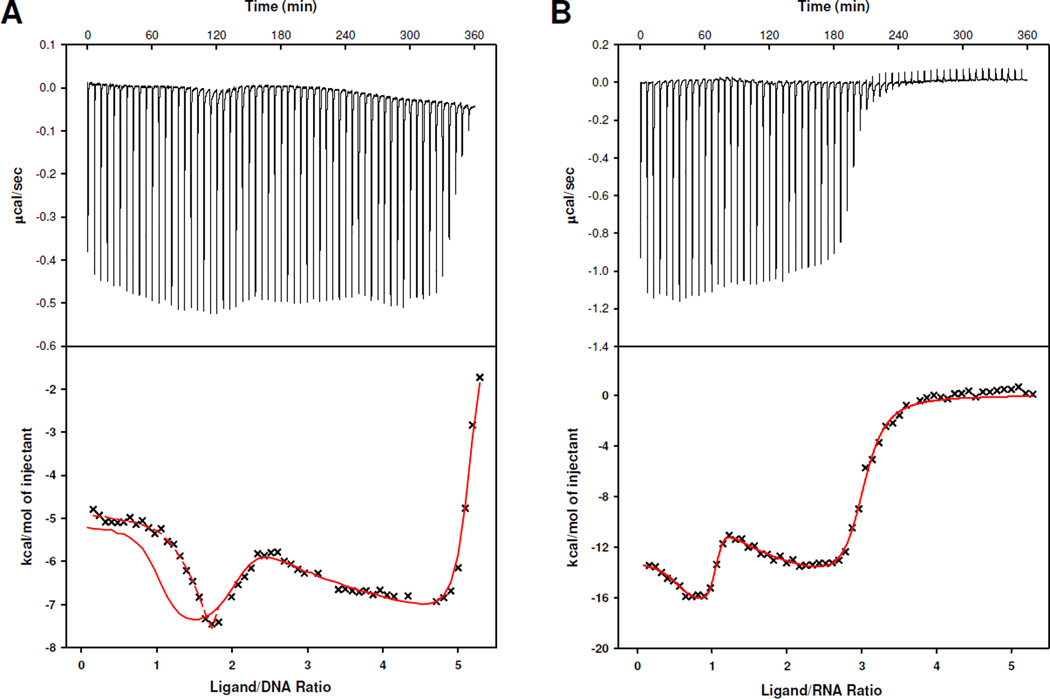
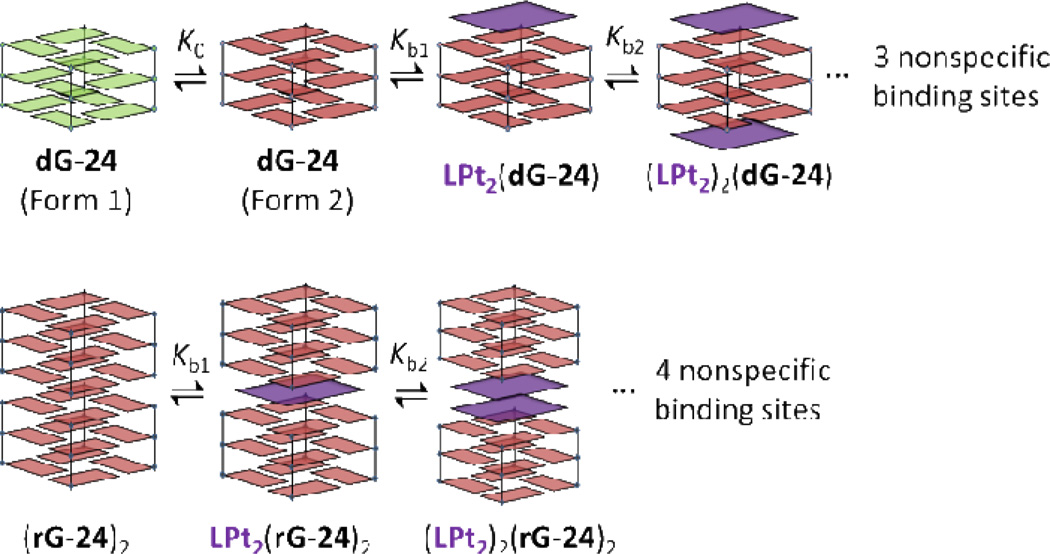
Due to the complexity of the ligand binding, attempts to estimate binding constants from CD spectral changes by fitting the titration curves in Figure 3E,F to conventional multi-site and tight-binding models54 were unsuccessful. Alternatively, ITC experiments were performed under the same conditions used in the CD titrations to deconvolute the binding processes and explore the energetics of association of compound 7 with dG-24 and rG-24. The raw data of heat change over time (top) and plots of the integrated, corrected molar heats versus the ligand-to-nucleic acid ratios (bottom) are given in Figure 5. The ITC profiles recorded for the telomeric DNA and RNA show pronounced differences. In both cases, the integrated ITC data were fit to the sequential binding model (see Supporting Information), which is based on the assumption that compound 7 binds to the G-quadruplexes in distinct, consecutive binding events to non-identical sites. Dimerization of compound 7 was considered negligible under the specific conditions of the ITC experiment and was not included in the modeling of the calorimetric data. Based on the changes in relative absorptivities observed in simple dilution experiments performed with compound 7 in the same buffer (Supporting Information), dimerization of this agent (Kdim ≈ 3 × 104 M−1) proves to be approximately three orders of magnitude weaker than the first two binding events with the G-quadruplexes (see below). This model produced the best fits for the data sets generated for dG-24 and rG-24. The results of the data fitting routines are shown as red traces in the bottom panels of Figure 5A and Figure 5B. As can be seen in Figure 5B, the curve fit generated for rG-24 describes the entire experimental data set well. By contrast, the same model does not produce a good fit for the first equivalent of added compound 7 in the case of dG-24, but is able to predict the data well in the later stages of the titration (Figure 5A). To account for this discrepancy, the sequential binding model had to be modified to include a pre-binding G-quadruplex equilibrium (Supporting Information). This was based on the following reasoning: as demonstrated previously by optical and high-resolution spectroscopies41–45 (and in agreement with the CD data acquired in this study), the sequence dG-24 folds into co-existing G-quadruplex structures in solution. Our CD titrations also suggest that multiple conformations exist, to one of which compound 7 binds selectively. A reaction scheme which includes a 70%/30% equilibrium distribution of two major conformations provided a good fit of the data (dashed red trace in Figure 5A, bottom panel). Clearly, the polymorphism of the G-quadruplex DNA complicates the interpretation of the calorimetric data acquired for the first binding event.
Figure 5.
ITC profiles for the titrations of dG-24 (A) and rG-24 (B) with compound 7. The top top panels show the raw data of heat flow vs. time and the bottom panels show the calculated molar heats of ligand binding. The solid red traces in (A) and (B) are best fits to the sequential binding site model. The dashed line in (A) represents a best-fit for the first binding event based on a mechanism involving a pre-binding G-quadruplex equilibrium.
Based on the titration data in Figure 5A, the telomeric sequence dG-24 binds five equivalents of compound 7. The first two binding events can be distinguished easily based on their differences in heat production, which is manifest as distinct up- and downward sloping of the titration curve at ligand-to-DNA ratios of 1 and 2. Three additional molecules associate with dG-24 until saturation is reached at a ligand-to-DNA ratio of 5. The shape of the titration curve suggests that the first two equivalents of ligand bind strongly to specific sites, whereas additional ligand associates with the G-quadruplex nonspecifically. By contrast, the telomeric RNA sequence rG-24 shows saturation behavior after addition of three equivalents of compound 7 (Figure 5B). According to the changes in reaction heat plotted for this titration, the binding process seems to involve one specific binding event and the non-specific binding of two additional ligands. Unfortunately, none of the binding models available in the ITC/Origin software, including the sequential binding model, was able to fit the experimental data. This problem was solved after taking into account observations in the recent literature48 and the results of the CD titration of rG-24, which suggest that compound 7 binds to the dimerized form of the RNA G-quadruplex. This led us to propose a modified model in which the first binding event occurs with one ligand molecule per RNA dimer, which corresponds to a ligand-to-RNA ratio of 0.5. According to this scheme, each rG-24 dimer binds two ligands with high specificity, as implicated by the curve shape at ligand-to-RNA ratios of ≤ 1, and four additional ligands nonspecifically at ligand-to-RNA ratios of 1–3.
For the first two (specific) binding events the thermodynamic parameters Kb, ΔH°, ΔS°, and ΔG° were calculated (Table 3). A comparison of the binding constants for the first binding step (Kb1) determined for dG-24 and rG-24 shows that compound 7 binds to the former sequence with an affinity that is at least one order of magnitude higher than that observed for the RNA. For the first binding event in dG-24 a Kb1 in the 108–109 M−1 range can be estimated (the accuracy of this value is limited by the large error in experimental binding heats for such high binding affinities55,56). By contrast, the Kb2 values for the second binding event show similar values for dG-24 and rG-24 in the range 107–108 M−1. Additional ligand binds with Kb values in the range 105–106 M−1 (not listed).
Table 3.
Summary of thermodynamic dataa for titrations of sequences dG-24 and rG-24 with LPt2(NO3)4 (7)
| First binding event | Second binding event | |||||||
|---|---|---|---|---|---|---|---|---|
| Sequence |
Kb1 (M−1) |
ΔH1° (kJ mol−1) |
−TΔS1° (kJ mol−1) |
ΔG1° (kJ mol−1) |
Kb2 (M−1) |
ΔH2° (kJ mol−1) |
−TΔS2° (kJ mol−1) |
ΔG2° (kJ mol−1) |
| dG-24 | > 108 | −23.5b | −29.1 | −52.6 | 4.4 × 107 | −31.7 | −12.8 | −44.5 |
| rG-24 | 7.8 × 107 | −55.3 | 10.3 | −44.4 | 2.7 × 107 | −75.1 | 32.8 | −42.2 |
For the first two binding events (n = 1, 2); extracted from ITC data acquired at T = 298 K and fitted with the sequential binding model.
Experimental error 10%, < 5% for all other enthalpies.
A comparison of the thermodynamic data calculated for the interactions of compound 7 with the telomeric DNA and RNA shows that the thermodynamic driving forces for ligand binding to the two sequences are very different. While the interaction of compound 7 with dG-24 is both enthalpically and entropically favorable, ligand binding to rG-24 is entropically disfavored, and the sole driving force for RNA–ligand association is the large binding enthalpy. Interestingly, the binding heat generated per mole of ligand binding to rG-24 is approximately twice as large as the heat observed for the titration of dG-24. These observations support the proposed binding mechanism involving the dimeric form of rG-24. An intercalative binding mode in the former complex, in which compound 7 is sandwiched between two RNA molecules, would maximize hydrophobic ligand–RNA interactions (large negative ΔH°) but not cause a significant gain in dehydration entropy.
DISCUSSION
The purpose of this study was to delineate the molecular mechanism by which compound 7, a platinum–perylene agent, recognizes the human telomeric G-quadruplex. The design of this type of compound was inspired by its potential therapeutic applications as a G-quadruplex-specific, telomere-targeted platinating agent. More specifically, the ultimate goal of this approach is to generate a hybrid agent that recognizes and stabilizes the G-quartets in the human telomeric G-quadruplex to deliver platinum selectively to the loop adenines in this DNA secondary structure. The most critical event in the binding mechanism of such an agent would be the reversible, first binding step. Thus, a suitably modified reversible binder was studied in order to shed light onto the recognition of the G-quadruplex structure by this type of conjugate.
The cationic platinum(II)-modified perylene derivative 7 shows excellent solubility in aqueous buffers and does not undergo aggregation beyond the dimeric state in the concentration range 10–200 µM. This is most likely a consequence of the pH-independent 4+ charge, which renders the formation of self-stacked multimers unfavorable. Using a combination of spectroscopic and biophysical methods we demonstrated that compound 7 shows a distinct preference for G-quadruplexes exhibiting an antiparallel strand orientation. One of the most striking features of compound 7 is its ability to shift the equilibrium that exists between the two G-quadruplex structures adopted by dG-12 from the parallel to the antiparallel form. Using the sequences dG-24 and rG-24 we demonstrated that the binding preference of compound 7 for the antiparallel G-quadruplex persists for the biologically more relevant unimolecular form of this nucleic acid secondary structure. The CD spectrum recorded for the 2:1 complex (Figure 3), which shows a highly symmetrical positive band in the DNA region around 290 nm, is highly reminiscent of the CD signature observed for the all-antiparallel (“basket”, [2+2]) structure (typically observed in Na+-containing solution)57. However, high-resolution structural studies are needed to substantiate this notion. Another important observation in this system is that compound 7 is unable to induce the (naturally disfavored) antiparallel form of the telomeric RNA G-quadruplex. This is a critical feature as it allows compound 7 to discriminate between the DNA- and RNA-form of the human telomeric repeat. It is now well established that the noncoding telomeric DNA is transcribed into UUAGGG-containing repeats (termed TERRA), but the cellular function of the transcripts and their role in telomere regulation remain elusive58. Selective recognition and/or induction of the antiparallel G-quadruplex by exogenous ligands designed to target the telomeres, therefore, would be an important feature in cells where high levels of telomeric transcript may compete with DNA binding. In this regard, the tendency of compound 7 to also strongly associate with double-stranded DNA would create a highly competitive situation for a telomere-targeted agent in a cellular context, which may require changes to the current design. It is unclear, however, if non-intercalative groove aggregation or electrostatic binding of the platinum–perylenes with B-form DNA will ultimately promote the (undesired) formation of irreversible adducts by the electrophilic metal in genomic DNA.
While the design of compound 7 and its derivatives as a truly selective telomere-targeted therapy may require improvements, the high aqueous solubility and low degree of self-stacking render this agent a useful probe for studying telomeric G-quadruplex structure in solution. On the basis of the experimental data acquired in this study, it is possible to propose binding mechanisms for compound 7 with the DNA and RNA forms of the human telomeric repeat, in which the platinum moieties contribute to the binding affinity and selectivity of the agent. The latter notion is consistent with previous reports demonstrating that the nature of the side chains in perylenediimides has a major influence on G-quadruplex affinity and conformation (21). In this regard, it is noteworthy to mention that other ligands have been studied for their interactions with the telomeric G-quadruplex (DNA) and their effect on the structure’s polymorphism in dilute solution using combined spectroscopic/calorimetric methods. TMPyP4, for instance, a tetracationic porphyrin derivative, has been demonstrated to induce the antiparallel basket form of the G-quadruplex in a process involving one strong (and structure-determining) and three weaker sequential binding events to produce a 4:1 ligand–DNA complex.59 While compound 7 shows a similar binding affinity (Kb > 10−8 M−1) and preference for the antiparallel form of the G-quadruplex (DNA), the stoichiometry and thermodynamic signature of the binding process show critical differences from that of the porphyrin ligand.
A plausible mechanism of complex formation between compound 7 and the telomeric G-quadruplex DNA involves end-on stacking to inequivalent binding sites of two ligand molecules with one specific antiparallel structure (Figure 6A). In particular, the absence of exciton coupling between the first two equivalents of bound ligand supports this binding mode and rules out ligand–ligand aggregation on the G-quadruplex. By contrast, the binding events in telomeric RNA are more complicated due to the aggregation state of this sequence in solution. A recent X-ray crystallographic study has shown that in the solid state telomeric RNA packs as dimers in which two G-quadruplexes are mutually stacked in a 5´ to 5´ fashion47. Such an arrangement would produce three high-affinity sites for perylene binding: two identical terminal end-on binding sites and one unique intercalation site at the hydrophobic interface of the two RNA molecules. Since the first binding event involves association of a single molecule of compound 7 with the RNA dimer, intercalation is the most likely binding mode (Figure 6B). The second equivalent of ligand then co-intercalates with the first molecule as a stacked chromophore dimer (ICD of exciton-coupled neighboring perylenes). This binding mode, in which a double-layer of ligands is sandwiched between two 5´ G-quartets, has recently been observed in the solid state of an RNA G-quadruplex–acridine complex60.
Figure 6.
Proposed mechanism of complex formation between compound 7 and the human telomeric DNA and RNA quadruplexes.
In conclusion, platinum–perylenes based on compound 7 provide a new platform for the design of G-quadruplex-binding agents. The emergence of novel topologies in the complex structural landscape of G-quadruplexes has spawned a search for small molecules with predictable binding preferences both for biochemical and biomedical applications61,62. By combining the intercalator-driven irreversible nucleobase interactions of platinum with the selective recognition of the antiparallel form of the human telomeric G-quadruplex, it should be possible to engineer hybrid agents for applications in telomere-directed chemotherapy and as chemical probes for trapping and detecting G-quadruplexes in live cells.
Supplementary Material
ACKNOWLEDGMENT
J.D.D. was supported by the Wake Forest University Summer Research Fellowship program. We thank Xiao Xu (WFU, Department of Physics) for assistance with the MATLAB computations and Amanda J. Pickard (WFU, Chemistry) for assistance with the acquisition of UV-visible spectra.
FUNDING
This work was funded in part by the U. S. National Institutes of Health (grant CA101880).
ABBREVIATIONS USED
- G
guanine
- COSY
correlated spectroscopy
- CD
circular dichroism
- ICD
induced circular dichroism
- SDS
sodium dodecyl sulfate
- ES-MS
electrospray mass spectrometry
- ITC
isothermal titration calorimetry
- MM/MD
molecular mechanics/molecular dynamics
- MM-PBSA
molecular mechanics-Poisson Boltzmann surface area
Footnotes
ASSOCIATED CONTENT
Supporting Information. COSY spectrum of compound 7; UV-visible and CD titrations of compounds 3·2HCl and 7; results of the equilibrium dialysis experiment; ligand binding models used to fit ITC data; experimental details, discussion, and energy calculations for MM/MD simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Parkinson GN. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, U.K.: RSC Publishing; 2006. pp. 1–30. [Google Scholar]
- 2.Patel DJ, Phan AT, Kuryavyi V. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neidle S. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn EH, Greider CW, Szostak JW. Nat. Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 6.Harley CB. Nat. Rev. Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 7.Kelland L. Clin Cancer Res. 2007;13:4960–4963. doi: 10.1158/1078-0432.CCR-07-0422. [DOI] [PubMed] [Google Scholar]
- 8.Phan AT. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 9.Chaires JB. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balasubramanian S, Neidle S. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgiades SN, Abd Karim NH, Suntharalingam K, Vilar R. Angew. Chem. Int. Ed. Engl. 2010;49:4020–4034. doi: 10.1002/anie.200906363. [DOI] [PubMed] [Google Scholar]
- 12.Di Antonio M, Doria F, Richter SN, Bertipaglia C, Mella M, Sissi C, Palumbo M, Freccero M. Journal of the American Chemical Society. 2009;131:13132–13141. doi: 10.1021/ja904876q. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand H, Bombard S, Monchaud D, Teulade-Fichou MP. J Biol Inorg Chem. 2007;12:1003–1014. doi: 10.1007/s00775-007-0273-3. [DOI] [PubMed] [Google Scholar]
- 14.Rao L, Bierbach U. J. Am. Chem. Soc. 2007;129:15764–15765. doi: 10.1021/ja077390a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury JR, Rao L, Bierbach U. J. Biol. Inorg. Chem. 2011;16:373–380. doi: 10.1007/s00775-010-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han HY, Cliff CL, Hurley LH. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 17.Fedoroff OY, Salazar M, Han HY, Chemeris VV, Kerwin SM, Hurley LH. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 18.Martins ET, Baruah H, Kramarczyk J, Saluta G, Day CS, Kucera GL, Bierbach U. J. Med. Chem. 2001;44:4492–4496. doi: 10.1021/jm010293m. [DOI] [PubMed] [Google Scholar]
- 19.Djuran MI, Lempers ELM, Reedijk J. Inorg. Chem. 1991;30:2648–2652. [Google Scholar]
- 20.Casagrande V, Alvino A, Bianco A, Ortaggi G, Franceschin M. J. Mass. Spectrom. 2009;44:530–540. doi: 10.1002/jms.1529. [DOI] [PubMed] [Google Scholar]
- 21.Pivetta C, Lucatello L, Krapcho AP, Gatto B, Palumbo M, Sissi C. Bioorg. Med. Chem. 2008;16:9331–9339. doi: 10.1016/j.bmc.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 22.Franceschin M, Lombardo CM, Pascucci E, D'Ambrosio D, Micheli E, Bianco A, Ortaggi G, Savino M. Bioorg. Med. Chem. 2008;16:2292–2304. doi: 10.1016/j.bmc.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 23.Samudrala R, Zhang X, Wadkins RM, Mattern DL. Bioorg. Med. Chem. 2007;15:186–193. doi: 10.1016/j.bmc.2006.09.075. [DOI] [PubMed] [Google Scholar]
- 24.Tuntiwechapikul W, Taka T, Bethencourt M, Makonkawkeyoon L, Lee TR. Bioorg. Med. Chem. Lett. 2006;16:4120–4126. doi: 10.1016/j.bmcl.2006.04.078. [DOI] [PubMed] [Google Scholar]
- 25.Kerwin SM, Chen G, Kern JT, Thomas PW. Bioorg. Med. Chem. Lett. 2002;12:447–450. doi: 10.1016/s0960-894x(01)00775-2. [DOI] [PubMed] [Google Scholar]
- 26.Kern JT, Kerwin SM. Bioorg. Med. Chem. Lett. 2002;12:3395–3398. doi: 10.1016/s0960-894x(02)00763-1. [DOI] [PubMed] [Google Scholar]
- 27.Kerwin SM. Curr. Pharm. Des. 2000;6:441–471. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 28.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 29.Kern JT, Thomas PW, Kerwin SM. Biochemistry. 2002;41:11379–11389. doi: 10.1021/bi0263107. [DOI] [PubMed] [Google Scholar]
- 30.Micheli E, Lombardo CM, D'Ambrosio D, Franceschin M, Neidle S, Savino M. Bioorg. Med. Chem. Lett. 2009;19:3903–3908. doi: 10.1016/j.bmcl.2009.03.106. [DOI] [PubMed] [Google Scholar]
- 31.Ragazzon P, Chaires JB. Methods. 2007;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White EW, Tanious F, Ismail MA, Reszka AP, Neidle S, Boykin DW, Wilson WD. Biophys. Chem. 2007;126:140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Wurthner F, Thalacker C, Diele S, Tschierske C. Chem. Eur. J. 2001;7:2245–2253. doi: 10.1002/1521-3765(20010518)7:10<2245::aid-chem2245>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 34.Donati F, Pucci A, Ruggeri G. Phys. Chem. Chem. Phys. 2009;11:6276–6282. doi: 10.1039/b903120k. [DOI] [PubMed] [Google Scholar]
- 35.Rossetti L, D'Isa G, Mauriello C, Varra M, De Santis P, Mayol L, Savino M. Biophys. Chem. 2007;129:70–81. doi: 10.1016/j.bpc.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paramasivan S, Rujan I, Bolton PH. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Phan AT, Patel DJ. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sponer J, Spackova N. Methods. 2007;43:278–290. doi: 10.1016/j.ymeth.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fadrna E, Spackova N, Stefl R, Koca J, Cheatham TE, 3rd, Sponer J. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phan AT, Luu KN, Patel DJ. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wlodarczyk A, Grzybowski P, Patkowski A, Dobek A. J Phys Chem B. 2005;109:3594–3605. doi: 10.1021/jp045274d. [DOI] [PubMed] [Google Scholar]
- 45.Chang C-C, Chien C-W, Lin Y-H, Kang C-C, Chang T-C. Nucl. Acids Res. 2007;35:2846–2860. doi: 10.1093/nar/gkm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. J Am Chem Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 47.Collie GW, Haider SM, Neidle S, Parkinson GN. Nucleic Acids Res. ASAP; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martadinata H, Phan AT. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Kaminaga K, Komiyama M. J. Am. Chem. Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 50.Remin M. J. Biomol. Struct. Dyn. 1997;15:251–264. doi: 10.1080/07391102.1997.10508190. [DOI] [PubMed] [Google Scholar]
- 51.Rangan A, Fedoroff OY, Hurley LH. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 52.Schalley CA, Springer A. Mass Spectrometry and Gas-Phase Chemistry of Non-Covalent Complexes. Hoboken, NJ: Wiley; 2009. Chapter 11. [Google Scholar]
- 53.Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. J. Am. Soc. Mass. Spectr. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 54.Premvardhan L, Maurizot JC. Eur. Biophys. J. 2010;39:781–787. doi: 10.1007/s00249-009-0550-x. [DOI] [PubMed] [Google Scholar]
- 55.Baranauskiene L, Petrikaite V, Matuliene J, Matulis D. Int. J Mol. Sci. 2009;10:2752–2762. doi: 10.3390/ijms10062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiseman T, Williston S, Brandts JF, Lin LN. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Patel DJ. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 58.Lopez de Silanes I, d'Alcontres MS, Blasco MA. Nat Commun. 2010;1:1–9. doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 59.Martino L, Pagano B, Fotticchia I, Neidle S, Giancola C. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 60.Collie GW, Sparapani S, Parkinson GN, Neidle S. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 61.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 62.Sparapani S, Haider SM, Doria F, Gunaratnam M, Neidle S. J. Am. Chem. Soc. 2010;132:12263–12272. doi: 10.1021/ja1003944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.