Abstract
Neural crest cell specification and differentiation to a sympathetic neuronal fate serves as an important model for neurogenesis and depends upon the function of both bHLH transcription factors, notably Hand2, and homeodomain transcription factors, including Phox2b. Here, we define a 1007 bp cis-regulatory element 5′ of the Hand1 gene sufficient to drive reporter expression within the sympathetic chain of transgenic mice. Comparative genomic analyses uncovered evolutionarily conserved consensus-binding sites within this element, which chromatin immunoprecipitation and electrophoretic mobility shift assays confirm are bound by Hand2 and Phox2b. Mutational analyses revealed that the conserved Phox2 and E-box binding sites are necessary for proper cis-regulatory element activity, and expression analyses on both Hand2 conditionally null and hypomorphic backgrounds demonstrate that Hand2 is required for reporter activation in a gene dosage-dependent manner. We demonstrate that Hand2 and Hand1 differentially bind the E-boxes in this cis-regulatory element, establishing molecular differences between these two factors. Finally, we demonstrate that Hand1 is dispensable for normal tyrosine hydroxylase (TH) and dopamine β-hydroxylase (DBH) expression in sympathetic neurons, even when Hand2 gene dosage is concurrently reduced by half. Together, these data define a tissue-specific Hand1 cis-regulatory element controlled by two factors essential for the development of the sympathetic nervous system and provide in vivo regulatory evidence to support previous findings that Hand2, rather than Hand1, is predominantly responsible for regulating TH, DBH, and Hand1 expression in developing sympathetic neurons.
Introduction
All neuronal cell types share a broad developmental program, whereby bHLH transcription factors, cooperatively with other factors, orchestrate the progressive specification of neuronal progenitors (Bertrand et al., 2002; Howard, 2005).
The paired-like homeodomain transcription factor Phox2b is required to define the sympathetic nervous system (SNS) differentiation program. As SNS precursor cells differentiate into sympathetic neurons (SNs), the bHLH protein Hand2 is upregulated in a Phox2b-dependent manner (Howard et al., 2000). The Hand2-related bHLH factor Hand1 is then upregulated in a Hand2-dependent manner (Morikawa et al., 2007). Collectively, Phox and Hand factors drive expression of the norepinephrine biosynthetic enzyme dopamine β-hydroxylase (DBH) (Pattyn et al., 1999; Rychlik et al., 2003; Xu et al., 2003).
Hand2 loss-of-function studies demonstrate its requirement for SN differentiation (Howard et al., 1999; Lucas et al., 2006; Morikawa et al., 2007; Hendershot et al., 2008) whereas Hand2 gain-of-function directs neural crest cells (NCCs) and parasympathetic ganglia to take on noradrenergic characteristics (Howard et al., 1999, 2000; Müller and Rohrer, 2002; Xu et al., 2003).
Hand1 is expressed in SNs, but no other neuronal subtype (Cserjesi et al., 1995; Morikawa and Cserjesi, 2004). siRNA knockdown experiments in cultured mouse superior cervical ganglion, which express both Hand1 and Hand2, indicate that these factors perform overlapping functions, such as the promotion of cell survival (Doxakis et al., 2008). Hand1 expression is lost in NCC-specific Hand2 knock-outs (Morikawa et al., 2007). Although Hand1 expression has not been assessed in Phox2b mutants, as Hand2 expression is dependent upon Phox2b (Howard et al., 2000; Hendershot et al., 2008), Hand1 expression would likely be absent from Phox2b mutant SNs as well. Because it is Hand2-dependent, it is unclear whether Hand1 could functionally compensate for a loss of Hand2 in the SNS; however, Hand2 ablation in differentiated SNs, which express Hand1, results in defective SNs (Schmidt et al., 2009), suggesting Hand2 is not redundant with Hand1.
Here, we investigate the regulatory and functional relationship between Hand1 and Hand2 in developing SNS. We define a 1007 bp evolutionarily conserved region (ECR) 5′ to the Hand1 locus sufficient to drive reporter gene expression throughout the developing and postnatal sympathetic ganglia (SG). Through chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay analyses (EMSAs), we demonstrate that Hand2 and Phox2b bind to evolutionarily conserved consensus sites within this ECR. Additionally, we demonstrate that Hand2 and Phox2b are necessary for Hand1 transcriptional regulation, and that transcriptional regulation is sensitive to Hand2 gene dosage. We demonstrate that, although Hand2 and Hand1 interact with Phox2b identically in vitro, Hand1 and Hand2 display different binding affinities for the conserved E-boxes within the Hand1 SG enhancer. Finally, we demonstrate that tyrosine hydroxylase (TH) and DBH protein expression within SNs is unperturbed by a loss of Hand1 function, either alone or on a Hand2 heterozygous background. Together, these data define a novel SNS-transcriptional module with which to model interactions between the transcription factors that regulate the SNS-specific developmental program. These data provide the first in vivo insights into cooperative Hand2 and Phox2b regulation of downstream genes in the SNS, and shows a true distinction between Hand1 and Hand2 function during SNS development.
Materials and Methods
Bioinformatics.
All sequences were obtained from Ensembl (http://www.ensembl.org) via BLASTN analyses using the mouse Hand1 sequence as a point of reference. Synteny to the Hand1 locus was validated for all conserved noncoding sequences. mVISTA alignment, PATTERNMATCH and CLUSTALW analyses were performed on the following web sites: (http://genome.lbl.gov/vista/index.shtml and http://workbench.sdsc.edu).
Cloning.
Genomic DNA flanking the Hand1 locus was isolated and used to generate reporter constructs corresponding to (−9559 to −7712 bp) common to Constructs #1, #3, #4, #6, and, in truncated form, #5, (+4300 to +8448 bp) common to Constructs #1 and #3, and (−7570 to +294 bp) common to Constructs #1 and #2. Sequencing validated all genomic regions. For Constructs #3-#6 an Hsp68 basal promoter was used.
To generate Constructs #6 and #7, point mutagenesis was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions with the following primers and their reverse complements: Phox2mut: 5′-TCTGTGATTTTATGGCAGAGGAGAGCAGGCAGCGCGCGCGCGTTCTGCTCTGTTTTCTG-GGACTTTTCCTTTC-3′; Ebox1mut: 5′-CTGTGCTTGTAATCTTGGTGTTCGGTTAGCTTGTTCACTCACAGCTGAAATTTAAA-3′; Ebox2mut: 5′-ATTTAAACAACAAAAAGCACGCGTGTGGACAGAAGCTGAGAAAGACACTCG-3′; and Ebox3mut: 5′-CGGGATAGGAGGAGGGTTCACCCCTCTCCTAGCCCC-3′. Mutagenized base pairs are in bold font.
To generate a FLAG-PHOX2b, PCR was used to flank human PHOX2b with SalI sites. This cDNA was subsequently cloned into the pCS2+FLAG expression construct (Vincentz et al., 2008).
Transgenic mice.
The Indiana University Transgenic and Knock-Out Mouse Core generated all transgenic mice. Yolk sac DNA was used to assess transgene integration using the following primer pairs: Constructs #1 and #2, (5′-TTTGGACGTCTGAACCCTTC-3′) and (5′-CGGAAACCAGGCAAAGCGCC-3′); Constructs #3, #4 and #6, (5′-ATTGTCGACCCTAAATCCCC-3′) and (5′-AGCACAGGCTCAAAGTTGC-3′); Construct #5, (5′-CCTAACACGGGATAGGAGGA-3′) and (5′-AGCACAGGCTCAAAGTTGC-3′).
Genotyping the Hand1tm1Eno and Hand1tm2Eno (Firulli et al., 1998; McFadden et al., 2005), Hand2tm1Cse and Hand2tm1Eno, (Srivastava et al., 1997; Morikawa et al., 2007), and Wnt1-Cre (Danielian et al., 1998) alleles have previously been described. Embryos of either sex were used for analyses. Hand1 conditional mice were a generous gift from Eric Olson (University of Texas Southwestern Medical Center). Hand2 conditional mice were graciously provided by Peter Cserjesi (IU Wells Center, Indianapolis, IN).
X-gal staining and histology.
X-gal staining was performed as previously described (Firulli et al., 1998). If stained embryos were to be sectioned and processed for immunohistochemistry, they were fixed for both prestaining and poststaining in 10% neutral buffered formalin, rather than, respectively, 2% paraformaldehyde-0.2%glutaraldehyde and 4% paraformaldehyde.
ChIP.
SK-N-BE(2) cells were transfected using Fugene HD (Roche) at a ratio of 7:2. Following 48 h of culture, cells were harvested and processed as previously described (Holler et al., 2010). Equal amounts of protein were immunoprecipitated overnight at 4°C with 50 μl of either α-FLAG- or α-Myc-conjugated agarose beads (Sigma). Eluted immunoprecipitated DNA was diluted in ddH2O and 5 μl of each dilution was used for PCR. As a control, 200 μl of cross-linked, sheared cell lysate was subjected to reverse crosslinking, proteinase K treatment, and RNase treatment. A dilution series, spanning 10 ng–1 × 10−6 ng at tenfold intervals, was used to generate a standard curve. ChIP primer pairs were as follows (forward, F; reverse, R): Ebox1 (F): 5′-CCTCAGCATTTTGTAACCTCTC-3′, Ebox1 (R): 5′-GCCATTCAACTGACATGTTTTT-3′; Ebox2 (F): 5′-TCCAGGTTATCATCATCTGTCA-3′, Ebox2 (R): 5′-CAAGTGTCTCTCCCAGCCAT-3′; Ebox3 (F): 5′-CAATCTGACGCAAAATGTTAGG-3′, Ebox3 (R): 5′-CTCTCTGAGCCTGGAGGAGA-3′; Phox2 (F): 5′-TTTATGGCAGGGGAGAAGAG-3′, Phox2 (R): 5′-GAGAACAGAAGGGAAAGCCC-3′; DBH (F): 5′-CCCACTGATGACGTCCATGT-3′, DBH (R) 5′-TGAATGGGGTGGGTCCAG-3′; and GAPDH (F): 5′-CTGCACCACCAACTGCTTAG-3′, GAPDH (R): 5′-CAGTGAGCTTCCCGTTCAG-3′.
Quantification of genomic DNA precipitates was performed in a LightCycler 480 II (Roche) using LightCycler FastStart DNA Master SYBR Green I under the following amplification parameters: 95°C, 10 s; 53°C, 30 s; and 72°C, 20 s for 45 cycles. Amplicons were verified through dissociation curve analyses. Equivalent DNA concentration was determined based on critical threshold (Ct) measurements, or changes in fluorescence per PCR cycle number at a given threshold. Using Abs/Quant second Derivative Max analyses in the LightCycler Software release 1.5.0, standard curves were generated for each primer set using the Ct values of the genomic DNA dilution series. From these standard curves, equivalent concentrations were calculated for each coimmunoprecipitated sample. Values are represented as equivalent to an amount of standard genomic DNA.
EMSA.
EMSAs were performed as previously described (Trochet et al., 2005; Firulli et al., 2007), using the following oligos annealed to their complements: Phox2, 5-GCAGGCAATTAGATTAATTTCTGCT-3; Phox2mut, 5-GCAGGCAgcgcgcgcgcgTTCTGCT-3; Ebox1, 5-GGTTAGCTTCATCTGTCACAGCTGA-3; Ebox2, 5-AGCACGCGTCAGGTGAGAAGCTGAG-3; E-box3, 5-AGGAGGAGGCATCTGCCCTCTCCTA-3. Oligos featuring mutated E-boxes (Ebox1mut, 5-GGTTAGCTTgtTCacTCACAGCTGA-3; Ebox2mut, 5-AGCACGCGTgtGGacAGAAGCTGAG-3; Ebox3mut, 5-AGGAGGAGGgtTCacCCCTCTCCTA-3) were used as nonspecific competitors for Hand factor binding assays. Densitometry analyses were performed using Bio-Rad Quantity One software.
Coimmunoprecipitation.
Coimmunoprecipitation (Co-IP) experiments were performed as previously described (Vincentz et al., 2008).
Immunohistochemistry.
Immunohistochemistry was performed as previously described (Vincentz et al., 2008). α-Tubb3 (Abcam), α-TH (Abcam), and α-DBH (Enzo) antibodies were used at dilutions of 1:1000, 1:400, and 1:200, respectively, in combination with biotinylated secondary antibodies, either α-Mouse or α-Rabbit (Vector Laboratories), diluted 1:250, then, in the case of Tubb3, SA-HRP (Vectastain ABC kit, Vector Laboratories) or, in the case of α-TH and α-DBH, SA-Fluorescein (Vector Laboratories) diluted 1:200. HRP color reaction was developed using the DAB Peroxidase Substrate kit (Vector Laboratories) according to the manufacturer's protocol.
Results
Hand1-reporter lines recapitulate gene expression in the SNS
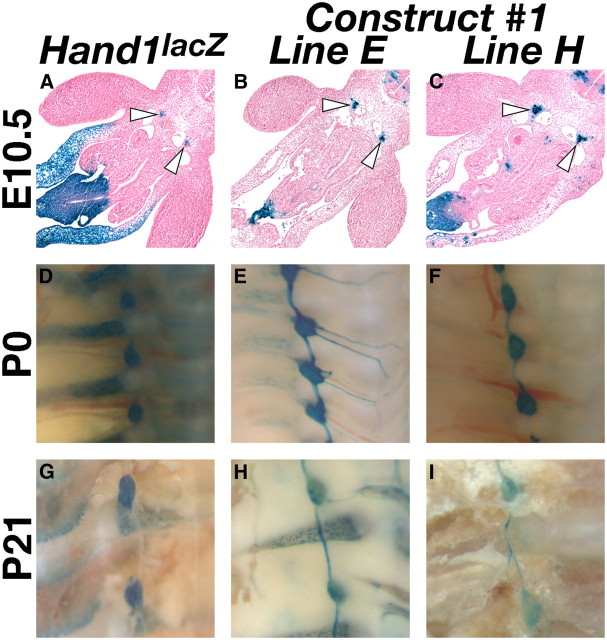
We first sought to establish the regulatory relationship between Hand1 and Hand2 by locating the cis-regulatory element(s) driving Hand1 gene expression in the SNS. To this end, we identified regions containing ECRs, or genomic sequences >75% conserved over at least 100 bp, proximal to the Hand1 transcription start site. We performed pairwise alignment of the 10 kb of genomic DNA respectively flanking mouse and opossum Hand1, identifying 10 discrete ECRs (Fig. 1A, red boxes). The genomic regions containing these conserved ECRs were isolated and used to synthesize a β-galactosidase (lacZ) reporter construct (Fig. 1A). Eight independent transgenic lines expressing this reporter construct were established, and lacZ expression was assessed. Of these 8 lines, 5 showed robust X-gal staining within the sympathetic ganglion at E12.5 (Fig. 1D,E). Further deletion analyses (Constructs #2-#5), directed by the presence of strong candidate consensus binding sites detailed below (see Fig. 3A) localized this enhancer to a region spanning −9559 to −8552 from the Hand1 transcription start site (Fig. 1A,F–M). Detailed spatiotemporal characterization of two independent transgenic lines expressing Construct #1, the full-length Hand1 reporter, revealed that, similar to the Hand1lacZ knock-in allele, expression in the sympathetic trunk in both transgenic lines was first detectable at E10.5 (Fig. 2A–C) and persisted postnatally to at least P21 (Fig. 2G–I).
Figure 1.
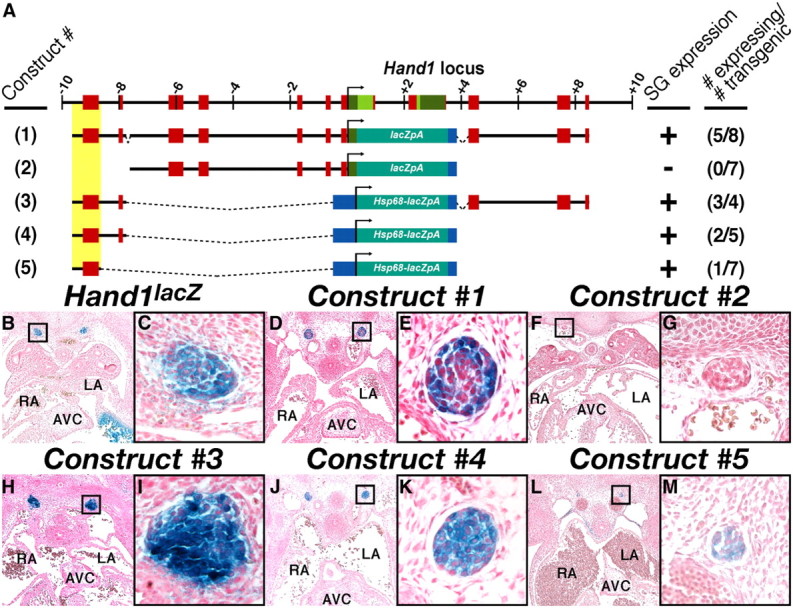
A 1007 bp genomic region 5′ of Hand1 is sufficient to drive SN-specific reporter transgene expression. A, Schematic representation of the Hand1 locus, including 10 kb 5′ and 3′ of the transcriptional start site and the reporter constructs used in these analyses. Exons, evolutionarily conserved regions, and the minimal Hand1 SG cis-regulatory element are highlighted in green, red, and yellow, respectively. Positive transgenic expressers within the SG, and the total number of transgenic embryos examined are indicated. B–M, Transverse sections of X-gal-stained E12.0-12.5 embryos show robust transgene expression in the SNS of transient transgenics expressing Constructs #1 (D, E), #3 (H, I), #4 (J, K), and #5 (L, M), but not #2 (F, G). A section of a stained E12.5 Hand1lacZ/+ embryo is included for comparison (B, C). High-magnification insets, denoted by boxes in B, D, F, H, J, and L, are featured in C, E, G, B, K, and M, respectively. AVC, Atrioventricular cushions; LA, left atrium; RA, right atrium; SG, sympathetic ganglion.
Figure 3.
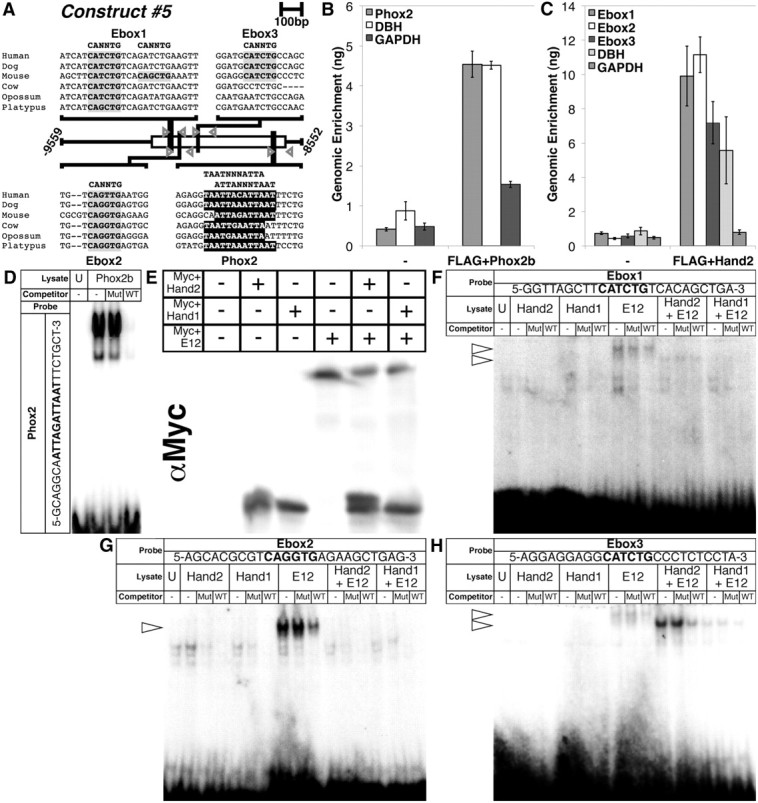
Phox2b and Hand2 bind to evolutionarily conserved consensus sites within the minimal Hand1 SN enhancer. A, A schematic shows three conserved E-box binding sites (highlighted in gray) and one conserved Phox2 binding site (highlighted in black) in the 1007 bp minimal Hand1 SN enhancer contained in Construct #5. Gray arrowheads denote ChIP primers flanking each of these binding sites. B, C, Immunoprecipitation of either FLAG+Phox2b or FLAG+Hand2 cross-linked complexes using α-FLAG antibody, produces enrichment of the Phox2 site-containing region (B) and three E-box-containing regions (C), respectively, in a manner similar to that of a positive control (DBH), but not a negative control (GAPDH). Error bars represent SE. D, EMSAs using lysate programmed with FLAG+Phox2b demonstrate binding to a radiolabeled oligonucleotide mimicking the Phox2 consensus site in the Hand1 SG enhancer. E, A Western blot for α-Myc verifies that Myc+Hand2 and Myc+Hand1 transcribed in vitro either in the presence or absence of Myc+E12 were synthesized in equivalent amounts. F–H, EMSAs using these lysates reveal that E-box1 (F) is weakly bound by E12 homodimers and Hand2-E12 heterodimers, but not Hand1-E12 heterodimers, nor Hand2 or Hand1 homodimers; that E-box2 (G) is bound solely by E12 homodimers; and that E-box3 (H) is not bound by Hand2 or Hand1 homodimers, is weakly bound by E12 homodimers and Hand1-E12 heterodimers, and is robustly bound by Hand2-E12 heterodimers. Binding in all EMSAs was competitively inhibited by unlabeled wild-type (WT) oligos, but not by unlabeled mutant (Mut) oligos. For all EMSAs, unprogrammed lysate (U) is used to control for nonspecific protein binding. Arrowheads denote specific protein–DNA complexes.
Figure 2.
The Hand1-reporter transgene recapitulates Hand1 expression in the SNS both prenatally and postnatally. A–C, Transverse sections of representative X-gal-stained E10.5 embryos from two independent transgenic lines, termed Line E (B) and Line H (C), show robust transgene expression in SNS progenitors (arrowheads), comparable to control Hand1lacZ embryos (A). D–I, Whole-mount preparations of X-gal-stained transgenic mice at P0 (E, F), and P21 (H, I) confirm that reporter expression persists in both lines in a manner comparable to that of age-matched Hand1lacZ mice (D and G, respectively).
Expression was also detected in the umbilicus, the outer curvature of the progress zone of the limb, and a ventromedial region of the neural tube at E10.5, and, perinatally, in the gastrointestinal tract. Although consistent among all five expressing lines, Hand1 is not endogenously expressed within any of these domains (Cserjesi et al., 1995; Barnes et al., 2010), and we conclude that this expression is artifactual. This aberrant expression is likely due either to the absence of a repressor element located outside of Construct #1, or to the integration of a high copy number of reporter transgenes, which may, through competitive binding, alter the transcriptional profile of a cell. Fortuitously, these artifactual expression domains provided a convenient internal control for subsequent mutational analyses, identifying F0 embryos in which reporter transgenes have integrated into transcriptionally active loci.
Phox2b and Hand2 bind to evolutionarily conserved sites within the minimal Hand1 SG enhancer
To identify transcription factors that might regulate Hand1 expression in the SNS, we searched for transcription factor binding sites that have been fixed in the Hand1 SG enhancer across evolution. Local alignments of the conserved ECRs corresponding to the 1007 bp minimal Hand1 SG enhancer contained in Construct #5 in mammalian species, using the CLUSTALW multiple sequence alignment program, uncovered one Phox2 binding site (Fig. 3A, highlighted in black) and two E-box binding sites (Fig. 3A, highlighted in gray) conserved among all mammalian orders, and a third E-box binding site (Fig. 3A, highlighted in gray) conserved among placentals.
To determine whether Phox2b and Hand2 bind these sites, respectively, we first performed ChIP assays. As commercially available antibodies for Phox2b and Hand2 are unreliable, and obtaining enough nuclear material from isolated embryonic sympathetic ganglia would be impractical, we performed our ChIP analyses using FLAG-epitope tagged variants of these transcription factors in a cell culture system. FLAG+Phox2b or FLAG+Hand2 mammalian expression constructs were transfected into human SK-N-BE(2) neuroblastoma cells. Following shearing and immunoprecipitation, relative amounts of precipitated DNA were quantified. Sheared genomic DNA was used to generate a standard curve for each set of amplicons. GAPDH, a housekeeping gene not regulated by either Phox2b or Hand factors, was used as a negative control, whereas the DBH promoter, to which both Phox2b and Hand2 directly bind (Rychlik et al., 2005; Doxakis et al., 2008), served as a positive control. Immunoprecipitation of FLAG+Phox2b cross-linked complexes using α-FLAG antibody resulted in significant enrichment of the Phox2 site-containing region compared with our negative control, similar to that observed for DBH (Fig. 3B). Similarly, immunoprecipitation of FLAG+Hand2 cross-linked complexes using α-FLAG antibody produced significant enrichment of all three E-box-containing regions, as well as the DBH promoter, when compared with control (Fig. 3C). Further ChIP analyses performed using a binding-deficient Hand2 variant (Myc+Hand2ΔB; McFadden et al., 2002) produced no significant enrichment (data not shown), confirming the specificity of this assay.
Our ChIP analyses suggested that Phox2b bound to its putative binding site in the Hand1 SG enhancer. However, immunoprecipitation of FLAG+Phox2b cross-linked complexes produced a slight, but nonetheless detectable, enrichment of our negative GAPDH control (Fig. 3B). We therefore further validated these results and confirmed specific binding of Phox2b to the Phox2 binding site through EMSA. In vitro translated FLAG+Phox2b formed complexes with radiolabeled oligos replicating the Phox2 site identified in the Hand1 SG enhancer (Fig. 3D). These complexes were specifically competed by an excess of unlabeled oligo, but not by unlabeled competitor in which the Phox2 site had been mutated. Additional EMSAs confirmed that FLAG+Phox2b does not bind to these mutagenized oligos (data not shown). EMSAs were further used to assess Hand2 binding to the three conserved E-boxes in the Hand1 SG enhancer. In vitro translated Myc-tagged Hand2 bound as a heterodimer with Myc-tagged E12 weakly, but specifically, to E-box1 (Fig. 3F), and, more robustly, to E-box3 (Fig. 3H) but failed to detectably bind to E-box2 (Fig. 3G). Hand2 homodimers did not bind at detectable levels to any of the three E-boxes assayed (Fig. 3F–H). As a whole, these results demonstrate that Phox2b and Hand2 bind to evolutionarily conserved sites within the Hand1 SG enhancer.
The Phox2 consensus-binding site within the Hand1 SG enhancer is necessary for sympathetic ganglia expression
We then assessed whether Phox2b is required for Hand1 SG enhancer transactivation in vivo. Sympathetic neurons undergo widespread apoptosis in Phox2b-null embryos (Brunet and Pattyn, 2002) making it impractical to test whether Hand1 SG enhancer activity would be altered on this genetic background. We therefore tested whether the Phox2 cis-binding element is required for Hand1 SNS gene expression. As demonstrated via EMSA, mutation of the Phox2 site eliminates Phox2b binding (Fig. 4B). We therefore point mutagenized, in an identical manner, the Phox2 binding site in Construct #4 (Fig. 4A), which spans −9559 to 7712 of the Hand1 transcription start site (Fig. 1A,J,K). Zero of 7 transgenic embryos generated using this mutagenized construct showed transgene expression in the SG at E11.5 (Fig. 4B–D) although artifactual ectopic expression was observed in the limb and neural tube (arrow, Fig. 4B,E,H,K) of three of these transgenics, suggesting that the integration locus was conducive to transcriptional activity. We conclude that the conserved Phox2 cis-binding element in the minimal Hand1 enhancer is necessary for Hand1 SG expression.
Figure 4.
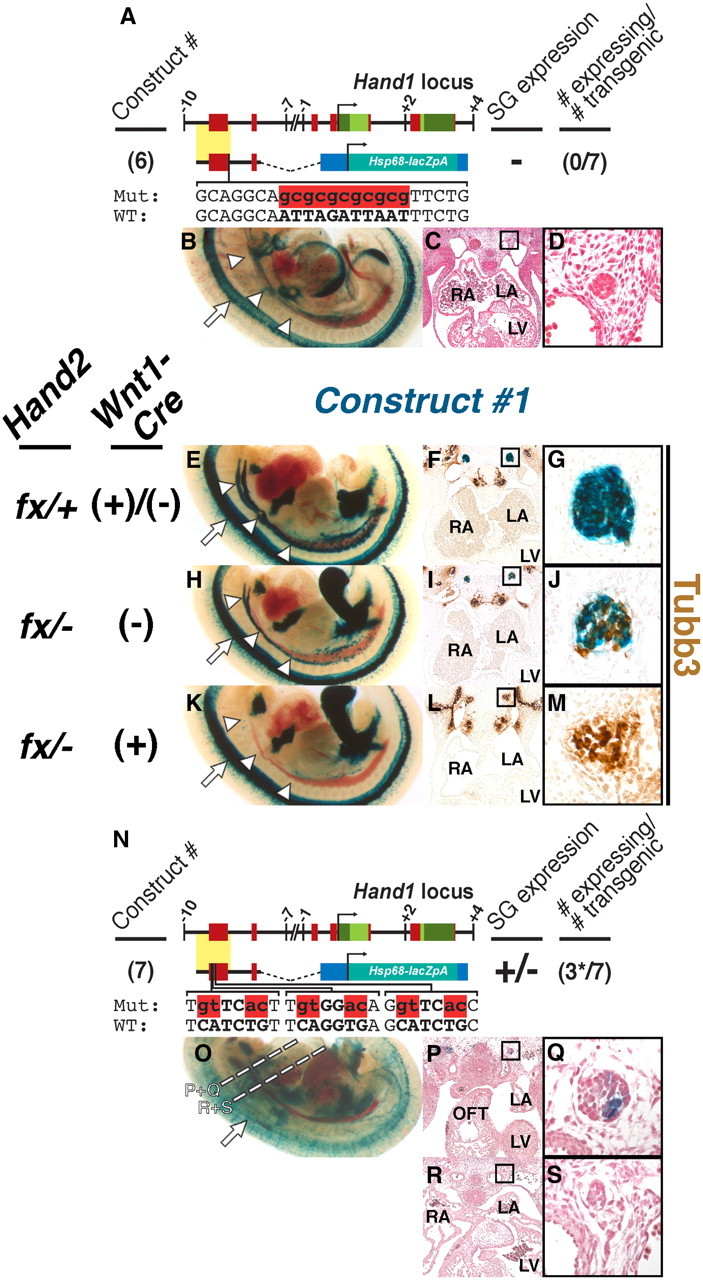
Phox2 and Hand2 are necessary for Hand1-reporter trans-activation in vivo. A, Schematic of Hand1-reporter Construct #6, in which the Phox2 binding site in Construct #4 has been mutated (red). B–D, Zero of 7 E11.5 transient transgenic embryos displayed SG expression. E–M, Expression of Construct #1 is reduced in E11.5 Hand2fx/−;Wnt1-Cre(−) hypomorphs and is lost or reduced in identically staged Hand2fx/−;Wnt1-Cre(+) conditional knock-outs. Cleared whole-mount preparations (E, H, K) show that, compared with control embryos (E), fewer cells within the Hand2 hypomorphic SG (H) express Construct #1 (n = 9 of 10). In the majority of Hand2 conditional knock-out embryos examined, Construct #1 expression is undetectable (K; n = 4 of 6). Transverse sections of Construct #1-expressing embryos, stained with X-gal (blue) in which neurons have been labeled with α-Tubb3 immunohistochemistry (brown), photographed at low (F, I, and L) and high (G, B, and K; denoted by boxes) magnifications show that, whereas SG in control embryos (F, G) are almost exclusively comprised of Hand1-reporter-positive neurons, and Hand2 conditional knock-out SG (L, M) are exclusively comprised of Hand1-reporter-negative neurons, the Hand2 hypomorphic SG (I, J) are comprised of a mix of both Hand1-reporter-positive and -negative neurons. n = 2 for each genotype. N, Schematic of Hand1-reporter Construct #7, in which the three evolutionarily conserved E-box binding sites in Construct #4 have all been mutated (red). O–S, Three of 7 E11.5 transient transgenic embryos displayed detectable SG expression. However, expression in these three embryos was not uniform, and was restricted to SG rostral to the OFT (P, Q). Expression was absent in SG caudal to the OFT (R, S). Dashed lines denote the planes of section for P–S. LA, Left atrium; LV, left ventricle; RA, right atrium.
Hand2 is necessary for SG expression of the Hand1 reporter
Hand2 ablation in SNS precursors causes an absence of Hand1 expression, establishing that Hand2 lies upstream of Hand1 during SG differentiation (Morikawa et al., 2007). To test whether Hand2 is necessary for Hand1 gene expression via its DNA binding to the SG enhancer (Fig. 3C,F,H), we bred Construct #1 mice onto a Hand2 NCC-specific knock-out background, incorporating the Hand2 conditional allele (Morikawa et al., 2007), and the Wnt1-Cre driver (Danielian et al., 1998). Importantly, this Hand2 conditional allele is reported to be hypomorphic, and Hand2fx/− mice display cleft palate resulting from defective cranial NCC differentiation (Morikawa et al., 2007). qRT-PCR indicates that the transcriptional output from a single copy of this hypomorphic allele generates 35% of total wild-type Hand2 expression (P. Cserjesi, personal communication). In most E11.5 Hand2 conditional knock-out embryos examined, Construct #1 expression was undetectable (n = 4 of 6; Fig. 4K–M). The remaining conditional knock-out embryos examined displayed markedly reduced reporter expression. Interestingly, Construct #1 expression was consistently reduced in Hand2fx/− hypomorphs (n = 9 of 10; Fig. 4H–J), compared with controls (Fig. 4E–G). Assessing cell morphology, examination of embryo tissue sections suggest that the Hand2fx/− hypomorphic SG contains both Construct #1-expressing and non-expressing cells. Immunohistochemistry for the neuronal marker Hu and the cell proliferation marker phospho-histone H3 at E10.5 failed to detect a significant difference between cell number or proliferation index in SG of Hand2fx/− and Hand2fx/+ embryos (data not shown), further suggesting that the diminished Construct #1 expression seen in Hand2fx/− hypomorphs reflects reduced gene expression, rather than a loss of SG cells.
We therefore sought to confirm whether the Hand1 SG enhancer displays mosaic expression in Hand2fx/− hypomorphic SG. E11.5 Construct #1-positive, Hand2 mutant embryos were sectioned and stained for the pan-neuronal marker Tubulin β-III (Tubb3). Tubb3 immunohistochemistry demonstrated that, as expected, control SG display almost uniform Hand1 reporter expression (Fig. 4F,G), whereas Hand2 conditional knock-out SG are devoid of Hand1 reporter expression (Fig. 4L,M). Hand2fx/− hypomorphic SG, however, presented as a mix of both Construct #1-positive and Construct #1-negative neurons (Fig. 4I,J). Together, these data show that the Hand1 SG enhancer is regulated by Hand2 in a gene dosage-dependent manner.
Although these results demonstrate that Hand1 enhancer cis-activity is dependent upon Hand2, it cannot be ruled out that Hand2 regulation is indirect. Sympathetic neurons, in the absence of Hand2, may simply differentiate into a neuronal cell type that does not express Hand1. To exclude this possibility, we mutagenized the three conserved E-boxes in the Hand1 SG enhancer in a manner identical to that detailed in the EMSAs described previously (Fig. 4N). Of the 7 transgenic mice generated with this mutagenized construct, 4 exhibited no significant expression within the SG. Interestingly, 3 of 7 E11.5 transgenic embryos generated showed weak reporter expression in the SG (Fig. 4O–S); however, unlike the wild-type Hand1 SG enhancer, expression in these transgenic lines was not uniformly observed throughout the cells of the sympathetic chain. Indeed, reporter expression was limited to a small subpopulation of SNs within ganglia at or rostral to the level of the cardiac outflow tract (OFT; Fig. 4P,Q). Caudal to the OFT, reporter gene expression in these three lines was undetectable. Collectively, SG reporter expression from Construct #7 constitutes a markedly reduced expression domain compared with wild-type reporter transgenics, suggesting that, like the Phox2 binding site, the conserved E-boxes in the minimal Hand1 enhancer are necessary for proper Hand1 SG enhancer cis-activity.
Hand1 and Hand2 display unique binding specificities for consensus binding sites in the Hand1 SG enhancer
The observation that Hand1 SG-specific cis-element activity is dependent upon E-boxes is not surprising; however, the finding that the Hand1 SG enhancer displays reduced activity on a Hand2 hypomorphic background is surprising. As Hand1 and Hand2 have been shown to function cooperatively in other developmental contexts (McFadden et al., 2005; Barbosa et al., 2007), including the survival of cultured neonatal sympathetic neurons (Doxakis et al., 2008), one would predict that an autoregulatory mechanism would allow Hand1 to maintain wild-type expression levels when Hand2 function is compromised. Although a significant body of evidence suggests that Hand1 and Hand2 might perform identical functions, the idea that these two factors are functionally distinct is not without precedent (Holler et al., 2010). Previous studies have shown that Phox2a and Hand2 can synergistically transactivate DBH (Rychlik et al., 2003; Xu et al., 2003). Additional studies have indicated that, while siRNA-mediated knockdown of Hand2 reduces DBH expression in newborn superior cervical ganglion, knockdown of Hand1 fails to produce a similar effect, indicating that these factors may perform unique functions with regard to DBH transactivation (Doxakis et al., 2008). We thus investigated whether Hand2 performs transcriptional functions unique from Hand1. Neither Hand1 nor Hand2 molecularly interact with Phox2a (Xu et al., 2003). However, Hand2 has been shown to molecularly interact with Phox2b in neuroblastoma cells (Reiff et al., 2010; M. J. Howard, unpublished results). We hypothesized that if Hand2, but not Hand1, molecularly interacts with Phox2b, this might reflect the unique requirement for Hand2 in developing sympathetic neurons, and we therefore tested, via Co-IP experiments in HEK293 cells, whether Hand1 and Phox2b molecularly interact. As expected, immunoprecipitation of FLAG+Phox2b coprecipitates Myc+Hand2 (Fig. 5A). Myc+Hand1 can also be coprecipitated with FLAG+Phox2b (Fig. 5A), indicating that both Hand factors can interact with Phox2b.
Figure 5.
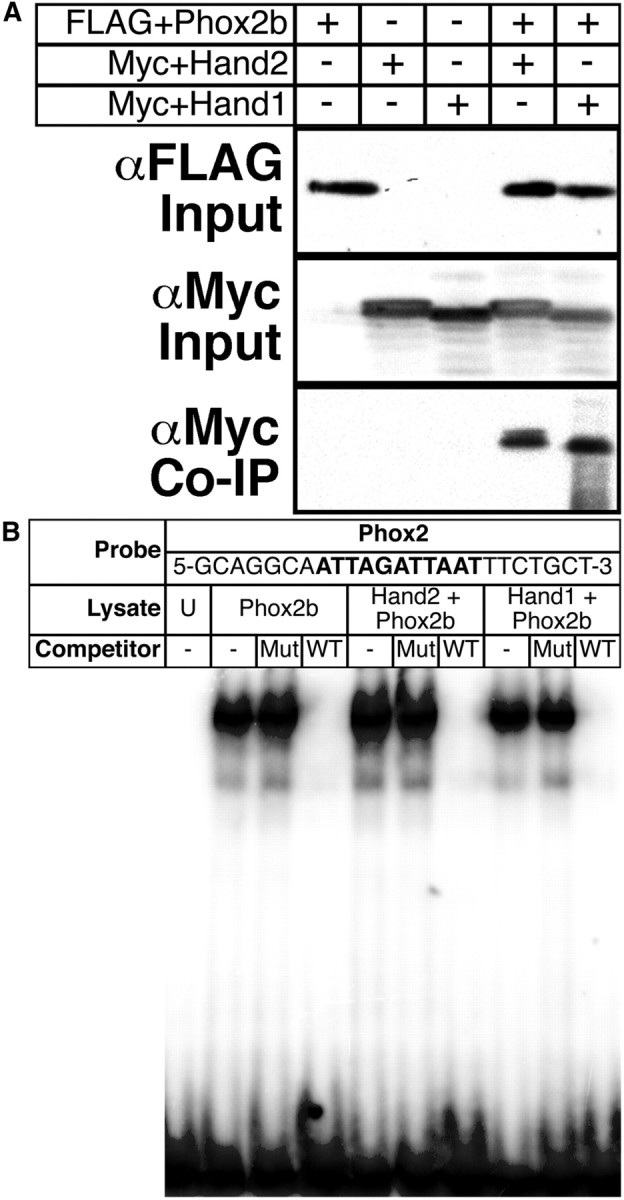
Hand2 and Hand1 interact with Phox2b in a similar manner. A, Coimmunoprecipitation experiments reveal that FLAG+Phox2b (α-FLAG Input) effectively precipitates both Myc+Hand2 and Myc+Hand1 (α-Myc Input) when the two are cotransfected (α-Myc Co-IP), illustrating protein–protein interactions between these factors. B, EMSAs using lysate programmed with FLAG+Phox2b either alone or with Myc+Hand2 or Myc+Hand1 demonstrate that formation of specific Phox2b-oligo complexes is not positively or negatively altered in the presence of either Hand factor.
We then tested, via EMSA, whether Hand1 and Hand2 might, through protein–protein interaction, differentially affect Phox2b DNA binding to its cognate consensus-binding site. In vitro translated FLAG+Phox2b was cosynthesized with either Myc+Hand2 or Myc+Hand1 or with an empty pCS2+MT vector. Western blot verified effective transcription and translation (data not shown). Lysates were incubated with radiolabeled oligos replicating the Phox2 site identified in the Hand1 SG enhancer. Gel-shift analyses revealed no detectable difference in Phox2b DNA-binding either alone or in the presence of Hand2 or Hand1 (Fig. 5B). Together, these results fail to uncover a Phox2b interaction unique to either Hand2 or Hand1.
We therefore assessed Hand2 and Hand1 binding to the conserved E-box binding sites in the Hand1 SG enhancer to test whether Hand2 and Hand1 possess different DNA binding affinities independent of Phox2b interaction. Like other Twist family bHLH transcription factors, Hand factors function through dimerization, either with other Hand factors, or with E-proteins such as E12. We therefore synthesized Myc+Hand2 or Myc+Hand1 in vitro, either alone or cosynthesized with Myc+E12. Myc+E12, transcribed alone, was also included as a control. Western blot was used to verify that the peptides were synthesized in equal amounts, and that the intensity of shifted DNA complexes represented differences in DNA binding affinities, as previously established (Firulli et al., 2007; Fig. 3E). Densitometry analyses revealed that the ratio of Myc+E12 cosynthesized Myc+Hand2 to Myc+Hand1 was 1.3:1.0. EMSA revealed that, like Hand2, Hand1 does not bind to any of the three E-box-containing oligos at detectable levels as a homodimer (Fig. 3F–H). Hand2-E12 heterodimers were found to weakly bind to E-box1 (Fig. 3F); however, Hand1-E12 heterodimers displayed no detectable binding to this E-box. Hand2-E12 binds most robustly to E-box3 (Fig. 3H), whereas Hand1-E12 binding to this E-box is comparatively weaker (Fig. 3H). Densitometry analyses revealed that Hand2-E12 binding was 4.9-fold more robust than that of Hand1-E12. E-box2 was bound only by E12 homodimers, and, interestingly, both Hand2 and Hand1 each seem to inhibit formation of this E12 homodimer protein–DNA complex (Fig. 3G). Collectively, these data suggest that Hand2-E12 heterodimers display a stronger DNA binding affinity for E-boxes within the Hand1 SG enhancer than Hand1-E12 heterodimers, a functional difference that may explain both the specific Hand2 gene dosage requirement for Hand1 SG enhancer activity and the inability of Hand1 to functionally replace Hand2 and thereby regulate its own transcription in the SG.
Hand1 is dispensable for sympathetic neurogenesis
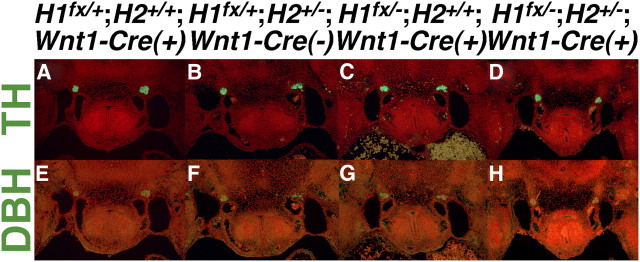
It has been established that Hand2 is required not only for the initial expression of the norepinephrine biosynthetic enzymes characteristic of noradrenergic neurons (Howard et al., 1999, 2000; Lucas et al., 2006; Morikawa et al., 2007; Hendershot et al., 2008), but that continual Hand2 transcriptional input is required to maintain expression of these factors (Schmidt et al., 2009). We observe that Hand1 reporter expression is diminished in the SG of Hand2 hypomorphs (Fig. 4H–J). Additionally, studies using an alternative Hand2 hypomorphic allele expressing 14% of wild-type Hand2 mRNA levels report not a reduction in sympathetic neuron cell number, but a general trend toward a decrease in the number of TH-expressing cells (Hendershot et al., 2008), These data suggest that expression of specific genes within the SG is sensitive to Hand2 expression levels. If Hand2 and Hand1 are functionally redundant in these neurons, it would follow that Hand1 would contribute to the overall level of Hand factor dosage, and might affect expression of these noradrenergic markers. However, our analyses of the Hand1 SG enhancer suggests that Hand2 might uniquely regulate genes in the sympathetic neurons. To test the requirement for noradrenergic marker expression during sympathetic neurogenesis, we performed immunohistochemistry upon E11.5 embryos in which Hand1 function has been specifically ablated in the neural crest and its derivatives. Targeted disruption of Hand1 causes embryonic lethality before specification of sympathetic progenitors (Firulli et al., 1998). We therefore used the Hand1 conditional allele (McFadden et al., 2005) and the Wnt1-Cre driver (Danielian et al., 1998) to disrupt Hand1 function in the SNS. Immunohistochemistry for TH and DBH protein failed to detect an obvious difference in expression between Hand1 conditional knock-outs (Fig. 6C,G) and control embryos (Fig. 6A,E). These results indicate that TH and DBH expression is unperturbed by a loss of Hand1 function. Hand2 heterozygotes, which have 50% of Hand2 expression compared with wild-type mice (Hendershot et al., 2008), lack apparent SNS phenotypes (Fig. 6B,F). When that genetic output is reduced to 35%, gene expression in the SG is altered (Fig. 4H–J). Thus, the critical threshold for Hand2 dosage appears to lie between 35% and 50% of wild-type dosage. We reasoned that, if Hand1 does indeed contribute to the overall Hand factor gene dosage requirement, this contribution might only become evident on a sensitized Hand2 genetic background. We therefore conditionally ablated Hand1 on a Hand2+/− background, and examined TH and DBH protein expression at E11.5. TH and DBH expression was not appreciably altered in Hand1fx/−;Hand2+/−;Wnt1-Cre(+) embryos (Fig. 6D,H) compared with controls (Fig. 6A,E) and Hand2 heterozygotes (Fig. 6B,F). We conclude that Hand1 function is not required to maintain noradrenergic marker expression in developing sympathetic neurons, and that Hand2 is thus uniquely required to perform this function.
Figure 6.
Hand1 function is not necessary for TH and DBH expression in the SG. A–H, Fluorescent immunohistochemistry upon transverse sections of E11.5 embryos reveals that TH (A–D) and DBH (E–H) protein expression is indistinguishable between control (H1fx/+;H2+/+;Wnt1-Cre(+); A, E) Hand2 heterozygous (H1fx/+;H2+/−;Wnt1-Cre(−); B, F), Hand1 conditionally null (H1fx/−;H2+/+;Wnt1-Cre(+); C, G), and doubly Hand1 conditionally null, Hand2 heterozygous (H1fx/−;H2+/−;Wnt1-Cre(+); A, E) mutants.
Discussion
Here, we investigate whether Hand1 and Hand2 perform unique functions in the developing SNS, or whether their respective functions are governed by their sequential expression patterns and describe a 1 kb evolutionarily conserved cis-regulatory element 5′ to the Hand1 transcriptional start site that is sufficient to drive gene expression within the SNS. This enhancer initiates expression at E10.5, consistent with what is observed both for Hand1 mRNA and in Hand1lacZ mice, and is expressed postnatally. Within this cis-regulatory element lie several evolutionarily conserved consensus-binding sites, including a Phox2 binding site and three E-box binding sites, which are bound by the transcription factors Phox2b and Hand2, respectively. We show that these Phox2 and E-box binding sites are necessary for activity of this cis-regulatory element, indicating that Phox2b and Hand2 are direct upstream regulators of Hand1. However, we also show that these E-box elements are uniquely responsive to Hand2 gene dosage, indicating that Hand1 cannot autoregulate in a manner that maintains its expression at wild-type levels when Hand2 function is diminished (Fig. 4E–M). We attribute this to a lower affinity of Hand1-E12 heterodimers to bind the conserved E-boxes within its own enhancer, in contrast to Hand2-E12 heterodimers, which bind two of these E-boxes efficiently in EMSA (Fig. 3F,H). As noradrenergic marker expression is unperturbed in Hand1-null SG, even on a sensitized Hand2 heterozygous background, we conclude that transcriptional properties unique to Hand2 render it uniquely necessary for sympathetic neurogenesis.
Interestingly, the Hand1 SG enhancer is prone to ectopic lacZ activity within the neural tube and limb, locations in which Hand1 is not endogenously expressed. This finding could reflect the loss of a critical Hand1 repressor element not present within the ECR or simply be a frequent artifact of transgene integration. Regardless of mechanism, this artifactual expression became a powerful tool to fully rule out the possibility that the loss of Hand1-SG enhancer activity via Phox2 cis-element mutation results from transgene integration into transcriptionally dead regions of the genome. Indeed, ChIP and EMSA analyses show that Phox2b directly binds to the conserved Phox2 element (Figs. 3D, 6B). Combinatorially, the observations that Hand2 binds E-box sequences within the Hand1 SNS element that are required for its proper expression, and that transgene expression is lost within Hand2 conditionally null mice, render it equally clear that Hand2 also directly regulates Hand1, supporting previous data (Morikawa et al., 2007). Additionally, while not necessary for SNS expression, loss of the 3′ regions of the Hand1 reporter (present in Constructs #1 and #3; Fig. 1A,D,E,H,I) reduces the frequency of reporter-expressing transient transgenics. This may indicate that the 3′ regions contain elements that promote euchromatin stability, better enabling Hand2, Phox2b, and other putative trans-activators to access the Hand1 minimal regulatory element.
It has been established that the specification and differentiation of NCC-derived neurons hinges upon the transcriptional activity of both bHLH transcription factors, such as Hand2, and homeodomain transcription factors, such as Phox2b (Howard, 2005). The molecular programs that govern sympathetic neuron differentiation do not follow a stepwise linear progression, whereby transcription factors sequentially upregulate one another's expression and ultimately upregulate markers of SNS differentiation, but rather an intricate transcriptional network, in which these factors not only regulate each other's expression, but also molecularly interact to regulate each other's transcriptional activity, cumulatively upregulating noradrenergic biosynthetic enzymes. Studies exploring transcriptional regulation of one of these enzymes, DBH, have revealed that complex transcriptional and protein–protein interactions between Hand2 and Phox2a in vitro enable these two factors to synergistically regulate DBH transcription (Rychlik et al., 2003; Xu et al., 2003). Here, we report that Phox2b and Hand2 bind to consensus binding sites within the Hand1 SG enhancer, and that this enhancer is dependent upon both Hand2 and Phox2b function in vivo. Moreover, epitope-tagged Hand2 and Phox2b physically interact when coexpressed in cultured cells (Reiff et al., 2010) (Fig. 5A). We provide evidence that Phox2b and Hand1 similarly interact (Fig. 5A), supporting previous findings that Phox and Hand factors play an essential integrated role in implementing the sympathetic neuronal program (Howard et al., 2000; Hendershot et al., 2007). It is still unclear whether these factors function as part of a larger, multimeric transcriptional complex containing additional essential factors, for example, Insm1 (Rychlik et al., 2003; Xu et al., 2003). Clearly neither Phox2b nor Hand2 alone is sufficient to drive Hand1 SG expression. The simplest explanation for this result is that both factors form a core complex that drives the SG program; however we cannot rule out that other factors, not identified in this study, are also required. Further characterization of this cis-regulatory element is needed to yield insight into the protein–protein interactions that govern its activity. Indeed, given the restricted nature of neuronal Hand1 expression, this cis-regulatory element provides a powerful tool with which to explore the transcriptional mechanisms specifically driving sympathetic neuron differentiation.
Previous studies have proposed that Hand2 gene dosage is important for proper SG development (Howard et al., 1999; Hendershot et al., 2008). These studies are the first to definitively demonstrate a gene dosage requirement for Hand2 in SG-specific gene expression in vivo and that Hand1 is directly sensitive to Hand2-dosage. As Hand2 is known to regulate DBH in the peripheral nervous system, these findings could have broader implications for norepinephrine biosynthetic programs during both development and disease.
As evidenced by the differing expression of the Hand2-responsive SG reporter described in this paper, among cells of the sympathetic chain, the cells of the Hand2 hypomorphic SNS are molecularly distinct. In contrast to Hand2 wild-type or Hand2-null sympathetic neurons, which either uniformly express or uniformly fail to express Hand1, Hand2 hypomorphs display SGs that are a mixture of both Hand1-SG reporter expression-positive and reporter expression-negative neurons. Differing responses to neurotrophic signals mediated, in part, by the nerve growth factor receptor TrkA, have been shown to be critical to sympathetic neuron cell survival and, by extension, the competitive cellular mechanism by which the peripheral nervous system matures (Deppmann et al., 2008). Cells expressing high levels of TrkA survive, while those expressing lower levels undergo apoptosis (Deppmann et al., 2008). Studies in sympathetic neuronal explants indicate that TrkA expression levels are directly regulated by Hand factors (Doxakis et al., 2008). It would therefore be of interest to determine whether neurotrophic responses are processed differentially by the cells of the Hand2-hypomorphic SG, and whether the Hand2-responsive SG reporter described here correlates with cells expressing high levels of these neurotrophic factor receptors, which will ultimately survive the developmental process.
The cumulative contributions of Hand1 and Hand2 are critical in certain developmental contexts (McFadden et al., 2005). In vitro studies have established that Hand1 and Hand2 can function similarly to promote SG cell survival, but may perform distinct functions in terms of transcriptionally regulating the noradrenergic gene program (Doxakis et al., 2008). The overlapping and distinct functions of Hand1 and Hand2 in an in vivo context are not clear. In this study, we establish that proper expression of Hand1 through the novel SG enhancer requires a critical threshold of Hand2 function, supporting the interpretations of previous reports. Our data suggest that, despite their similarities, Hand1 cannot in fact functionally compensate for Hand2 within the SNS in vivo, and that distinct properties of these two factors render Hand1 dispensable and Hand2 critical in this developmental context. A possible confirmation of this hypothesis is observed in Hand2Hand1 knock-in mice, in which a Hand1 cDNA has been targeted to the Hand2 locus. Homozygous Hand2Hand1/Hand1 embryos die at E12.5–13.5. Although SG marker gene expression has not been assessed in these mutants, they establish that Hand1 cannot replace Hand2 past mid-gestation (D. Srivastava and H. Yamagishi, personal communication). Collectively, these studies not only provide valuable insight into the molecular interactions between Phox2 and Hand factors during sympathetic neurogenesis, but also draw the first concrete functional distinction between Hand1 and Hand2 factors during sympathetic neuron differentiation.
Footnotes
Support for this work was provided by NIH Grants RO1HL061677-12 and 1P01HL085098-05 (A.B. Firulli) and R01NS040644 (M.J.H.). We thank Danny Carney for technical assistance. We thank the Herman B Wells Center Cardiac Developmental Biology Group for helpful discussions. Infrastructural support at the Herman B Wells Center is partially supported by the Riley Children's Foundation and Division of Pediatric Cardiology.
References
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, Conway SJ, Vincentz JW, Firulli AB. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev Dyn. 2010;239:3086–3097. doi: 10.1002/dvdy.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Brunet JF, Pattyn A. Phox2 genes—from patterning to connectivity. Current opinion in genetics and development. 2002;12:435–440. doi: 10.1016/s0959-437x(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Lyons GE, Olson EN. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Mihalas S, Sharma N, Lonze BE, Niebur E, Ginty DD. A model for neuronal competition during development. Science. 2008;320:369–373. doi: 10.1126/science.1152677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxakis E, Howard L, Rohrer H, Davies AM. HAND transcription factors are required for neonatal sympathetic neuron survival. EMBO reports. 2008;9:1041–1047. doi: 10.1038/embor.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, Howard MJ. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Clouthier DE, Shepherd IT, Coppola E, Studer M, Firulli AB, Pittman DL, Howard MJ. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Developmental biology. 2008;319:179–191. doi: 10.1016/j.ydbio.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler KL, Hendershot TJ, Troy SE, Vincentz JW, Firulli AB, Howard MJ. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev Biol. 2010;341:291–304. doi: 10.1016/j.ydbio.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MJ. Mechanisms and perspectives on differentiation of autonomic neurons. Dev Biol. 2005;277:271–286. doi: 10.1016/j.ydbio.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- Howard M, Foster DN, Cserjesi P. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev Biol. 1999;215:62–77. doi: 10.1006/dbio.1999.9450. [DOI] [PubMed] [Google Scholar]
- Lucas ME, Müller F, Rüdiger R, Henion PD, Rohrer H. The bHLH transcription factor hand2 is essential for noradrenergic differentiation of sympathetic neurons. Development. 2006;133:4015–4024. doi: 10.1242/dev.02574. [DOI] [PubMed] [Google Scholar]
- McFadden DG, McAnally J, Richardson JA, Charité J, Olson EN. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development. 2002;129:3077–3088. doi: 10.1242/dev.129.13.3077. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development. 2004;131:2195–2204. doi: 10.1242/dev.01091. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, D'Autréaux F, Gershon MD, Cserjesi P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol. 2007;307:114–126. doi: 10.1016/j.ydbio.2007.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Rohrer H. Molecular control of ciliary neuron development: BMPs and downstream transcriptional control in the parasympathetic lineage. Development. 2002;129:5707–5717. doi: 10.1242/dev.00165. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Reiff T, Tsarovina K, Majdazari A, Schmidt M, del Pino I, Rohrer H. Neuroblastoma Phox2b variants stimulate proliferation and dedifferentiation of immature sympathetic neurons. J Neurosci. 2010;30:905–915. doi: 10.1523/JNEUROSCI.5368-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik JL, Gerbasi V, Lewis EJ. The interaction between dHAND and Arix at the dopamine beta-hydroxylase promoter region is independent of direct dHAND binding to DNA. J Biol Chem. 2003;278:49652–49660. doi: 10.1074/jbc.M308577200. [DOI] [PubMed] [Google Scholar]
- Rychlik JL, Hsieh M, Eiden LE, Lewis EJ. Phox2 and dHAND transcription factors select shared and unique target genes in the noradrenergic cell type. J Mol Neurosci. 2005;27:281–292. doi: 10.1385/JMN:27:3:281. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Lin S, Pape M, Ernsberger U, Stanke M, Kobayashi K, Howard MJ, Rohrer H. The bHLH transcription factor Hand2 is essential for the maintenance of noradrenergic properties in differentiated sympathetic neurons. Dev Biol. 2009;329:191–200. doi: 10.1016/j.ydbio.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Trochet D, O'Brien LM, Gozal D, Trang H, Nordenskjöld A, Laudier B, Svensson PJ, Uhrig S, Cole T, Niemann S, Munnich A, Gaultier C, Lyonnet S, Amiel J. PHOX2B genotype allows for prediction of tumor risk in congenital central hypoventilation syndrome. Am J Hum Genet. 2005;76:421–426. doi: 10.1086/428366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli BA, Conway SJ, Firulli AB. Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev Dyn. 2008;237:3809–3819. doi: 10.1002/dvdy.21803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Firulli AB, Zhang X, Howard MJ. HAND2 synergistically enhances transcription of dopamine-beta-hydroxylase in the presence of Phox2a. Dev Biol. 2003;262:183–193. doi: 10.1016/s0012-1606(03)00361-0. [DOI] [PubMed] [Google Scholar]