Abstract
We have adapted the Ba2+ ion-catalyzed concurrent Michael addition reaction to solid-phase derivatization on ZipTipC18 pipette tips using 2-aminoethanethiol as a nucleophile. This approach provides several advantages over the classical in-solution-based techniques, including ease of operation, completeness of reaction, improved throughput, efficient use of dilute samples, and amenability to automation. Phosphoseryl and phosphothreonyl peptides, as well as phosphoserine peptides with adjoining prolines, were used to optimize the reaction conditions, which proved highly compatible with the integrity of the samples. The analyte was recovered from the silica-based C18 resin at minimal sample loss. The use of the protocol for improved phosphopeptide detection by signal enhancement was demonstrated with low-level amounts of proteolytic digests from model proteins and experimental samples, an effect found especially prominent with multiple phosphorylated species. The reaction products proved highly suitable for structural characterization by collisionally induced dissociation (CID), and the resultant increased spectral information content, greatly facilitating mapping of the site of phosphorylation. In select cases, the method enables phosphorylation site localization within known protein sequences on the basis of single-stage data alone. The solid-phase strategy presented here provides a simple, versatile, and efficient tool for phosphopeptide structural characterization equipment readily available in most biological laboratories.
Keywords: ZipTipC18 pipette tips, derivatization, phosphopeptide identification, phosphorylation site determination
INTRODUCTION
Protein phosphorylation is recognized as a critical event in modulation of cellular processes, including cellular signaling, cell cycle progression, and differentiation.1
Existing approaches to map this important post-translational modification rely predominately on the use of mass spectrometry (MS) methods to identify and sequence the peptide of interest.2–5 MALDI-TOF MS is commonly used to detect phosphopeptides by monitoring the indicative, metastable postsource decay (PSD) products or the characteristic mass shift after alkaline phosphatase treatment.2,3 Various strategies, using electrospray ionization tandem MS coupled with liquid chromatography (LC), are often used, including precursor ion and neutral loss scanning for diagnostic ions generated under the conditions of CID.4,5 However, these methods are often challenged by the ionization inefficiency of the phosphopeptides, their low stoichiometry relative to the unphosphorylated counterparts, and the limited information content of the MS/MS spectra, as a result of the intrinsic lability of the phosphate group upon CID. Immobilized metal ion affinity chromatography (IMAC) is a frequently reported technique to address these problems by phosphopeptide enrichment.6
Titanium oxide microcolumns have been advocated more recently as an alternative to IMAC to minimize variable peptide recovery and to provide for improved selectivity of phosphopeptide enrichment.7 MALDI-matrix additives, such as ammonium salts or phosphoric acid, have been shown to promote phosphopeptide ionization during MALDI-MS.8,9 Although the above strategies afford improved phosphopeptide detection, issues still persist with regard to neutral loss of phosphate, because as the favored fragmentation event, it dominates over peptide backbone cleavage. Subjecting the resultant neutral loss product ion from the MS/MS spectrum to a further cycle of fragmentation (MS/MS/MS) may improve the peptide sequence information content and thus, aid in phosphorylation site determination.10 Electron transfer dissociation of phosphopeptides has emerged as a viable method to minimize phosphate gas-phase β-elimination. The method proved informative on large, multiply protonated peptides, predominantly contained in endopeptidase Lys-C protein digests, and has been used more recently in combination with CID in a complementary manner.11
Chemically induced β-elimination of phosphate from serine and threonine residues, coupled with Michael addition, provides a chemical strategy to afford improved MS/MS sequence information by precluding neutral loss of phosphate. One approach advocates the use of ethanethiol as nucleophile for concurrent β-elimination/Michael addition.12 The derivatization proved to enhance ionization of the modified peptides during LC-MS analysis and afforded improved MS/MS spectral information. As applied to digests of PKA, this advantage has been exploited to map completely the phosphorylation sites of the protein in a single LC-MS/MS experiment.13 In contrast, multiple experiments were required for each of the optional techniques used in this study to provide complementary data. However, the relatively stringent conditions used in these protocols raised concerns about chemical contamination, peptide bond hydrolysis, and unwanted chemistry side reactions. Typically, sodium hydroxide in high concentrations is used as an elimination reagent, fortified with organic solvents, and the mixtures are incubated for several hours or overnight—conditions known to promote protein degradation. Reagent excess is commonly removed by ultrafiltration, dialysis, or precipitation—manipulations known to incur substantial, adsorptive sample loss, which may limit the general usefulness of these methods. Several approaches using the β-elimination/Michel addition reaction have been advocated to detect phosphopeptides by signal enhancement.14–16 The MALDI-TOF response of phosphoseryl and phosphothreonyl peptides, after reaction with alkenethiols of increasing carbon chain length, was examined, of which, octanethiol proved most effective to promote phosphopeptide signal enhancement.14 Improved MALDI-TOF detection was demonstrated using guanidinoethanethiol as a Michael addition reagent.15 The signal-enhancing effect observed in the MALDI-MS spectra was most noticeable for the two lysine-terminated phosphorylated peptides contained in tryptic digests of α-S1 casein. However, the tag proved to be only partially stable upon MALDI-TOF/TOF fragmentation. Recently, (2-ME) trimethylammonium hydrochloride has been used as a Michael donor in the addition reaction, resulting in markedly enhanced detection of a model phosphopeptide during electrospray MS.16 This label was also susceptible to fragmentation during CID, thereby complicating MS/MS data interpretation.
Noticeably milder reaction conditions were reported, which use barium hydroxide as an elimination reagent and 2-aminoethanethiol as a Michael donor, eliminating the use of sodium hydroxide and organic solvent mixtures and allowing the reaction to proceed in pure aqueous solution.17 Barium hydroxide has been reported earlier to preferentially catalyze the β-elimination of phosphoserine.18 With these developments, the chemistry became amenable to solid-phase adaptation, as demonstrated earlier, using ZipTipC18 pipette tips as analyte sorbent for the in situ reaction.19 This derivatization format offers several compelling advantages over the classical in-solution-based methods, the predominant sample preparation technique in current proteomic studies. Samples are cleaned up in situ, eliminating interfering matrix components and excess reagent. The process inherently concentrates the analyte, providing for completeness of reaction, improved throughput, and efficient use of dilute samples, and is highly amenable to automation. Since the introduction of this concept to phosphoprotein characterization,19 its apparent benefits have been exploited in only a few proteomics studies aimed at the facilitation of de novo sequence interpretation. Lysine-containing peptides bound to Cleanup C18 pipette tips were reacted to completion with 2-methoxy-4,5-dihydro-1H-imidazole at a threefold faster reaction rate.20 Near-quantitative, solid-phase peptide amino group sulfonation, using 3-sulfopropionic acid N-hydroxysuccinimide ester, was accomplished in <3 min, whereas the analogous solution-phase reaction typically required up to 30 min for completion.21 It should be noted here that modern analytical sample preparation techniques are based almost exclusively on solid-phase derivatization, coupled online with separation systems, and in these applications, have found widespread use for automated trace analysis of a bioorganic compound.22
In the expanded evaluation of the chemical strategy reported here, various reaction conditions were explored using a panel of model phosphopeptides and the optimized protocol then applied to low-level protein model digests. As demonstrated in the report, the conversion products were readily recognized in the MALDI-TOF spectra of unfractionated digests by the resultant signal enhancement and their characteristic mass shifts. In addition, sequencing on CID proved to be substantially more informative than for the corresponding chemically unmodified peptides. To determine the efficiency of the chemical strategy to detect phosphorylated, proteolytic fragments from experimental proteins, the method was applied to a tryptic digest of α-catenin, immunoprecipitated from EGF-stimulated A431 human epidermoid carcinoma cells. EGF stimulation of A431 human epidermoid carcinoma cells results in disruption of the α-catenin/β-catenin intercellular adhesion complex, promoting β-catenin transactivation. This finding was the starting point for comprehensive biochemical studies to reveal α-catenin's role in the intercellular adhesion complex and its correlation to tumor cell invasion.23 As demonstrated in our report, the method provided evidence of a phosphorylation event localized at serine 641, which is implicated in the protein's dissociation from the adhesion complex. An additional application involved the yeast Maf1 transcriptional repressor protein, which had been expressed in Escherichia coli and subsequently phosphorylated in vitro. Phosphorylation of yeast Maf1 on consensus PKA sites regulates its distribution between the cytoplasm and the nucleus, and simultaneous mutation of serine residues 90, 101, 177, 178, 209, and 210 at the PKA consensus sites (6SA mutant) leads to nuclear accumulation. Relocation of MAF1 from the cytoplasm to the nucleus is thought to provide one level of control over its inhibitory interaction with RNA polymerase III.24 As demonstrated in our report, the protocol enabled unambiguous identification of five phosphorylated fragments, which could be mass-matched to the known protein sequence. These fragments were contiguous with, or contained, PKA recognition sites and allowed identification of two previously assigned and two novel sites of phosphorylation.
MATERIALS AND METHODS
TFA, GelCode Blue Stain Reagent, was obtained from Pierce (Rockford, IL, USA). Formic acid, 2-aminoethanethiol hydrochloride, and barium hydroxide octahydrate were from Sigma-Aldrich (Milwaukee, WI, USA). Ammonium hydrogen carbonate was purchased from Fluka (Ronkonkoma, NJ, USA). N-octyl glucoside (OGS) was obtained from Roche Diagnostics (Indianapolis, IN, USA). Methanol and acetonitrile were from Burdick & Jackson (Muskegon, MI, USA). Bis(2-ME) (BMS) was obtained from Calbiochem (La Jolla, CA, USA). Isopropyl-1-thio-β-d-galactopyranoside was from United States Biochemical (Cleveland, OH, USA). Tris (hydroxymethyl) aminomethane, magnesium chloride, sodium chloride, sodium fluoride, DTT, iodoacetamide, angiotensin I human acetate salt hydrate, HSA, horse heart myoglobin, β-lactoglobulin from bovine milk, carbonic anhydrase from bovine erythrocytes, OVA, bovine β-casein, its monophosphorylated peptide T6, anhydrous methanol, and anhydrous-deuterated methanol (CD3OD) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ni2+-nitrilotriacetic acid (NTA)-agarose beads were obtained from Qiagen (La Jolla, CA, USA). E. coli Rosetta 2 (DE3) and pET-30a(+) were purchased from Novagen (San Diego, CA, USA). The protease inhibitors leupeptin and pepstatin were obtained from Roche Diagnostics. Resource Q resin, [γ-32] ATP and unlabeled ATP were from GE Healthcare (Piscataway, NJ, USA). Murine rPKA was purchased from New England Biolabs (Ipswitch, MA, USA). ZipTipC18 pipette tips (0.6 μL bed volume) and ZipTipμ-C18 pipette tips (0.2 μL bed volume) were purchased from Millipore (Billerica, MA, USA). α-Cyano-4-hydroxycinnamic acid was from Agilent Technologies (Palo Alto, CA, USA). Polyacrylamide gels (Criterion precast gel, 1 mm, 10%) were from Bio-Rad (Hercules, CA, USA). The following model phosphopeptides were synthesized in-house and purified by HPLC: P1, RADpSHEGEVA, m/z 1150.4; P2, SHNSALYpSQVQK, m/z 1441.2; P3, GIKSHNpSALYSQVQK, m/z 1739.3; P4, TATGpSGIKSHNSAL, m/z 1423.6; P5, RGApSPVE, m/z 795.5; P6, RRQpSPVA, m/z 835.5; P7, KRpTIRR, m/z 909.9. The following phosphopeptides were purchased from AnaSpec (San Jose, CA, USA): β-casein phosphopeptide T6, FQpSEEQQQTEDELQDK, m/z 2061.8; PKA regulatory subunit II substrate phosphopeptide, DLDVPIPGRFDRRVpSVAAE, m/z 2192.4; UOM9, PKC substrate-3 phosphopeptide, KRPpSQRHGSKY amide, m/z 1422.5; DAM1, outer kinetochore protein DAM1 phosphopeptides, SFVLNPTNIGMpSKSSQGHVTK, m/z 2312.6. Trypsin (modified sequencing grade) was purchased from Promega (Madison, WI, USA). Pepsin from porcine gastric mucosa was purchased from Sigma-Aldrich (St. Louis, MO, USA). Materials and methods used to generate and purify phosphorylated α-catenin were described recently by Ji et al.23
Maf1 Protein Purification and In Vitro Phosphorylation
A full-length cDNA clone of Saccharomyces cerevisiae Maf1 was inserted into pET-30a(+) to yield a C-terminal hexahistidine-tagged protein. The plasmid was transformed into E. coli Rosetta 2 (DE3) and protein expression induced by adding 0.1 mM isopropyl-1-thio-β-d-galactopyranoside at 15°C, followed by incubation overnight. The protein was purified under native conditions using Ni2+-NTA-agarose, according to the manufacturer's recommendations. Maf1, in 50 mM Tris-HCl (pH 7.0), 75 mM NaCl, 10% glycerol, 1 mM DTT, and protease inhibitors (leupeptin and pepstatin, each at 1 μg/ml), was purified further by LC at a flow rate of 0.5 ml/min using a Resource Q column (7×114 mm). A linear gradient, from 75 mM to 1 M NaCl, was used, and the protein eluted at 300 mM NaCl. The Maf1 concentration of pooled fractions was determined by absorbance measurement at 280 nm using a molar extinction coefficient calculated from the protein sequence. Aliquots of Maf1 were stored at −70°C until use. In vitro phosphorylation of native rMaf1 (5 μg) was performed with murine rPKA (1.25 units) in a 50-μL reaction volume, following the manufacturer's recommendations. The reaction mixture containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM NaF, and 1 mM [γ-32P] ATP (22 dpm/pmol) or 1 mM cold ATP was incubated at 30°C for 30 min and an aliquot analyzed by SDS-polyacrylamide electrophoresis. After staining with GelCode Blue Stain Reagent, the gel was dried and exposed to a phosphor-storage screen, together with aliquots of [γ-32P] ATP, spotted on filter paper as a standard. Signal intensities of labeled Maf1 and ATP standard were quantified using ImageQuant software obtained from Molecular Dymamics (Sunnyvale, CA, USA). The stoichiometry of phosphorylation, as observed in separate experiments, ranged from 2 to 3 moles phosphate, incorporated/mole Maf1. Phosphorylated protein, prepared in parallel reactions with unlabeled ATP, was stored at −70°C until use.
Protein In-Gel Proteolytic Digestion
GelCode Blue-stained bands, from 5 to 50 pmoles protein loaded onto the gels, were excised and destained twice with 200 μL 25 mM ammonium bicarbonate in 50% aqueous acetonitrile for 30 min at 37°C. Bands were dried briefly in a SpeedVac and carboxyamidomethylated, essentially as described by Speicher,25 except that BMS was used as a reducing agent. Bands were dehydrated for 10 min in 100 μL acetonitrile, briefly dried in a SpeedVac. Dried gel bands were swollen at room temperature in 20 μL 25 mM ammonium bicarbonate/0.01% OGS or in 20 μL 10 mM hydrochloric acid/0.01% OGS, supplemented with Promega-modified trypsin or pepsin, respectively, to an enzyme:substrate ratio of 1:10 (w:w). After 20 min, 40 μL 25 mM ammonium bicarbonate or 40 μL 10 mM hydrochloric acid/0.01% OGS was added, and the digestion continued for 18 h at 37°C. After incubation, 50 μL 0.1% TFA was added. The supernatant was removed, and another 50 μL 0.1% TFA was added. The gel bands were incubated for 30 min at 37°C. The combined extracts were reduced in volume to 35 μL, acidified by addition of 5 μL 8% TFA, to a final concentration of 1% TFA, and immobilized on ZipTipC18 pipette tips as described below.
Protein In-Solution Proteolytic Digestion
In-solution tryptic digestion of 2–20 pmoles protein was performed in 35 μL 25 mM ammonium bicarbonate/0.005% OGS at an enzyme:substrate ratio of 1:100 (w:w). After 18 h at 37°C, the digests were acidified with 5 μL 8% TFA to a final concentration of 1% TFA and immobilized on ZipTipC18 pipette tips as described below.
Sample Immobilization
ZipTipC18 pipette tips or ZipTipμ-C18 pipette tips were wetted six times with 10 μL methanol, followed by six washes with 10 μL 0.1% TFA. Model peptides (2–20 pmoles) were loaded from 10 μL aliquots of 0.2% TFA/0.01% OGS solutions onto ZipTipC18 pipette tips and subjected to 10 sample load/dispense cycles. The ZipTipC18 pipette tips were then washed three times with 10 μL 0.1% TFA. For high-volume sample enrichment (40 μL), peptides (0.125–20 pmoles) were sequentially loaded in 10 μL aliquots onto ZipTipC18 pipette tips or ZipTipμ-C18 pipette tips and dispensed into a 0.5-ml microfuge tube. The sample was then transferred back in this step-wise mode to the original collection tube. This alternating load/dispense enrichment cycle was repeated five times. The ZipTip pipette tips were then washed five times with 10 μL 0.1% TFA. Optionally, 200 μL disposable pipette tips can be press-fitted into ZipTipμ-C18 pipette tips or ZipTipC18 pipette tips, as suggested by the manufacturer and operated with a 200-μL pipettor to pass the sample slowly, 10 times over the resin. Up to 200 μL sample can be processed for binding in this manner (www.millipore.com/publications).
On-Resin β-Elimination and On-Resin β-Elimination with Concurrent Michael Addition
An aqueous, 66-mM barium hydroxide solution was used for β-elimination (7.49 mg/mL). A 2/1 mixture of aqueous 100 mM barium hydroxide (31.54 mg/mL) and aqueous 100 mM 2-aminoethanethiol hydrochloride (11.36 mg/mL; 66 mM barium hydroxide/33 mM 2-aminoethanethiol hydrochloride, pH 12.1) was used for β-elimination with concurrent Michael addition. Barium hydroxide was finely ground to facilitate dissolution. Residuals, mainly carbonates, were removed by centrifugation (1 min at 13,000 rpm). Freshly prepared solutions of the elimination base and of the nucleophile were used for each experiment. The analyte was bound to ZipTipC18 pipette tips or ZipTipμ-C18 pipette tips, which were then flushed twice to waste with 10 μL reagents—then loaded onto the solid supports from 60 μL, which had been placed into 0.5 ml microcentrifuge tubes. Alternatively, the incubation was carried out in 1 ml-capped microcentrifuge tubes, which had been loaded with 80 μL reagents. The samples were incubated in the tubes for predetermined times at 37°C or 55°C, while immersed in the reagent solutions. After incubation, the tips were washed with 100 μL 0.1% TFA, passed over the resin in 10 μL aliquots. Peptides were eluted in 5–10 μL 50% acetonitrile/0.1% TFA/0.01% OGS, of which, 1 μL was used for MALDI analysis. In some experiments, ZipTipμ-C18 pipette tips were eluted directly in 1 μL matrix containing 0.1% TFA onto the MALDI plate.
In-Solution β-Elimination with Concurrent Michael Addition
For in-solution derivatization, 40 μL aliquots of solutions containing 50 fmol/μL peptide were taken to complete dryness by SpeedVac evaporation. The peptides were then resuspended in 40 μL 66 mM barium hydroxide/33 mM 2-aminoethanethiol hydrochloride reagent mixture and incubated for predetermined times at 55°C. After derivatization, the preparations were acidified with 2 μL 20% TFA and submitted to ZipTipμ-C18 pipette tip purification. Peptides were eluted onto the target in 1 μL matrix containing 0.1% TFA.
ZipTipC18 Pipette Tip Alkali Compatibility Evaluation
Methanolic HCl solutions were prepared freshly by adding drop-wise 40 μL acetyl chloride to 250 μL methanol or deuterated methanol kept on ice. Aliquots of the model peptide angiotensin I (2 nmoles) were taken to dryness in a SpeedVac, followed by addition of 25 μL methanolic HCl reagents. After 90 min incubation at 12°C, the reaction mixtures were dried. The methyl-d0-esterified peptide and the methyl-d3-esterified peptide were dissolved in 50% acetonitrile/0.1% TFA and stored at −21°C prior to use. In the stable isotope experiments described below, the methyl-d0-esterified preparation was used as a reference peptide after appropriate dilution.
Efficiency and reproducibility of relative peptide recovery from the support were assessed as follows: aliquots of angiotensin I solutions in 0.2% formic acid/0.01% OGS (20 or 100 pmoles) were subjected to 10 sample load/dispense cycle. The ZipTipC18 pipette tips were washed three times with 10 μL 0.1% TFA to remove unbound material and eluted twice with 10 μL 50% acetonitrile/0.1% TFA/0.01% OGS. Eluates were dried in a SpeedVac, dissolved in 25 μL isotopically labeled, methanolic HCl, incubated for 90 min at 12°C, and subsequently dried. The methyl-d3-esterified peptides were then dissolved in 10 μL 50% acetonitrile/0.1% TFA, and 5 μL peptide solutions were mixed with 5 μL methyl-d0-esterified reference peptide, followed by addition of 10 μL MALDI matrix. The final mixture (1 μL) was then applied to the MALDI target for subsequent relative quantitation of the eluted peptides.
To assess the impact of derivatization with respect to peptide retention, ZipTip-bound peptides (20 or 100 pmoles) were rinsed three times with 10 μL 0.1% TFA. The ZipTipC18 pipette tips were then loaded with 10 μL β-elimination/Michael addition reaction mixture and incubated while immersed in the reagent for 2 h at 37°C or for 1 h at 55°C. The ZipTip tips were desalted by passing 100 μL 0.1% TFA in 10 μL aliquots over the resin and eluted twice with 10 μL 50% acetonitrile/0.1% TFA/0.01% OGS. The eluates were dried and methyl-d3-esterified under the conditions described above. The reaction mixture was dried and dissolved in 10 μL 50% acetonitrile/0.1% TFA. In parallel experiments, ZipTip-bound peptides (20 or 100 pmoles) were washed and subsequently eluted. The eluates were taken to dryness, methyl-d0-esterified, and followed by vacuum evaporation of the reaction mixture. The reaction product was dissolved in 10 μL 50% actonitrile/0.1% TFA and used as a reference peptide. The differentially labeled preparations (5 μL) were combined and mixed with 10 μL MALDI matrix. The mixture (1 μL) was applied onto the MALDI target. Potential alkali-induced sample loss from the reversed-phase support was determined by measuring the ratio of the relative abundance of the methyl-d3- and methyl-d0-esterified peptides in the mass spectra.
MS
A Voyager-DE STR Biospectrometry (Applied Biosystems, Foster City, CA, USA) was used and operated in the reflector mode at an accelerating voltage of 20 kV. Laser intensity was typically set at 1690–2400, and spectra were acquired using 100–200 laser shots/spectrum. The data shown are based on three accumulated acquisitions. Some spectra were obtained in the linear mode using 80 laser shots/spectrum, and five to 10 spectra acquired from spots at different positions were averaged. For peptide fragmentation, a 4700 Proteomics Analyzer was used and operated in the MS and MS/MS modes; typical laser power for MS was ∼3700; for MS/MS, ∼4500. Usually, 1000–2000 shots were acquired for MS; 2000–20,000 for MS/MS.
RESULTS AND DISCUSSION
Sample Handling
The solid-phase procedure is exceedingly simple in implementation. Peptides or protein digests are bound to ZipTipC18 pipette tips or ZipTipμ-C18 pipette tips, followed by a brief solvent wash to remove contaminants from the support, which may interfere with the reaction. The ZipTips are then loaded with the reagent, left immersed in the reaction mixture during the subsequent incubation, and desalted prior to MS analysis (Fig. 1B, inset). This sample handling format offers some notable advantages over the classical in-solution-based methods, the predominant sample preparation technique in current proteomic studies, as follows: (1) The process of sample adsorption inherently concentrates the analyte on the support, allowing the chemistry to take place in an environment, which is essentially free of reagent dilution. As a result, the reaction proceeds in situ at higher efficiency and faster kinetics than in solution, as demonstrated by the experiments described below. (2) The digests can be enriched effectively on the solid phase from dilute solutions, in which the chemical reaction inherently proceeds at slow reaction rates. In these sample limited situations, ZipTipμ-C18 pipette tip is preferentially used, enabling product elution directly onto the target in as little as 0.5 μL matrix with consequent improved mass detection. (3) The solid-phase strategy obviates the need for sample dry-down, commonly practiced in proteomics studies to concentrate peptide mixtures prior to derivatization in solution. As noted for low-level samples (<2 pmole), this sample-handling step can cause substantial adsorptive peptide loss, ranging up to 50% or more of the starting solution.25 (4) The solid-phase procedure should be readily amenable to automation on a ZipTipC18 pipette tip/ZipTipμ-C18 pipette tip-compatible liquid-handling station.
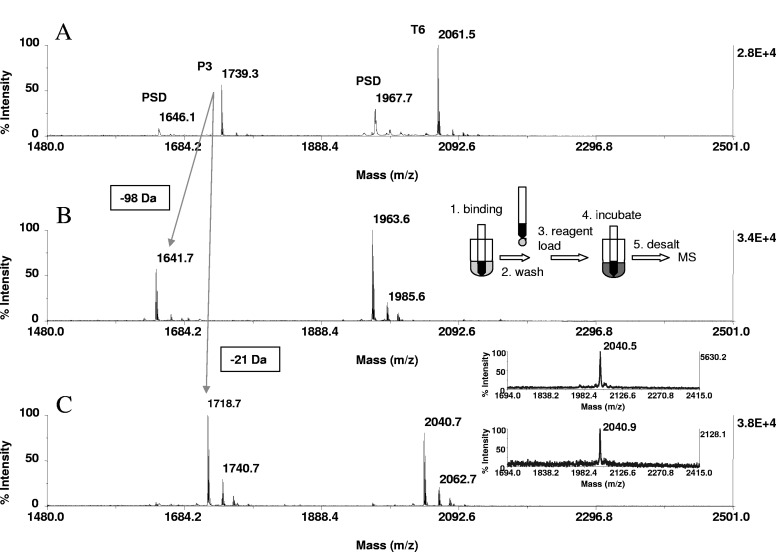
FIGURE 1.
Solid-phase β-elimination and β-elimination with concurrent Michael addition of phosphoseryl peptides. A peptide mixture containing 20 pmoles P3 (GIKSHNpSALYSQVQK) and T6 (FQpSEEQQQTEDELQDK) was bound to ZipTipC18 pipette tips and incubated with 66 mM aqueous barium hydroxide or aqueous barium hydroxide/2-aminoethanethiol reaction mixture composed of 66 mM base and 33 mM nucleophile. After 1 h at 37°C, the reactions were terminated by a solvent wash. MALDI-TOF spectra of (A) unmodified peptides, (B) after β-elimination, and (C) after β-elimination with concurrent Michael addition. Reactions were accompanied by a 98-Da mass shift/eliminated phosphate and by a 21-Da mass shift/derivatized phosphoamino acid, as indicated by cross arrows in the spectrum. Minor ions represent sodium and potassium adducts. PSD in A denotes PSD products. One-tenth of the eluates corresponding to ∼2 pmoles peptide was applied to the target. (C, inset) The MALDI mass spectra of the Michael addition products of T6 at sample loads of 250 fmole (upper panel) and 125 fmole (lower panel). Samples were derivatized on ZipTipμ-C18 pipette tips, washed, and eluted directly in matrix onto the target. (B, inset) A schematic representation of the derivatization handling steps.
As has been reported, inclusion of trace amounts of the nonionic detergent OGS in the in-gel digestion buffers affords improved peptide recovery by enhancing peptide solubility and minimizing peptide adsorption associated with sample handling. The detergent proved highly compatible with MALDI-TOF analysis and with LC-MS, owing to its strong retention on reversed-phase columns.26 To take advantage of this favorable effect, in-gel and in-solution protein digestion was performed in the presence of the additive, and test peptide solutions evaluated in this study were supplemented with OGS prior to immobilization.
On-Resinβ-Elimination and β-Elimination with Concurrent Michael Addition
A panel of peptides, phosphorylated at serine and at serine C-terminally adjacent to proline, was used to define the experimental conditions for efficient β-elimination and β-elimination with concurrent Michael addition. Serine-proline sequence motifs, often mapped by protein mutational analysis, have attracted much interest, as they are in a subset of phosphoproteins, specifically isomerized by the prolyl isomerase Pin1, a catalyst for tumorigenesis of commonly occurring cancers.27 Moreover, according to a recent report, a significant portion of cellular phosphorylation events is regulated by proline-directed kinases.28
Representative MALDI-TOF data from the derivatization experiments are shown in Figs. 1 and 2. The phosphoseryl peptides P3 (GIKSHNpSALYSQVQK) and T6 (FQpSEEQQQTEDELQDK) were subjected β-elimination or to β-elimination with concurrent Michael addition for 1 h at 37°C. Under these conditions, the peptides were essentially quantitatively converted to their dehydroalanyl and S-2-aminoethylcysteine counterparts (Fig. 1A–C), as were the additional, six phosphoseryl peptides listed in Materials and Methods. Consequently, this protocol was adopted as a standard procedure to examine proteolytic digests from model proteins and experimental samples. As indicated in Fig. 1, the reactions were accompanied by the characteristic mass shifts of 98 Da and 21 Da, which provides a means to identify phosphopeptides in the mass maps of protein digests, as discussed later in the text. Under the same reaction conditions, a substantial portion (>50%) of the peptide P5 (RRQpSPVA), in which the phosphoseryl residue is adjoined by proline, remained in its dehydroalanyl form (Fig. 2B, inset). The slower reaction kinetics of the addition reaction is expected, as proline shields the β-carbon of the residue from the nucleophilic attack. Similar results were obtained with the phosphothreonyl peptide P7 (RGApSPVE), consistent with the common perception that the methyl group of the residue protects the β-carbon from nucleophilic attack (data not shown). Conversion yields improved when the peptides were incubated at increased temperatures. As shown in Fig. 2B, the S-2-aminoethylcysteine derivative of P5 was formed efficiently after 1 h incubation at 55°C. Comparable results were obtained with P6 (RGApSPVE; results not shown) and as would be expected, with the phosphoseryl peptides listed in Materials and Methods. Under these more stringent reaction conditions, the phosphothreonyl peptide P7 (KRpTIRR) was converted with an estimated 70% efficiency to its β-methyl-S-2-aminoethylcysteine derivative (Fig. 2D). At the same temperature, the β-elimination product of this peptide was fully formed already after 30 min incubation, indicating that the concurrent addition phase limits the rate of the overall reaction (Fig. 2C, inset). This trend became considerably more pronounced when the same reaction was performed in solution (see below).
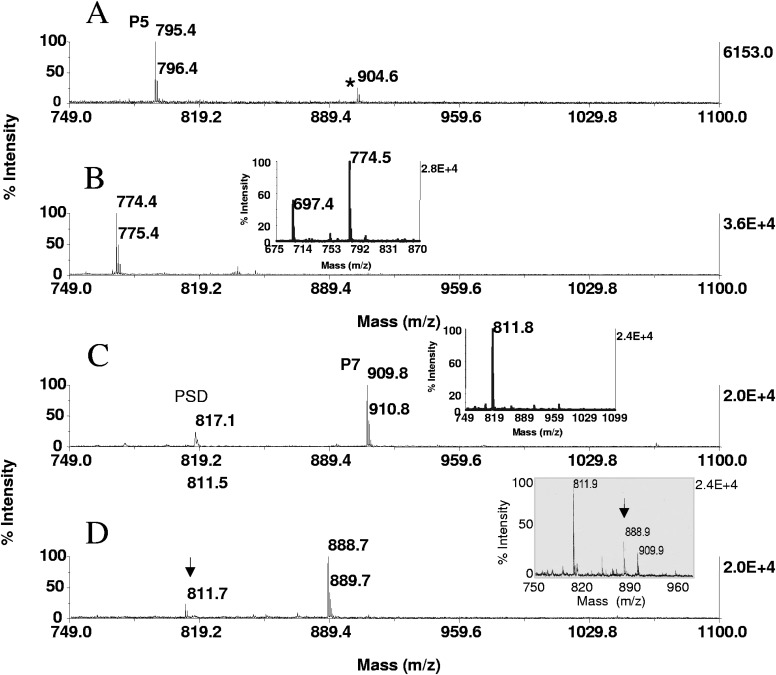
FIGURE 2.
Solid-phase β-elimination with concurrent Michael addition of the phosphoserine/proline- and phosphothreonine-containing peptides. P5 (RGApSPVE) and P7 (KRpTIRR) were enriched on ZipTipμ-C18 pipette tips from 40 μL aliquots of a 50-fmol/μL solution and incubated in the 66-mM barium hydroxide/33-mM 2-aminoethanethiol hydrochloride reagent mixture for 1 h at 55°C. Equimolar amounts of P7 were taken to dryness, reacted for 1 h at 55°C in 40 μL alkaline nucleophile solution or for 30 min at 55°C in 40 μL elimination base, followed by ZipTipμ-C18 pipette tip purification. The samples were eluted in matrix directly onto the target. MALDI-TOF spectra of (A) unmodified peptide P5, (B) after the concurrent reaction, (C) unmodified peptide P7, and (D) after the concurrent reaction. (B, inset) The MALDI-TOF spectrum of P5 after incubation for 1 h at 37°C. Note residual β-elimination product at m/z 697.4, owing to incompleteness of the addition reaction. (A) *Unidentified, minor species. (C, inset) The MALDI-TOF spectrum of P7 after 30 min incubation at 55°C in 66 mM barium hydroxide. (D, inset) MALDI-TOF spectrum of P7 after concurrent in-solution derivatization. Note incompleteness of thiol adduct formation (arrow).
In the experiments, 40 μL aliquots of the phosphopeptide solution (50 fmol/μL) were processed for binding on ZipTipμ-C18 pipette tips, providing for ∼200-fold analyte enrichment on the 200-nL sorbent beds. The resultant, locally increased peptide concentration should, in principle, allow the chemistry to proceed more efficiently and faster on the solid-phase than in solution. To test the validity of this precept, we subjected the phosphothreonyl peptide to in-solution derivatization. In this procedure, aliquots of the phosphopeptide solution were subjected to SpeedVac evaporation, followed by addition of 40 μL alkaline nucleophile reagent to reconstitute the dried peptide to its original concentration. After incubation for 1 h at 55°C, the in-solution preparations were purified on ZipTipμ-C18 pipette tips and deposited directly in matrix onto the MALDI plate. The MALDI-TOF spectra of the samples revealed that the β-elimination product was accumulated to near completion, whereas the concurrent addition reaction was markedly retarded (Fig. 2D, inset). Similarly, under conditions that afforded complete β-elimination on the solid-phase (Fig. 2C, inset), ∼50% of the peptide remained intact when the treatment was performed in solution, and an additional 30 min incubation was needed to complete the reaction (results not illustrated). The comparison data clearly identified the peptide preconcentration effect on the solid phase as a critical determinant of the accelerated reaction rate and of the relatively high efficiency of the overall reaction. We note that the moderate reactivity of phosphothreonine has been recognized in earlier studies using 2-aminoethanethiol or other nucleophiles in concurrent in-solution reaction schemes and has been addressed in these protocols by prolonged incubation (>4 h).14,29 However, experimentation to optimize the in-solution protocol was not pursued in our study. Our data are in general accord with those reported earlier in proteomics studies, exploiting accelerated in situ peptide amine-directed chemical modification to facilitate de novo sequence interpretation.20,21 It should be noted here that current analytical sample preparation techniques, such as used for automated, on-line trace analysis of bioorganic compounds, rely on solid-phase derivatization and have, because of the considerably improved mass detection, replaced the analogous in-solution derivatization.22
With these developments, MALDI-TOF maps, prepared from digest before and after derivatization, can be scanned for serine and threonine phosphorylation, as discussed later in the text. However, we recognize the need for an expanded data set to demonstrate the general use of the method for derivatization of phosphothreonyl peptides and have, to this purpose in recent work, extended the evaluation of the solid-phase chemical strategy to a panel of phosphothreonyl peptides, including phosphothreonyl peptides preceded by proline. A manuscript of this detailed study, describing the optimal reaction conditions for comprehensive derivatization of phosphopeptides, is currently in preparation.
Reliability of Derivatization
The reliability of derivatization was assessed in a semiquantitative mode with the synthetic β-casein peptide T6. The peptide (1 pmole) was loaded from 40 μL aliquots of a 25 fmole/μL peptide solution onto eight separate ZipTipμ-C18 pipette tips and subjected to β-elimination/Michael addition under the conditions described in Fig. 1. The samples were then eluted directly in matrix onto the MALDI plate. Eighty laser shots/spot were typically required to obtain stable signals. A total of 640 laser shots, sampled from eight different spots, was summed for each spectrum. The spectra were devoid of the unmodified species, and signal variations observed between the individual spectra were, at most, 15%. The comparable MALDI response observed in the spectra was interpreted to reflect high reliability of derivatization.
Derivatization at the Femtomole Level of Peptide
To explore the overall mass sensitivity of the method, aliquots from serial dilutions of the β-casein peptide T6 were immobilized on ZipTipμ-C18 pipette tips and converted to the Michael addition analogues, as described in Fig. 1. The eluates were deposited directly in matrix onto the MALDI plate, and spectra were acquired in the linear mode. As illustrated in Fig. 1C, inset, 125 fmoles starting material could be carried through the β-elimination/Michael addition reaction effectively. The spectrum demonstrates that low quantities of peptide are amenable to derivatization.
ZipTipC18 Pipette Tip Alkali-Compatibility Evaluation
Alkali-induced silica resin decomposition has been reported to diminish, to a minor extent, the retention of the silane-bonded stationary phase during long-term C18 reversed-phase column testing (>1 week) with mobile phases at pH 9–12.3.30 This effect was attributed to mechanical attrition around the covalently attached silanes, eventually causing splitting off of the bonded organic phase, and could therefore potentially reduce the analyte recovery from the silica-based reaction bed. However, our data show that peptides in general exhibited comparable signal intensities in the MALDI spectra prepared from the derivatized and the untreated samples, suggesting that the analyte was retained efficiently on the solid-phase when exposed to the alkaline reaction conditions (see Figs. 5 and 6). In support of this interpretation, we assessed the relative recovery of a model peptide (angiotensin I) from ZipTips, before and after the chemical treatment, using a stable isotope dilution technique. To this purpose, the peptide (2 nmoles) was treated with methanolic HCl or with deuterated methanolicHCL. The differentially labeled peptide stock solutions were adjusted to a concentration of 1 pmole/μL, mixed in equimolar amounts, and applied to the MALDI target. The spectrum shows that the methyl-d0- and methyl-d3-esterified peptides were observed at nearly equal abundance. The products were formed in quantitative yields, and there were no side-reactions (Fig. 3A). The methyl-d0-esterified peptide was used as an internal reference to determine the relative amount of peptide in the eluted fractions. In this experiment, 20 pmoles model peptide was then bound to ZipTipC18 pipette tips in triplicate, washed to remove unbound material, methyl-d3-esterified after elution, supplemented with the methyl-d0-esterified reference peptide, and subjected to MALDI-MS. As assessed from the spectral abundance ratios of the individual replicates, an estimated 82.2%, 84.3%, and 86.0% (average 84.1%) of the bound peptide was recovered from the solid -phase supports (Fig 3B). The reproducibility and efficiency of peptide recovery noted here are in accord with an earlier evaluation of ZipTipc18 pipette tips using 14C-accelerator MS for peptide quantitation.31
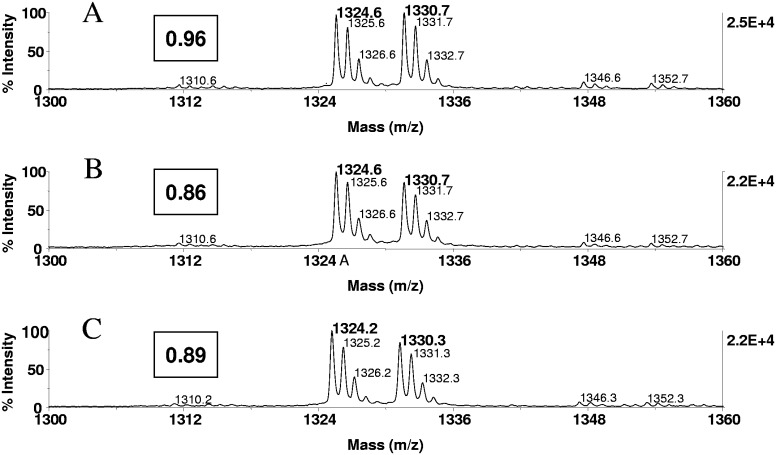
FIGURE 3.
ZipTipC18 pipette tip alkali-compatibility evaluation. Angiotensin I (20 pmoles) was immobilized on a ZipTipC18 pipette tip, which was then washed to remove unbound material and subsequently eluted. The eluate was methyl-d3-esterified and supplemented with the methyl-d0-esterfied reference peptide, which had been prepared in solution. An equimolar amount of peptide was immobilized on a ZipTipC18 pipette tip, which was exposed to the β-elimination/Michael conditions after a solvent wash, as described in Fig. 2, followed by methyl-d3 esterification of the eluate. Untreated samples were methyl-d0-esterified after elution and used as an internal reference. Experiments were in triplicate. MALDI-TOF spectra were generated using 400 laser shots/spectrum, accumulated from five different positions: MALDI-TOF spectra of (A) an equimolar mixture of the differentially labeled peptide prepared in solution (1 pmol/μl), (B) methyl-d3-esterified peptide from untreated sample mixed with methyl-d0-esterified internal reference peptide, and (C) methyl-d3-esterified peptide from alkali-exposed sample mixed with methyl-d0-esterified reference peptide from untreated sample. (A–C) The relative spectral abundance ratios. As estimated from the relative spectral abundance ratio, 86% of bound peptide was recovered from the C18 supports (average 84.1%), and 11% (average 13.5) of the peptide was lost during the alkali treatment. Approximately 0.5 pmoles peptides was applied to the target.
To assess the potential impact of the alkali treatment on peptide retention, ZipTipC18 pipette tips were loaded with 20 pmoles peptide and washed briefly. Samples were exposed for 1 h at 55°C to the β-elimination/Michael addition reaction mixture or left untreated. Eluates from the chemically treated samples were methyl-d3-esterified. Eluates from the untreated counterparts were methyl-d0-esterified and used as an internal reference in the mixture of the differentially labeled peptide samples. As estimated from the spectral abundance ratio calculated for the replicates, 11.0%, 13.7%, and 15.8% (average 13.5%) of peptide were lost as a result of the chemical treatment (Fig. 3C). A comparable, minor sample loss was observed when the incubation was extended for 1 additional h or after 2 h at 37°C (data not shown). The data indicate that the chemical treatment had only a minor impact on the utility of the reversed-phase support in this application.
Phosphopeptide Detection by Signal Enhancement
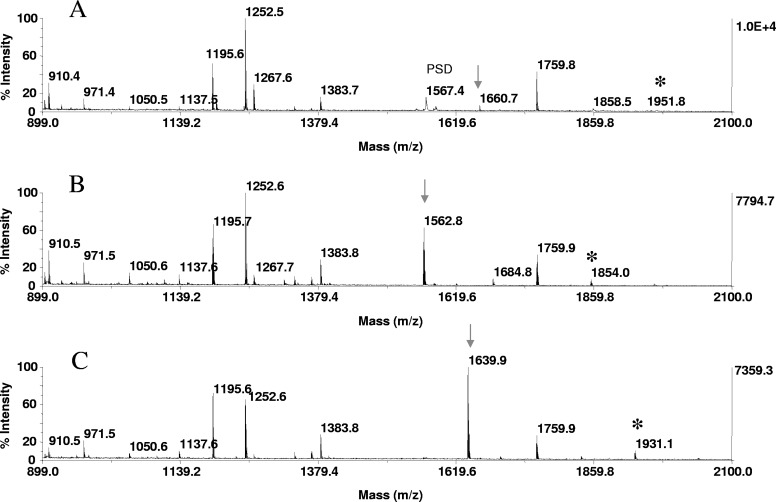
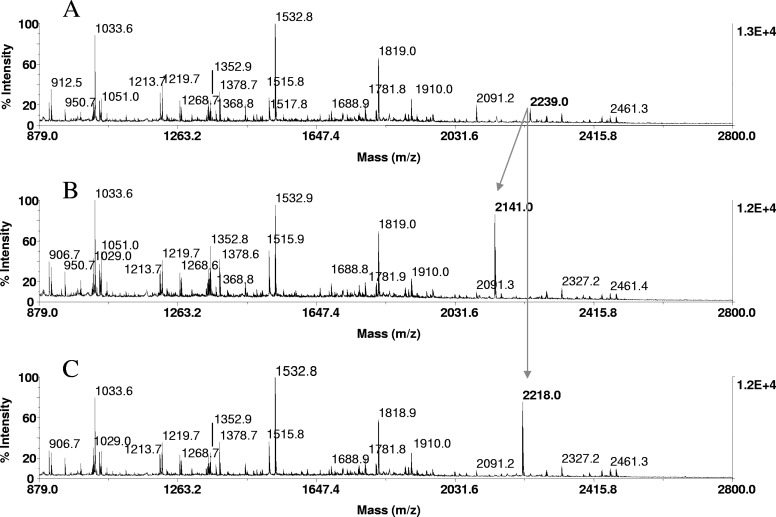
In these experiments, 2 picomoles of a resin-bound tryptic digest of β-casein, enriched from 50 femtomol/μL solutions onto ZipTipC18 pipette tips, was subjected to β-elimination and to β-elimination with concurrent Michael addition for 1 h at 37°C. An equimolar amount of the digest was immobilized and left untreated as a control. Comparison of the MALDI spectra prepared from these samples revealed the formation of a prominent ion at m/z 3038.6, representing the thiol adduct (Fig. 4C) of the tetraphosphorylated peptide at theoretical m/z 3122.2 (residues 1–25), which was undetectable in its native form (arrow in Fig. 4A). Analogous to its synthetic counterpart and unlike the other model peptides (Fig. 1), the monophosphorylated peptide observed at m/z 2061.8 (residues 33–48) exhibited a decreased signal intensity after derivatization. In contrast, an estimated nine- and 11-fold increase in relative signal intensity was observed for the substoichiometrically, monophosphorylated peptide at m/z 1660 (VPQLEIVPNpSAEER, residues 121–134) in the mass maps of an in-gel tryptic digest of α-S1 casein after β-elimination and β-elimination with concurrent Michael addition, respectively (Fig. 5 A–C). Equally prominent signal enhancements were noted for the tryptic peptide at m/z 2088.8 (EVVGpSAEAGVDAASVSEEFR, residues 240–359), derived from an in-gel digest of OVA (data not shown), and for the tryptic peptide at m/z 2239.0 (TPEELDDpSDFETEDFDVR, residues 634–651), derived from an in-gel digest of α-catenin (Fig. 6 A–C). The results verify the use of the on-resin derivatization strategy to facilitate phosphopeptide detection in unfractionated digests.
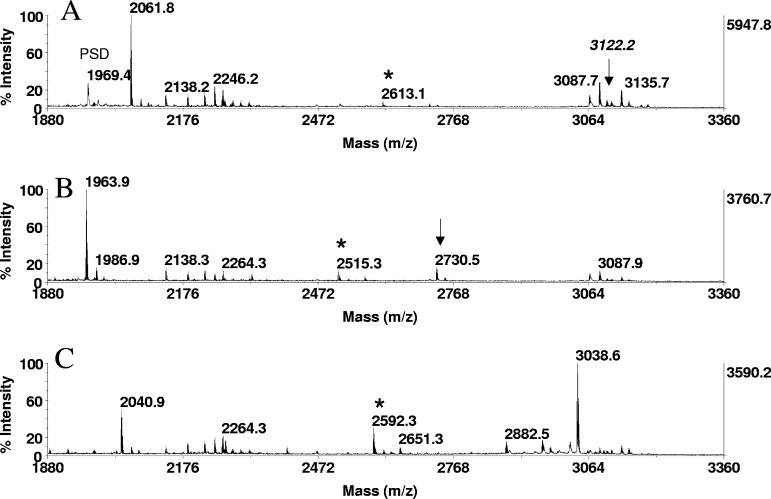
FIGURE 4.
Phosphopeptide detection by signal enhancement. In-solution tryptic digests of β-casein, derived from 2 pmoles protein, were immobilized onto ZipTipC18 pipette tips and subjected to β-elimination and to β-elimination with concurrent Michael addition under the conditions described in Fig. 1. One-fifth of the eluate corresponding to ∼400 fmoles digest was applied to the target. MALDI-TOF spectra of (A) untreated digest, (B) after β-elimination, and (C) after β-elimination with concurrent Michael addition. (A–C) Expanded sections of the spectra containing phosphorylated species, the dehydroalanyl, and S-2-aminoethylcysteine derivatives. Note the prominent ion of thiol adduct at m/z 3038.6 corresponding to the tetraphosphorylated species, spanning residues 16–40 (RELEELNVPGEIVEpSLpSpSpSEESITR), undetectable in A (arrow). (A–C) *Unidentified singly phosphorylated fragment and reaction products. (B) Arrow designates the dehydroalanyl derivative of the tetraphosphorylated tryptic fragment.
FIGURE 5.
Phosphopeptide detection by signal enhancement. In-gel tryptic digests of α-S1 casein, derived from 20 pmoles protein, loaded onto the gels, were bound to ZipTipC18 pipette tips and subjected to on-resin β-elimination and β-elimination with concurrent Michael addition under the conditions described in Fig. 1. One-tenth of the eluates corresponding to ∼2 pmoles digest was applied to the target. MALDI-TOF spectra of (A) untreated digest, (B) after β-elimination, and (C) after β-elimination with concurrent Michael addition. (A–C) Arrows indicate the phosphoserine peptide at m/z 1660.7, residues 121–134 (VPQLEIVPNpSAEER), its dehydroalanyl derivative at m/z 1562.8, and its thiol adduct at m/z 1639.9, which are observed at an estimated 11- and ninefold higher signal intensity, respectively, relative to the native counterpart. (A–C) *A miscleavage product at m/z 1951.8 spanning residues 119–134, its dehydroalanyl, and thiol derivative.
FIGURE 6.
Phosphopeptide identification in proteolytic digests of α-catenin, immunoprecipitated from EGF-stimulated A431 cells. Tryptic in-gel digests of α-catenin, derived from 20 pmoles protein loaded onto the gels, were bound to ZipTipC18 pipette tips and subjected to β-elimination and β-elimination with concurrent Michael addition under conditions described in Fig. 1. One-tenth of the eluates corresponding to ∼2 pmoles digests was applied to the target. MALDI-TOF spectra of (A) untreated tryptic digest, (B) after β-elimination, and (C) after β-elimination with concurrent Michael addition. Cross arrows indicate a phosphopeptide at m/z 2239.0, its β-elimination product at m/z 2141.0, and its thioladduct at m/z 2218.0. The diagnostic mass signature of 21 Da, accompanying the reactions, identified the peptide at m/z 2239.0 as the monophosphorylated species recognized as a phosphoseryl peptide, as it reacted under conditions that selectively promote phosphoserine conversion. The peptide mass-matched to a tryptic fragment within the known protein sequence (TPEELDDpSDFETEDFDVR), spanning residues 634–651. As indicated in bold, the sole serine at position 641 was tentatively assigned as the site of phosphorylation. This sequence-dependent assignment was subsequently confirmed by MALDI-TOF/TOF and LC-MS/MS.
The signal enhancement noted in our study appears to be attributable to the replacement of the negatively charged phosphate by the S-2-aminoethyl group, which incorporates a protonable hydrophobic moiety into the peptides, consistent with the observation that basic and hydrophobic residues in peptides tend to enhance the desorption/ionization process.32 The enhanced MALDI response, upon derivatization of the digests, was found most prominent with phosphopeptides containing a high proportion of glutamic and/or aspartic residues (Figs. 5 and 6) and with multiple phosphorylated species (Fig. 4). In contrast, phosphopeptides bearing multiple basic residues, such as derived from the basophilic substrates of Maf1 (e.g., WEQKRRIpSF), were moderately enhanced by the nucleophilic substitution (Fig. 7). Evidently, the additional, protonable moiety had only a minor impact on the favorable ionization behavior of these highly basic peptides. Accordingly, the native peptides and their modified counterparts were readily detectable in the spectrum, enabling facile identification of the phosphorylated species by interrogating the MALDI mass maps for the characteristic derivatization mass shift of 21 Da/phosphoamino acid.
FIGURE 7.
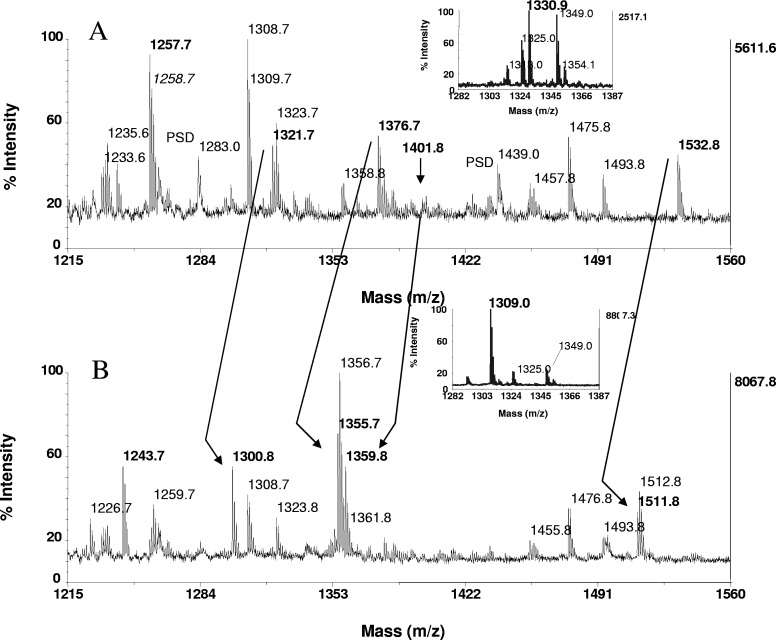
Phosphopeptide identification in proteolytic digests of Maf1. Tryptic and peptic in-gel digests of Maf1, derived from 15 pmoles protein, loaded onto the gels, were bound to ZipTipC18 pipette tips and subjected to β-elimination with concurrent Michael addition under the conditions described in Fig. 1. Expanded section of the MALDI-TOF spectra of (A) untreated tryptic digest and (B) after β-elimination with concurrent Michael addition. Arrows across the spectra indicate fragments recognized as phosphorylated by the characteristic mass shift of 21 Da/phosphoamino acid. Note the prominent ion of the thiol adduct at m/z 1359.8, recognized as a doubly phosphorylated species, undetectable in A (arrow) and mass-matched to RRpSpSSSSISSFK (residues 175–185). The known and assigned phosphorylation sites at serine 177 and 178, respectively, are marked in bold in the sequence. The ion at m/z 1321.7 corresponds to the singly phosphorylated form of the peptide. Additional ions identified in A were derived from monophosphorylated species and include the fragments at m/z 1376.7 and m/z 1532.8, corresponding to the sequences RDpSNSFWEQK (residues 88–97) and RDpSNSFWEQKR (88–98), in which, serine 90 was assigned by MS/MS as the site of phosphorylation. The peptide in A at m/z 1257.7 was identified as a carbamidomethylated species by the mass shift of 14 Da and mass-matched to VAYLYLICSR (residues 333–342) in the known protein sequence. Expanded section of the MALDI-TOF spectra of (A, inset) untreated peptic digest and (B, inset) after β-elimination with concurrent Michael addition. The peptic fragment at m/z 1330.0 was identified as a monophosphorylated species mass-matched to WEQKRRIpSF (residues 94–102) and hence, assigns serine in position 101 as a novel site of phosphorylation. One-tenth of the eluates of both digests, corresponding to ∼1.5 pmoles digest, was consumed for MALDI-TOF analysis.
Chemistry Side Reactions
Alkali-catalyzed peptide bond hydrolysis is of concern, as it could introduce unwanted complexity into the spectra, confounding phosphopeptide identification. This side-reaction has been studied in some detail, and the amide bonds in model peptides, most susceptible at pH 12.6, were identified as those linking glycine or serine N-terminally to glycine.33 As assessed by MALDI-TOF, fibrinogen I and the OVA tryptic peptide at m/z 2008.9, containing these sequence features, remained intact when exposed to the elimination/addition reagent mixture (pH 12.2) for 1 h at 37°C or 55°C (data not illustrated). To examine a wide range of peptides, we prepared a tryptic digest from an equimolar mixture of HSA, bovine carbonic anhydrase, bovine lactoglobulin, and bovine myoglobin. Inspection of the resultant complex tryptic mass maps, generated from the untreated sample and after the incubations, revealed only minor ion intensity changes in the spectral profiles, indicating that the peptides were unaffected by the chemical treatment (data not shown). Mass spectral signals, corresponding to dehydroderivatives of serine or threonine, were not observed in the spectra. This side reaction is promoted by reaction conditions considerably more stringent than used in our protocols.12,13 The data show that the reaction conditions used in our study were highly compatible with the integrity of the samples, providing a significant advantage in sample-limited situations.
Identification of Phosphopeptides in Proteolytic Digests of Experimental Samples
As reported recently, EGF stimulation of A431 human epidermoid carcinoma cells results in disruption of the α-catenin/β-catenin complex, which subsequently promotes β-catenin transactivation.23 In the early stage of this study, we examined whether dissociation of α-catenin from β-catenin involves a possible unknown, post-translational modification on α-catenin. To this purpose, we applied our method to in-gel tryptic digests of α-catenin, immunoprecipitated from EGF-stimulated A431 cells. In the experiments, digests immobilized on ZipTipC18 pipette tips were subjected to β-elimination and β-elimination with concurrent Michael addition at 37°C for 1 h and analyzed by MALDI-TOF (Fig. 6A–C). These reactions resulted in prominent signals at m/z 2141.0 and at m/z 2218.0, representing the β-elimination and the Michael addition products, respectively. The unique mass signatures of 21 Da, accompanying the addition reaction, identified the presence of a single phosphoamino acid in a peptide recognized at m/z 2239.0 in its native form, therefore providing evidence of the nature of the modification. The phosphopeptide peptide mass matched to a tryptic fragment within the known protein sequence, spanning residues 634–651 (TPEELDDSDFETEDFDVR), containing the phosphoryatable serine 641, threonine 634, and threonine 645. As this peptide is effectively derivatized under a reaction condition known to selectively promote phosphoseryl conversion,18 serine 641 was assigned as the most likely kinase target. This assignment was confirmed by MALDI-TOF MS/MS and LC-MS/MS analysis.23 Subsequent biochemical studies revealed casein kinase α-subunit II as the protein kinase involved in phosphorylation of α-catenin at serine 641 and that this phosphorylation event disrupts the α-catenin/β-catenin complex and consequently, promotes β-catenin transactivation.23
We then applied the chemical strategy to preparations of yeast rMaf1, which had been expressed in E. coli under native conditions and phosphorylated in vitro by murine rPKA. Maf1 contains six PKA recognition motifs that render the protein susceptible to modification at serine 90, 101, 177, 178, 209, and 210.24 Of these, serine 90, 177, 209, and 210 have been assigned previously as phosphorylated by LC-MS/MS during large-scale phosphoproteome mapping of S. cerevisiae.34–36 In the experiments, in-gel tryptic or peptic digests of Maf1 bound to ZipTipC18 pipette tips were subjected to β-elimination with concurrent Michael addition and subsequently analyzed by MALDI-TOF, along with untreated digests as a control (Fig. 7). This reaction results in a mass shift of 21 Da/phosphoamino acid and is accompanied by a variable signal enhancement of the derivatized peptides (1.5- to threefold). Accordingly, the tryptic peptides at m/z 1321.7, m/z 1376.7, and m/z 1532.8 in Fig. 7A and the peptic peptide at m/z 1330.0 (Fig. 7A, inset) were identified as a monophosphorylated species. Of these, the peptic peptide at m/z 1330.0 could be mass-matched by a database search of the sequence WEQKRRIpSF (residues 94–102), and hence, the sole serine in position 101 was assigned as a novel site of phosphorylation. This result highlights the potential of the method to locate sites of phosphorylation in known protein sequences, solely on the basis of single-stage MS data. The tryptic peptide at m/z 1376.7 mass-matched to a pair of isobaric fragments spanning residues 88–97 or residues 89–98. MALDI-TOF/TOF analysis of this peptide selected the former sequence, as produced by the enzyme corresponding to RDpSNSFWEQK, and unambiguously assigned serine 90 as the site of phosphorylation (data not shown). Serine 90 has been identified as a modification site in a large-scale phosphoproteome profiling study of S. cerevisiae.35 The presence of the N-terminal arginine in this peptide was expected, because of the neighboring aspartic acid, which is known to inhibit trypsin cleavage.
As illustrated in Fig. 7A, the tryptic peptide at m/z 1359.8, recognized as a doubly phosphorylated species, was detectable solely as the Michael addition product in contrast to its monophosphorylated counterpart at m/z 1321.7. This fragment, mass-matched to RRpSpSSSISSF (residues 175–185), aligns with the corresponding, larger fragment NDERIRRRpSSSSISSFK (residues 169–185), which had been sequenced earlier to localize serine 177 as the site of modification.34 Assignment of serine 178, as a likely novel site of phosphorylation, relies on the presence of PKA recognition motifs overlapping each other and the fact that rPKA was used for in vitro phosphorylation of Maf1.
The monophosphorylated fragments were readily detectable in the initial spectra attributable to the presence of the guanidino side chain(s), which tend to promote the ionization/desorption process (Fig. 7A).32 Consequently, the substitution of the permanently, negatively charged phosphate by the protonable hydrophobic moiety had only a minor impact on the relatively high ionization efficiency of these peptides. As noted above, the doubly phosphorylated peptide was observed only after derivatization, highlighting the benefit of the method to reveal this class of peptides for subsequent structural characterization. Charge neutralization by intrapeptide salt-bridge formation between the oppositely charged proximal residues can be invoked as the most likely cause to account for the ionization inefficiency of the native peptide.37 We note that phosphorylation at adjacent serines, induced by PKA, as well as by other members of the AGC kinase family (e.g., ribosomal protein S6 kinase), has been shown to play a pivotal role in modulation of a variety physiological processes, such as neuronal excitability and cardiac muscle contractility.38,39 A potential, physiological significance of phosphorylation at the proximal serine residues 177 and 178 in Maf1, in context with this protein's inhibitory interaction with RNA polymerase III, warrants future investigation.24
As further illustrated in Fig. 7, the peptide at m/z 1257.7 was identified as a carbamidomethylated species by the characteristic mass shift of 14 Da and as such, is clearly distinguishable from the phosphoamino acid modification. This fragment mass-matched to VAYLYLICSR (residues 333–342) in the known protein sequence. The result was unexpected and in contrast with earlier reports.18,40 Alternative alkylation procedures, such as carboxymethylation and 4-vinyl S-pyridylethylation, proved equally ineffective to protect thiols from β-elimination (Waleed Nasser, personal communication).
Phosphorylation Site Mapping by MALDI TOF-TOF MS/MS Analysis
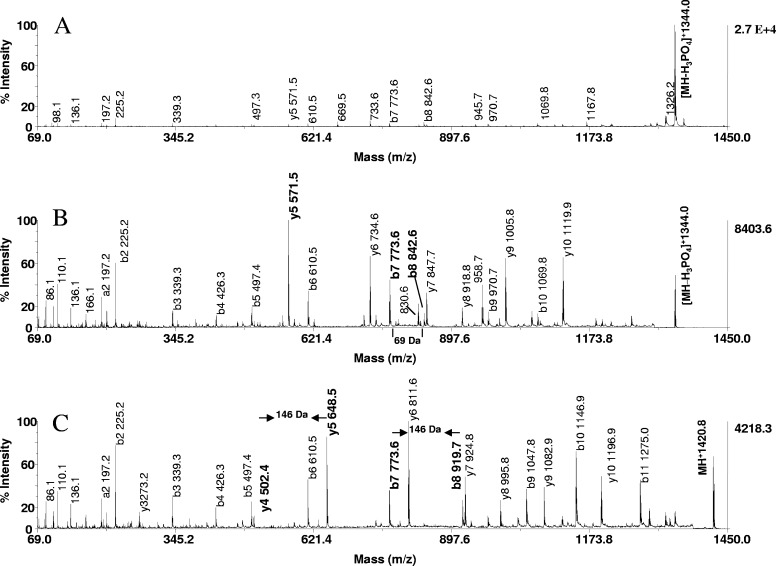
We next explored the use of the on-resin protocol for phosphorylation site mapping. In the experiments, aliquots of control, β-elimination, and additional products of model peptides (20 pmoles) were subjected to MALDI-tandem sequence analysis. Representative data obtained from the tryptic phosphoseryl peptide (SHNSALYpSQVQK) are illustrated in Fig. 8. The MALDI-MS/MS spectrum of the underivatized species shows a predominant ion at m/z 1344.0, formed by a neutral loss of the phosphate as the primary mode of fragmentation (Fig. 8A). The product ion spectrum is complicated by the presence of diverse, low-intensity ions, including those carrying the phosphate group and their dehydro derivatives. In contrast, the dehydroalanyl peptide at m/z 1344.0 and the thiol adduct at m/z 1420.8 fragmented to a noticeably higher extent (Fig. 8B and C). As exemplified by the resultant product ion spectrum illustrated in Fig. 8C, a nearly uninterrupted b and y ion series was produced in high abundance, including y5 and b8, representing the first y and b ions that contain the modification, as well as y4 and b7, which are contiguous to the modification. The location of the modification could be readily identified by its unique residue mass (146 Da) and hence, serine in position 8 as the site of phosphorylation. Similarly, β-elimination with concurrent Michael addition, using 2-aminoethanethiol of a tryptic digest of OVA, provided significantly more sequence information of the derivatized phosphopeptide at m/z 2067.8 than data obtained from its unmodified counterpart. Whereas both spectra shared y15 as a prominent ion, the product ions y16, y17, y18, and y19, which afforded an unambiguous phosphorylation site assignment of serine 345, were only observed after derivatization (data not shown). The data show that our derivatization protocol provides material highly suitable for unambiguous phosphorylation-site determination.
FIGURE 8.
Phosphorylation site mapping by MALDI-tandem MS. MALDI-TOF/TOF spectra of P2 (SHNSALYpSQVQK, m/z 1441) of (A) unmodified peptide, (B) dehydroalanyl derivative, and (C) S-2-aminoethylcysteine derivative. Note that the prominent neutral loss of phosphate from the parent in A as the major fragmentation pathway is precluded in B and C by β-elimination, without or with Michael addition. The derivatization markedly increased peptide backbone fragmentation and discriminates the site of phosphorylation as the unique residue masses of 69 and 146 Da, respectively. These unique mass signatures are contained in the product ions, y5 and b8, indicated in bold in the spectra, as are y4 and b7, which are contiguous to the modifications. The presence of the ion pair b7, b8 and the ion pair y4, y5 in the spectra afforded unambiguous assignment of serine in position 8 as the site of phosphorylation. Peptide (2 pmoles) and its derivatives were applied to the target.
CONCLUSIONS
A method for β-elimination and β-elimination with concurrent Michael addition on C18 reversed-phase supports has been developed using a panel of model phosphopeptides to demonstrate the feasibility of the approach. The protocol described in this study facilitates high-sensitivity MS detection of phosphorylated peptides in protein digests and enables facile assignment of phosphorylated residues from MS/MS sequencing information. The approach combines simplicity of operation with improved throughput and completeness of derivatization. The reaction conditions proved highly compatible with the integrity of the samples. Although MALDI-TOF tandem MS was used in this study for phosphorylation-site determination, ZipTipC18 pipette tips or ZipTipμ-C18 pipette tips can be readily interfaced with nano spray MS techniques, or the eluates can be optionally analyzed by LC-MS/MS. As applied to α-catenin immunoprecipitated from EGF-stimulated A431 cells, the method provided evidence of a phosphorylation event localized at serine 641. As applied to Maf1, the method afforded conclusive identification of peptides as phosphorylated species containing known and novel PKA targets. Two novel phosphorylation sites could be assigned in a sequence-dependent manner after single-stage MS alone. The solid-phase chemical strategy provides a versatile tool of general use for high-sensitivity applications using equipment readily available in most biological laboratories.
ACKNOWLEDGMENTS
This work was funded by NIH grants R33CA101150 to R.H.A. and GM42728 to I.M.W. and NCI grant CA16672 to John Mendelson. The authors thank The University of Texas-MD Anderson Cancer Center for its generous support, Karen Puglia for technical assistance, and Mr. Edward Nieves for valuable discussions and critical review of the manuscript.
Footnotes
The authors declare no conflict of interest associated with financial support.
REFERENCES
- 1. Helmbrecht K, Zeise F, Rensing L. Capherones in cell cycle regulation and mitotic signal transduction: a review. Cell Proliferation 2000; 33: 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Annan RS, Carr SA. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem 1996; 68: 3413–3421 [DOI] [PubMed] [Google Scholar]
- 3. Liao PC, Leykam J, Andrews PC, Gage DA, Allison J. An approach to locate phosphorylation sites in a phosphoprotein: mass mapping by combining specific enzymatic degradation with matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem 1994; 219: 9–20 [DOI] [PubMed] [Google Scholar]
- 4. Annan RS, Huddleston MJ, Verma R, Deshaies RJ, Carr SA. A multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal Chem 2001; 73: 393–404 [DOI] [PubMed] [Google Scholar]
- 5. Tholey A, Reed J, Lehmann W. Electrospray tandem mass spectrometric studies of phosphopeptides and phosphopeptide analogues. J Mass Spectrom 1999; 34: 117–123 [DOI] [PubMed] [Google Scholar]
- 6. Posewitz MC, Tempst P. Immobilized gallium (III) affinity chromatography of phosphopeptides. Anal Chem 1999; 71: 2883–2892 [DOI] [PubMed] [Google Scholar]
- 7. Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen JD. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide micro columns. Mol Cell Proteomics 2005; 4: 873–886 [DOI] [PubMed] [Google Scholar]
- 8. Yang X, Wu H, Kobayashi T, Solaro RJ, van Breemen RB. Enhanced ionization of phosphorylated peptides during MALDI-TOF mass spectrometry. Anal Chem 2004; 76: 1532–1536 [DOI] [PubMed] [Google Scholar]
- 9. Stensballe A, Jensen ON. Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides. Rapid Commun Mass Spectrom 2004; 18: 1721–1730 [DOI] [PubMed] [Google Scholar]
- 10. Beausoleil SA, Jedrychowki M, Schwartz D, et al. , Large-scale characterization of Hela cell nuclear phosphoproteins. Proc Natl Acad Sci USA 2004; 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 2007; 104: 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jaffe H, Veeranna Harish CP. Characterization of serine and threonine phosphorylation in β-elimination/ethanethiol addition-modified proteins by electro spray tandem mass spectrometry and database searching. Biochemistry 1998; 37: 16211–16224 [DOI] [PubMed] [Google Scholar]
- 13. Shen J, Smith RA, Stoll VS, et al. , Characterization of protein kinase A phosphorylation: multi-technique approach to phosphate mapping. Anal Biochem 2004; 324: 204–218 [DOI] [PubMed] [Google Scholar]
- 14. Molloy MP, Andrews PC. Phosphopeptide derivatization signatures to identify serine and threonine phosphorylated peptides by mass spectrometry. Anal Chem 2001; 73: 5387–5394 [DOI] [PubMed] [Google Scholar]
- 15. Ahn YH, Ji ES, Cho K, Yoo JS. Arginine-mimic labeling with guanidinoethanethiol to increase mass sensitivity of lysine-terminated phosphopeptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 2007; 21: 2204–2210 [DOI] [PubMed] [Google Scholar]
- 16. Chen M, Su X, Yang J, Jenkins CM, Cedars AM, Gross RW. Facile identification and quantitation of protein phosphorylation via β-elimination and Michael addition with natural and stable isotope labeled thiocholine. Anal Chem 2010; 82: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson AJ, Hart SR, Franz C, Barnouin K, Ridley A, Cramer R. Characterization of protein phosphorylation by mass spectrometry using immobilized metal ion affinity chromatography with on-resin β-elimination and Michael addition. Anal Chem 2003; 75: 3232–3243 [DOI] [PubMed] [Google Scholar]
- 18. Byford MF. Rapid and selective modification of phosphoserine residues catalysed by Ba2+ ions for their detection during peptide micro sequencing. Biochem J 1991; 280: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nika H, Hawke DH, Kobayashi R. Derivatization on reversed-phase supports for enhanced detection of phosphorylated peptides. 52nd ASMS Conference on Mass Spectrometry and Allied Topics. Nashville, TN, USA, 2004;Abstr. ThPN 292. [Google Scholar]
- 20. Cindric M, Cepo T, Skrlin A, Vuletic M, Bindila L. Accelerated on-column lysine derivatization and cysteine methylation by imidazole reaction in a deuterated environment for enhanced product ion analysis. Rapid Commun Mass Spectrom 2006; 20: 694–702 [DOI] [PubMed] [Google Scholar]
- 21. Conrotto P, Hellman U. Sulfonation chemistry as a powerful tool for MALDI TOF/TOF de novo sequencing and posttranslational modification analysis. J Biomol Tech 2005; 16: 441–452 [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson ME, Carpenter TS. The use of solid-phase supports for derivatization in chromatography and spectroscopy. Appl Spectrosc Rev 2005; 40: 391–412 [Google Scholar]
- 23. Ji H, Wang J, Nika H, et al. , EGF-induced ERK activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol Cell 2009; 36: 547–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moir RD, Lee JH, Haeusler RA, Desai N, Engelke DR, Willis IM. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA 2006; 103: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Speicher DT, Kolbas O, Harper D, Speicher DW. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J Biomol Tech 2000; 11: 74–86 [PMC free article] [PubMed] [Google Scholar]
- 26. Katayama H, Nagasu T, Oda Y. Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 2001; 15: 1416–1421 [DOI] [PubMed] [Google Scholar]
- 27. Ryo A, Liou Y-C, Lu KP, Wulf G. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J Cell Sci 2003; 116: 773–783 [DOI] [PubMed] [Google Scholar]
- 28. Ubersax JA, Ferrrell JE. Mechanism of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 2007; 8: 530–541 [DOI] [PubMed] [Google Scholar]
- 29. Rusnak F, Zhou J, Hathaway GM. Identification of phosphorylated and glycosylated sites in peptides by chemically targeted proteolysis. J Biomol Tech 2002; 13: 228–237 [PMC free article] [PubMed] [Google Scholar]
- 30. Kirkland JJ, van Straten MA, Claessens HA. High pH mobile phase effects on silica-based reversed-phase high-performance liquid chromatography. J Chromatog A 1995; 691: 3–19 [DOI] [PubMed] [Google Scholar]
- 31. Palmblad M, Vogel JS. Quantitation of binding, recovery and desalting efficiency of peptides and proteins in solid phase extraction micropipette tips. J Chromatogr B 2005; 814: 309–313 [DOI] [PubMed] [Google Scholar]
- 32. Baumgart S, Lindner Y, Kuhne R, Oberemm A, Wenschu H, Krause E. The contributions of specific amino acid side chains to signal intensities of peptides in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom 2004; 18: 863–868 [DOI] [PubMed] [Google Scholar]
- 33. Noll BW, Jarboe CJ, Hass LF. Kinetic studies on the alkali-catalyzed hydrolysis and epimerization of model alkyl and hydroxyalkyl di-and tripeptides. Biochemistry 1974; 13: 5164–5169 [DOI] [PubMed] [Google Scholar]
- 34. Chi A, Huttenhower C, Geer LY, et al. , Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA 2007; 104: 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X, Gerber SA, Rudner AD, et al. , Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J Proteome Res 2007; 6: 1190–1197 [DOI] [PubMed] [Google Scholar]
- 36. Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteome 2008; 7: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hider RC, Ragnarsson U, Zetterquist O. The role of phosphate group for the structure of phosphopeptide products of adenosine 3′, 5′-cyclic mono phosphate-dependent protein kinase. Biochem J 1985; 229: 485–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDonald BJ, Amato A, Connolly CN, et al. , Adjacent phosphorylation sites on GABAA receptor β subunits determine regulation by cAMP-dependent protein kinase. Nat Neurosci 1998; 1: 23–28 [DOI] [PubMed] [Google Scholar]
- 39. Zhang R, Zhao J, Potter JD. Phosphorylation of both serine residues in cardiac troponin I is required to decrease the Ca 2+ affinity of cardiac troponin C. J Biol Chem 1995; 270: 30773–30780 [DOI] [PubMed] [Google Scholar]
- 40. Simpson DL, Hranisavljevic J, Davidson EA. β-Elimination and sulfite addition as a means of localization and identification of substituted seryl and threonyl residues in proteins and proteoglycans. Biochemistry 1972; 11: 1849–1856 [DOI] [PubMed] [Google Scholar]