Abstract
We have used 19F NMR to analyze the metal ion-induced folding of the hammerhead ribozyme by selective incorporation of 5fluorouridine. We have studied the chemical shift and linewidths of 19F resonances of 5-fluorouridine at the 4 and 7 positions in the ribozyme core as a function of added Mg2+. The data fit well to a simple two-state model whereby the formation of domain 1 is induced by the noncooperative binding of Mg2+ with an association constant in the range of 100 to 500 M−1, depending on the concentration of monovalent ions present. The results are in excellent agreement with data reporting on changes in the global shape of the ribozyme. However, the NMR experiments exploit reporters located in the center of the RNA sections undergoing the folding transitions, thereby allowing the assignment of specific nucleotides to the separate stages. The results define the folding pathway at high resolution and provide a time scale for the first transition in the millisecond range.
Keywords: RNA structure, RNA catalysis, metal ions
RNA-mediated catalysis is important in the processing of cellular RNA molecules in some systems and in the synthesis of proteins. The origins of catalytic activity are not yet fully understood, but the observed rate enhancements probably are generated by a number of factors, including metal ion and nucleobase catalysis and local stereochemical effects. To create an environment in which catalysis can occur, the RNA molecules must be folded into a precise three-dimensional structure. In general, metal ions play an important role in RNA folding, because electrostatic interactions are very significant due to the polyelectrolyte character of nucleic acids. In particular, the binding of divalent ions usually is required to facilitate the folding of RNA species into their active conformations.
Folding has been extensively studied in the group I intron ribozyme (1–7). It is found that folding of this relatively large RNA is complex, with kinetic traps to complicate the analysis. It is therefore advantageous to examine a simpler system where the elementary steps are more accessible. The hammerhead ribozyme (8–10) provides such a system, with a number of specific virtues. It is one of the nucleolytic ribozymes that catalyses site-specific RNA cleavage, requiring only Mg2+ or similar cations for activity (11). It is relatively small, comprising just three helical sections organized around a central core as a three-way [HS1HS7HS3 (12)] junction. The fully folded structure has been determined crystallographically (13–16), and thus the folding steps can be related to this known structure. In addition, the folding process can be correlated with ribozyme activity.
We previously have studied the folding of the hammerhead ribozyme by using two methods that examine aspects of the global shape, i.e., comparative gel electrophoresis (17) and fluorescence resonance energy transfer (FRET) (18). In the absence of divalent metal ions, the hammerhead structure is extended, with a disordered core, but upon addition of metal ions folding occurs in two distinct steps. In the presence of 500 μM Mg2+ the RNA adopts a structure in which there is a large angle between helices II and III. As the Mg2+ concentration is increased over the range of 1 to 20 mM Mg2+ there is a further folding in which helix I changes position from being oriented in a similar direction to helix III to being close to helix II. Both events are well described by two-state transitions induced by the noncooperative binding of Mg2+, with Hill coefficients close to unity.
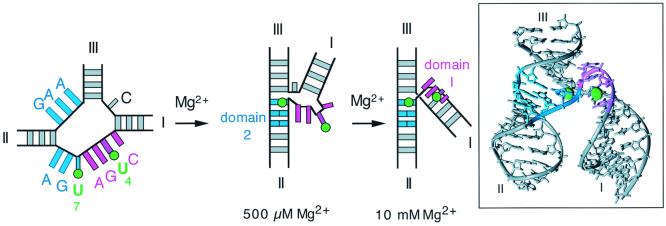
The structure of the folded ribozyme observed in the crystal is organized around two prominent features formed from the central core (13–16). The sequence C3U4G5A6 adopts the geometry of a turn found in the anticodon stem of tRNA and is termed domain 1. It is close to the scissile bond at C17 and is the probable catalytic center. Domain 2 is formed by pairing between G12A13A14 and U7G8A9 and mediates coaxial alignment between helices II and III in the structure. We previously have interpreted our results in terms of the initial formation of domain 2 at low (<500 μM) Mg2+ concentration, followed by the formation of domain 1 in the 1- to 10-mM range (Fig. 1). The range of Mg2+ concentration that induces the second transition corresponds well to that required for catalytic activity (11).
Figure 1.
The structure and folding of the hammerhead ribozyme. Schematic illustration of a two-stage folding scheme for the hammerhead ribozyme, with the crystallographic structure (Right). The same color scheme is used in both pictures. The final structure in the folding model is consistent with the three-dimensional structure of the ribozyme observed in the crystal (13, 14) and in solution (17, 37, 38). The crystallographic structure is therefore a valuable guide in the interpretation of this scheme at the atomic level. According to this model, the structure of domain 2 is formed in the first transition, from the nucleotides colored blue. Domain 1 forms in a second transition, occurring as the Mg2+ concentration rises above 1 mM, and involves the CUGA sequence colored magenta. The fluorine atoms introduced at the 5 positions of uridine 4 and 7 are shown as green spheres. Note that the fluorine atom of FU4 is centrally located in domain 1, whereas that of FU7 lies at the edge of domain 2 adjacent to domain 1. The molecular structure of the hammerhead ribozyme was drawn by using crystallographic coordinates deposited by McKay and coworkers (13), Protein DataBank ID number 1HMH.
The dissection of the folding process has been guided largely by information on the global shape of the ribozyme, i.e., by the angles subtended between the helical arms at different Mg2+ concentrations. The assignment of the two transitions into the folding of specific domains was essentially conjectural, based on consideration of the folded structure in the crystal, although supported by the effects of sequence variations (19, 20). To test and extend our ideas on the sequential formation of domains 2 and 1 we required further information coming directly from the regions proposed to be involved. NMR can provide precise structural information in solution (21, 22), but 1H NMR studies of the hammerhead ribozyme have not provided a full solution structure. To simplify the problem we turned to an alternative nucleus, that of fluorine. Like the proton, the 19F nucleus has a spin of 1/2, is 100% abundant and relatively sensitive, and has been used to some extent in nucleic acids (23–26). We can incorporate the NMR-active nucleus into RNA by site-specific substitution of uridine by 5-fluorouridine (FU). The fluorine atom is quite small (covalent radius = 0.72 Å) and thus its introduction should be relatively nonperturbing. We would expect that the selectively introduced fluorine nucleus would respond to structural changes in the surrounding RNA, primarily by changes in chemical shift. We have made two substituted hammerhead species with 5-FU incorporated into the core in place of U4 or U7. The positions of the fluorine atoms superimposed onto the crystal structure of the ribozyme is shown in Fig. 1. U4 is part of the CUGA sequence that forms domain 1, and the fluorine is located at the center of the uridine turn. We therefore would anticipate that this should respond in some manner to the formation of this domain; according to our folding model this would occur in the millimolar range of Mg2+ concentration. By contrast, U7 lies within domain 2; it therefore would be expected to respond to the formation of this structure, which occurs at submillimolar Mg2+ concentrations according to our previous experiments. However, inspection of Fig. 1 shows that the U7 fluorine atom is located very much at the interface between domains 1 and 2, and its magnetic environment could well be affected by structural transitions in both sections of RNA. The 19F NMR approach places conformationally sensitive probes directly into the participating elements in the RNA structure, giving a view of the folding process from the inside, in contrast to earlier methods that provide an essentially external view of the global structure.
Materials and Methods
Chemical Synthesis of RNA-Containing Oligonucleotides and Standards.
Oligonucleotides containing both DNA and RNA sections were synthesized by using phosphoramidite chemistry implemented on Applied Biosystems 394 DNA/RNA synthesizers. RNA sections were synthesized by using ribonucleotide phosphoramidites with 2′-t-butyldimethylsilyl (t-BDMS) protection (PrOligo, Hamburg, Germany). Five 200 nmol-scale syntheses of each hammerhead species were used for the NMR experiments. 5′ dimethoxytrityl-protected oligonucleotides were dissolved in 0.5 ml 1 M tetrabutylammonium fluoride (Aldrich) in tetrahydrofuran to remove t-BDMS groups and agitated at 20°C in the dark for 16 h. before desalting by G25 Sephadex (Amersham Pharmacia) and ethanol precipitation. They then were applied to NENSORB PREP columns (DuPont) to remove failure sequences lacking 5′ dimethoxytrityl groups. Fully deprotected oligonucleotides were purified by electrophoresis in 15% polyacrylamide containing 7 M urea in 90 mM Tris⋅borate (pH 8.3), 10 mM EDTA. The oligonucleotides were electroeluted into 8 M ammonium acetate and recovered by ethanol precipitation.
Design of Hammerhead Ribozyme Species and Control Molecules for NMR Studies.
The following sequences were synthesized (written 5′ to 3′, with deoxyribose nucleotides underlined and 5-FU indicated by F): HhRz-FU4, CCCGGAGCGUCCGACGAAAGUCGCFGAUGAGGCCGAAAGGCCGAAACGCUCCGGG; + ribo C17, CCCGGAGCGUCCGACGAAAGUCGCFGAUGAGGCCGAAAGGCCGAAACGCUCCGGG; HhRz-FU7, CCCGGAGCGUCCGACGAAAGUCGCUGAFGAGGCCGAAAGGCCGAAACGCUCCGGG; + ribo C17, CCCGGAGCGUCCGACGAAAGUCGCUGAFGAGGCCGAAAGGCCGAAACGCUCCGGG; Hairpin, CCGGFACCGUAAGGUACCGG.
5-FU was coupled as a 2′-O-methyl-5-FU cyanoethyl phosphoramidite (Glen Research, Sterling, VA). For NMR experiments, HhRz-FU4 and HhRz-FU7 were synthesized with deoxyribocytidine at position 17. Corresponding sequences with ribocytidine at position 17 were used to study ribozyme cleavage activity. The hairpin species contains a single 5-FU nucleotide in the center of the potential duplex stem.
Analysis of Self-Cleavage by the Modified Ribozyme Species.
Competence in ribozyme cleavage was examined by using a combined phosphorylation and cleavage assay. Purified HhRz-FU4 and HhRz-FU7 were incubated for 45 min in a total volume of 10 μl containing 10 units of T4 polynucleotide kinase (Biolabs, Northbrook, IL) and radioactively γ-32P-labeled ATP in 70 mM Tris⋅HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT at 37°C. This allowed both hammerhead ribozyme self-cleavage and end-labeling of the substrate and both products to proceed in one experiment. Uncleaved and cleaved molecules were separated by electrophoresis in 15% polyacrylamide gels in 90 mM Tris·boric acid/10 mM EDTA, pH 8.3 buffer containing 7 M urea and visualized by phosphorimaging (BAS-1500, Fuji).
NMR Sample Preparation.
Oligoribonucleotides for NMR experiments were heated to 94°C in the presence of 25 or 10 mM Tris⋅HCl (pH 7.5), 50 or 10 mM NaCl, followed by slow cooling to 4°C. D2O then was added to 10% (vol/vol). RNA concentrations were 30–40 μM for the hammerhead derivatives and 5–10 μM for stem-loop control species, in a total volume of 600 μl. At the end of a series of MgCl2 (99.999%, puratronic, Alfa Aesar, Karlsruhe, Germany) titrations, an aliquot of the oligoribonucleotide was analyzed on 15% denaturing polyacrylamide gels for nonspecific degradation during the NMR experiments. Less than 2% of the RNA had been degraded in each case, and the oligoribonucleotides were used in subsequent Mg2+ titrations without further gel purification. Removal of MgCl2 between the series of titrations was achieved by repeated ethanol precipitations, including two in the presence of EDTA.
19F NMR Spectroscopy.
NMR data were acquired at 300 K on a Bruker AM500 NMR spectrometer operating at 470.562 MHz, equipped with a 5-mm 19F/1H probe. Data were accumulated with a relaxation delay of 1 s over a spectral width of 5,618 Hz by using 16-k points. A total of 5,000 or ≥15,000 transients were accumulated and averaged for each experiment. Before Fourier transformation the data were apodised by exponential multiplication, using a line broadening of 20 Hz. Chemical shifts were referenced to external 5-FU (Sigma). Data processing and analysis was carried out by using felix (Molecular Simulations). Chemical shift values and linewidths were measured by using the real time peak fitting of a Lorentzian peak shape by simulated annealing.
Results
Construction and Activity of Hammerhead Ribozyme Species.
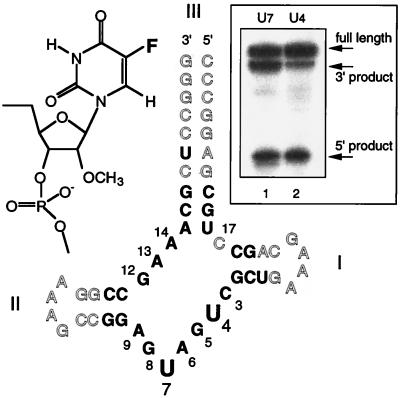
We used a cloverleaf-form hammerhead ribozyme for NMR experiments, formed from a single chemically synthesized strand (Fig. 2). The core was made of RNA, except for 2′ deoxyribose substitution at C17 to prevent self-cleavage. The molecules synthesized for the cleavage assay, however, contained a ribocytidine at position C17. The helical arms contained RNA for the core-proximal 2 or 3 bp and DNA elsewhere to maximize the efficiency of synthesis; these positions do not affect ribozyme folding. Helix III was extended to 10 bp to ensure a stable secondary structure. 5-FU was incorporated at either the 4 or 7 position in the core of the ribozyme (called HhRz-FU4 and HhRz-FU7, respectively) to provide the NMR-active nuclei in this study; these were introduced as 2′-O-methylribose-substituted nucleotides.
Figure 2.
The cloverleaf hammerhead construct used in these experiments and its ribozyme activity. The cloverleaf hammerhead species is made from a single strand, forming helix III of 10 bp, and helices I and II, each of which comprise 4 bp closed by a tetraloop. DNA residues are shown open, and RNA residues are shown in bold letters. For ease of synthesis, ribouridine replaced thymine in the otherwise-DNA sections, leading to two rU-dA base pairs. The numbering system follows Hertel et al. (39). Positions U4 and U7 were substituted by 2′-O-methyl-5-FU, the structure of which is shown (Left Inset). (Right Inset) The result of cleavage reactions of ribo-C17-containing FU-substituted hammerhead ribozymes, carried out at 37°C in the presence of 10 mM Mg2+.
The 5-FU-substituted hammerhead ribozyme derivatives were modified in both the base (5-fluoro) and the sugar (2′-O-methyl) rings. It has been shown that there are relatively few positions in the hammerhead core where the presence of ribose is essential, and these do not include U4 or U7 (27–30) although ribose modification at these positions would be expected to result in some lowering of activity. The effect of 5-fluorouracil modification has not been studied previously. We therefore examined the effect of these modifications on the cleavage activity of the cloverleaf ribozyme shown in Fig. 2. We used a simple assay in which radioactive labeling and cleavage could proceed together. Purified C17-ribose forms of HhRz-FU4 and HhRz-FU7 were incubated in buffer containing 10 mM MgCl2, in the presence of T4 polynucleotide kinase and radioactively γ-32P-labeled ATP. By this approach, both ribozyme cleavage and radioactive 5′ labeling of the substrate and the two cleavage products was achieved in a single experiment. The results are shown in Fig. 2. The two expected products of ribozyme cleavage are seen in both hammerhead species, showing that the substituted ribozymes retain site-specific cleavage activity. We have not attempted to quantify the rate of cleavage in these species, although some impairment of cleavage rate is apparent in the modified ribozymes.
5-FU Substituted at U4 Is Sensitive to a Change in Environment Occurring in the Millimolar Range of Mg2+ Ion Concentration.
U4 is part of the CUGA sequence that forms domain 1 in the folded ribozyme observed by crystallography (13, 14). We therefore replaced this nucleotide with 5-FU (HhRz-FU4; Fig. 2) to see whether the magnetic resonance of the 19F nucleus at this position would be sensitive to ion-induced folding. We anticipated that changes in chemical shift and/or linewidth might accompany the formation of domain 1. 19F NMR spectra were acquired at 470.6 MHZ as a function of Mg2+ concentration in 16 steps between 0 and 50 mM MgCl2. Spectra were recorded at 300 K in a buffered solution containing either 10 or 50 mM NaCl.
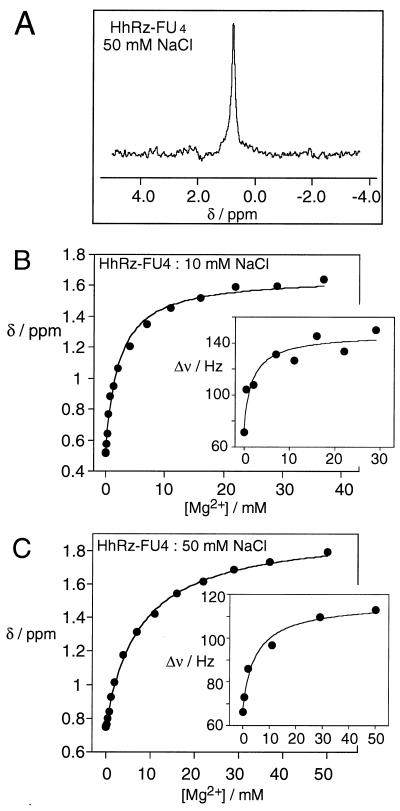
Fig. 3A shows a 19F NMR spectrum of HhRz-FU4 in 50 mM NaCl, externally referenced to 5-FU. In the absence of added Mg2+, the 19F nucleus of HhRz-FU4 resonates as a single peak at 0.7 ppm with a linewidth of 66 Hz. Very little change in chemical shift or linewidth was observed at Mg2+ concentrations below 0.5 mM. However, at higher Mg2+ concentrations the resonance exhibited a marked downfield shift of 1.2 ppm, eventually reaching 1.9 ppm. In the same single transition, the resonance linewidth broadened to 113 Hz. For comparison, we recorded 19F NMR spectra of an 8-bp stem-loop RNA species, containing 5-FU in the center of the duplex region. The fluorine nucleus resonated at −2.0 ppm, with a linewidth of 45 Hz, which shifted 0.4 ppm downfield over the same Mg2+ concentration range with a constant linewidth (data not shown). The changes in chemical shift and linewidth observed in the U4-substituted hammerhead species were clearly different from those of a simple duplex. The chemical shift changes suggested that the 19F nucleus experiences an altered magnetic environment due to a change in the local conformation induced by the binding of one or more Mg2+ to the hammerhead RNA.
Figure 3.
19F NMR experiments on HhRz-FU4. (A) A 19F NMR spectrum of HhRz-FU4 in 25 mM Tris⋅HCl (pH 7.5), 50 mM NaCl. A total of 26,800 transients were collected from a 40 μM solution of the RNA. All spectra were recorded at 27°C and referenced externally to 5-FU in 25 mM Tris⋅HCl (pH 7.5). (B and C) Plots of the chemical shift (ppm) of the FU4 19F resonance as a function of added Mg2+ concentration in 10 mM Tris⋅HCl (pH 7.5) with either 10 (B) or 50 mM (C) NaCl. The lines are the fits according to the two-state transition model. (Insets) The linewidth (Hz) of the 19F resonances (selecting spectra recorded with ≥15,000 transients) as a function of Mg2+ concentration. The lines are the fits according to the two-state model.
The Chemical Shift and Linewidth of the U4 Fluorine Nucleus Are Well Described by a Simple Two-State Transition.
Chemical shifts and linewidths were determined by a peak optimization procedure as a function of added Mg2+ concentration in the presence of 10 and 50 mM NaCl. They are plotted in Fig. 3 B and C, respectively. We previously had found that ion-dependent structural transitions in the hammerhead ribozyme observed by using FRET can be well fitted by using a simple two-state folding model,
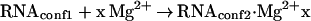 |
and we therefore applied such a model to the 19F NMR data. Thus the observed change in chemical shift as a function of Mg2+ concentration (δobs) is given by:
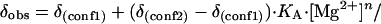 |
1 |
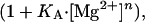 |
where KA is the apparent association constant for Mg2+ and n is a Hill coefficient.
Good fits were obtained (r > 0.997) for both series of titrations (shown by the lines in Fig. 3 B and C). The shape of these curves are clearly indicative of noncooperative binding, which is confirmed by the calculated Hill coefficients of unity within experimental error, i.e., n = 1.0 ± 0.2 and 1.0 ± 0.1 in 10 and 50 mM NaCl, respectively. The apparent association constants depended on the NaCl concentration, with values of KA = 460 M−1 and 120 M−1, corresponding to Mg2+ concentrations for half-completion of the transition [[Mg2+]1/2 = (1/KA)1/n]of 2.2 and 7.7 mM in 10 and 50 mM NaCl, respectively. Thus an increased background of monovalent metal ion concentration weakens the affinity of the RNA for the divalent metal ion.
We also attempted to fit the linewidth to a two-state model. To economize on instrument time, some spectra had been recorded with a reduced number of transients. Because the accuracy of measured linewidth depends on the signal-to-noise ratio, we used only high-quality spectra with a comparable number of transients (>15,000) for deriving linewidth data, for which the fits are shown in the insets of Fig. 3 B and C. The data exhibited more scatter than those for the chemical shifts, which was expected. Nevertheless, it was clear that as with the chemical shifts, the shapes of the isotherms indicate noncooperative binding of Mg2+ (e.g., n = 0.9 ± 0.4 in 50 mM NaCl). The apparent affinity for Mg2+ calculated from the linewidth data was closely similar to those calculated by fitting the chemical shift data, with [Mg2+]1/2 = 1.9 and 5.1 mM in 10 and 50 mM NaCl, respectively. The observation that the peaks broaden and do not sharpen again indicates that the increased linewidth is caused by altered tumbling behavior of the molecule after folding rather than by an exchange process between two or more conformations, which ultimately would be expected to result in a sharpened peak.
Taken together, the variation of chemical shift and linewidth indicate that the fluorine located on the FU4 base responds to a structural transition induced by the binding of Mg2+ in the millimolar range. This is very similar to the second transition observed by FRET measurements of the global structure of the ribozyme (18).
5-FU Substituted at U7 Responds to Two Mg2+ Ion-Induced Folding Events.
In a second series of experiments we introduced 5-FU at the 7 position of the hammerhead ribozyme (HhRz-FU7; Fig. 2). In the crystal (13, 14) this nucleotide forms part of domain 2, but the fluorine atom lies at the interface between domains 1 and 2. 19F NMR spectra of HhRz-FU7 were acquired as a function of Mg2+ concentration in the range 0 to 50 mM MgCl2 in 17 steps. Spectra were obtained at 300 K in a buffered solution with a constant NaCl background concentration of either 10 or 50 mM.
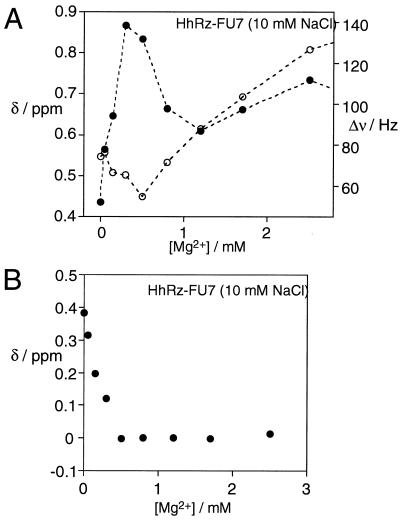
In contrast to the U4-substituted species, the fluorine nucleus of HhRz-FU7 exhibited two different transitions in response to addition of Mg2+ in different concentration ranges. In the range of 1 to 50 mM Mg2+, the 19F resonance of FU7 shifted a total of 1.0 ppm downfield to 1.65 ppm and broadened to a line width of 115 Hz. However, in contrast to FU4, the FU7 resonance also exhibited changes in the 0 to 1-mM range of added Mg2+ concentration. This transition was characterized by a small up-field shift to 0.60 ppm and an increase in linewidth to 103 Hz.
The Low Mg2+ Transition of HhRz-FU7.
Fig. 4A shows the chemical shift and linewidth variation of HhRz-FU7 in the low-millimolar Mg2+ concentration range in the presence of 10 mM NaCl. The 19F resonance of FU7 initially underwent a small, but reproducible, up-field shift together with a significant line broadening and subsequent narrowing below 1 mM Mg2+. Titrations in the presence of a background concentration of 50 mM NaCl showed the same behavior (data not shown). The changes in chemical shift were small, but they occurred systematically over the 0 to 500-μM range of added Mg2+ concentration. This finding, coupled with the large increase in linewidth in exactly the same range, strongly indicates the existence of an initial folding event in the submillimolar Mg2+ concentration range to which the fluorine of FU7 is sensitive. A folding event in this low range of Mg2+ concentration also was deduced from changes in the global structure observed by comparative gel electrophoresis (17) and FRET (18). We have not attempted to fit these NMR data to a binding model. However, the dependence of chemical shift changes on Mg2+ concentration indicates that an apparent association constant in excess of 2,000 M−1 would be required to fit these data. The narrowing of the 19F resonance in the 500 μM to 1 mM Mg2+ concentration range demonstrates that the line broadening is probably due to exchange processes. We interpret the linewidth changes as indicative of dynamic interconversion on an intermediate time scale relative to the NMR process, discussed below.
Figure 4.
19F NMR experiments on HhRz-FU7 at low Mg2+ concentrations. (A) Plots of the chemical shift (ppm, ○) and linewidth (Hz, ●) of the FU7 19F resonance as a function of added Mg2+ concentration (in the range 0 to 2.5 mM) in 10 mM Tris⋅HCl (pH 7.5), 10 mM NaCl. (B) Plot of the chemical shift of the FU7 19F resonance as a function of Mg2+ concentration in the presence of 10 mM NaCl, after subtraction of the transition occurring at higher Mg2+. This was achieved by using the equation of the line used to fit the data shown in Fig. 5, extrapolated into the low Mg2+ concentration region. A corresponding subtraction was made for the data obtained in the presence of 50 mM NaCl (not shown).
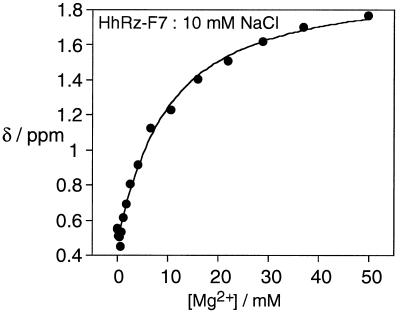
The High Mg2+ Transition of HhRz-FU7.
After the small, but systematic, up-field shift of the 19F resonance of FU7 in the submillimolar range of Mg2+ ions, further increases in concentration brought about a shift in resonant frequency in the opposite direction, with an order of magnitude greater amplitude. Starting at about 0.6 ppm in 1 mM Mg2+ ions, the peak shifted a total of 1.0–1.2 ppm downfield by 50 mM. In this region of Mg2+ concentration, the resonance of FU7 behaved in a similar manner to that of FU4. Chemical shifts measured in 1–50 mM Mg2+ plus 10 or 50 mM NaCl were well fitted to the two-state Mg2+ binding model (Fig. 5). The overall shapes of the apparent binding isotherms are indicative of noncooperative binding, and values of the Hill coefficient close to unity have been obtained from the fits, i.e., n = 1.1 ± 0.2 and 1.0 ± 0.1 in 10 and 50 mM NaCl, respectively. The association constants measured from the data corresponded to half-Mg2+ concentrations of 7.0 and 10.0 mM, respectively. In general, the dependence of the chemical shift of the FU7 fluorine nucleus in the high Mg2+ concentration range was very similar to that of the corresponding FU4 species, suggesting that they were responding to the same structural change under these conditions.
Figure 5.
19F NMR experiments on HhRz-FU7 at higher Mg2+ concentrations. Plot of the chemical shift (ppm) of the FU7 19F resonance as a function of added Mg2+ concentration (in the range 0 to 50 mM) in 10 mM Tris⋅HCl (pH 7.5), 10 mM NaCl. The line is the fit to the two-state model by using the data obtained at Mg2+ concentrations >1 mM.
In the same range of Mg2+ concentration, the linewidth of the 19F resonance of FU7 broadened (data not shown). In contrast to the effects observed in the submillimolar concentration range, there was no subsequent narrowing of the linewidth, which we therefore interpret in terms of reduced molecular tumbling due to the altered conformation.
The equation of the line used to fit the Mg2+ dependence of the FU7 chemical shift in the presence of 10 mM NaCl was used to subtract the effects due to the second transition from the up-field shifts observed in 0–500 μM Mg2+. This left the chemical shifts caused by the first transition alone, shown in Fig. 4B. This deconvolution makes the simplifying assumption that the two transitions occur independently.
Discussion
19F NMR signals from 5-FU bases strategically placed in the core of the hammerhead ribozyme provide insight into the Mg2+ ion-induced folding of this species. We find that the chemical shift of the 19F nuclei are sensitive to structural changes resulting from the binding of divalent metal ions, enabling us to dissect the folding process into its component steps. The 19F resonances from 5-FU placed at the 7 and 4 positions in the ribozyme change quite differently as a function of Mg2+ concentration, showing that these nucleotides participate in different stages of the folding process.
The Mg2+ dependence of the 19F NMR signals arising from FU7 and FU4 bases is in excellent agreement with the two-stage folding scheme outlined in Fig. 1. According to this model, the first stage involves the formation of domain 2, occurring in the 0 to 500-μM range of Mg2+ concentration. This stage of the folding process changes the magnetic environment (up-field shift) of FU7, while leaving that at FU4 essentially unaffected. The range of Mg2+ inducing the spectral change at FU7 is closely similar to that which causes the initial shortening of the end-to-end distance for helices I and III as observed by FRET (18). It is therefore probable that the NMR parameters and the analysis of the global shape are reporting on the same internal structural change. An ion-induced structural transition also has been observed by a change in the fluorescent quantum yield of 2-aminopurine substituted at position 7 (31). According to the folding model, domain 1 is unfolded at the low Mg2+ concentration stage, and thus the resonance of FU4 would not be expected to be significantly perturbed by the formation of domain 2, which is exactly what we observe. This first stage of the folding creates the scaffold on which the ribozyme is built, but the catalytic core remains unfolded, consistent with the lack of cleavage activity at this low Mg2+ concentration (11). As the Mg2+ concentration is raised into the millimolar region we observe that the 19F resonances of both FU7 and FU4 bases undergo pronounced downfield shifts, together with an increase in linewidth. The pronounced shift changes at FU4 show that this base is strongly affected by the second transition, consistent with the formation of domain 1 of the ribozyme at millimolar Mg2+ concentrations. These changes can be fitted to a simple two-state transition induced by ion binding with a Hill coefficient of close to unity and a half-Mg2+ concentration in the range 2 to 10 mM. This value depends on the background concentration of monovalent cations; clearly there are general polyelectrolyte effects operating in addition to more specific binding events, as we would expect. In principle we might expect to observe specific site binding of paramagnetic metal ions by 19F NMR, and we have observed relaxation broadening of the FU4 and FU7 resonances in the presence of Mn2+ (data not shown). Previous paramagnetic broadening studies by 31P NMR have identified specific ion binding sites in domain 2 (32), and a high affinity ion-binding site also has been observed by electron paramagnetic resonance (33). We also have observed changes in the solvent accessibility of the fluorine atoms of FU4 and FU7 during the folding process by means of altered solvent isotope shifts (data not shown), but we have not pursued this further.
The increase in 19F linewidths in the second transition can be fitted to the same two-state folding model and is consistent with line broadening due to changes in spin-spin relaxation arising from an altered correlation time because of a change in global shape. In the first transition, however, the resonance of FU7 is seen to broaden significantly, then revert to the initial linewidth before increasing again in the second transition. It is therefore most probable that the broadening that accompanies the first transition is due to exchange processes. At the transition midpoint, the intermediate exchange rate that would generate a single broadened resonance should conform to the condition:
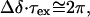 |
2 |
where Δδ is the change in chemical shift (in Hz) and τex is the lifetime. Using the calculated chemical shifts for the first transition (deconvoluted from the second transition as shown in Fig. 4B) gives values of τex = 0.8 and 1.7 ms in the presence of 10 and 50 mM sodium ions, respectively. These correspond to kinetic rates in the region of k = 1,000 s−1 for this transition.
The two-stage pathway for the folding of the hammerhead ribozyme is consistent with all of the available data and makes sense in terms of the crystallographic structure of the ribozyme (13–16). All of the evidence indicates two sequential transitions in which domains 2 and 1 are formed, each induced by the noncooperative binding of divalent metal ions. Sequence changes in the nucleotides forming domain 2 (such as G8U and A14G) interfere with the first transition (19, 20). By contrast, variants in the CUGA sequence (such as G5C and 2′-deoxyribo-G5) fail to affect the first transition, but prevent the high Mg2+ transition in which domain 1 becomes folded. An ion-binding site at P5 detected by uranyl-induced photocleavage is not formed when the second transition is blocked (20). The two-stage folding mechanism is also in good agreement with calorimetric analysis, whereby the folding of natural and variant hammerhead species was studied by isothermal titration with Mg2+ (34). It has been recently suggested that further structural rearrangement is required before an active conformation is achieved (35, 36). However, these would be related to the transition state structure, and because our data refer to the ground state of the folded ribozyme we cannot comment further on this possibility.
The local information from the 19F NMR allows us to assign the participating nucleotides to the two folding transitions unambiguously. It also puts a time scale on the processes involved in the formation of domain 2. These studies show that 19F NMR can be used as a relatively nonperturbing probe to generate valuable information on RNA folding.
Acknowledgments
We thank Anke Scholz for preliminary experiments, Dr. Z. Zhao for the chemical synthesis of RNA, Dr. T. Wilson for discussion, and the Cancer Research Campaign and Biotechnology and Biological Sciences Research Council for financial support.
Abbreviations
- FRET
fluorescence resonance energy transfer
- FU
5-fluorouridine
References
- 1.Rook M S, Treiber D K, Williamson J R. J Mol Biol. 1998;21:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 2.Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 3.Treiber D K, Rook M S, Zarrinkar P P, Williamson J R. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 4.Silverman S K, Cech T R. Biochemistry. 1999;38:14224–14237. doi: 10.1021/bi991333f. [DOI] [PubMed] [Google Scholar]
- 5.Deras M L, Brenowitz M, Ralston C Y, Chance M R, Woodson S A. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 6.Pan J, Deras M L, Woodson S A. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang X, Bartley L E, Babcock H P, Russel R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 8.Forster A C, Symons R H. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 9.Hazeloff J P, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 10.Epstein L M, Gall J G. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 11.Dahm S C, Uhlenbeck O C. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 12.Lilley D M J, Clegg R M, Diekmann S, Seeman N C, von Kitzing E, Hagerman P. Eur J Biochem. 1995;230:1–2. doi: 10.1111/j.1432-1033.1995.tb20526.x. [DOI] [PubMed] [Google Scholar]
- 13.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 14.Scott W G, Finch J T, Klug A. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 15.Scott W G, Murray J B, Arnold J R P, Stoddard B L, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 16.Murray J B, Terwey D P, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott W G. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- 17.Bassi G, Møllegaard N E, Murchie A I H, von Kitzing E, Lilley D M J. Nat Struct Biol. 1995;2:45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- 18.Bassi G S, Murchie A I H, Walter F, Clegg R M, Lilley D M J. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassi G S, Murchie A I H, Lilley D M J. RNA. 1996;2:756–768. [PMC free article] [PubMed] [Google Scholar]
- 20.Bassi G S, Møllegaard N E, Murchie A I H, Lilley D M J. Biochemistry. 1999;38:3345–3354. doi: 10.1021/bi982985r. [DOI] [PubMed] [Google Scholar]
- 21.Simorre J P, Legault P, Hangar A B, Michiels P, Pardi A. Biochemistry. 1997;36:518–525. doi: 10.1021/bi9620520. [DOI] [PubMed] [Google Scholar]
- 22.Simorre J P, Legault P, Baidya N, Uhlenbeck O C, Maloney L, Wincott F, Usman N, Beigelman L, Pardi A. Biochemistry. 1998;37:4034–4044. doi: 10.1021/bi972493z. [DOI] [PubMed] [Google Scholar]
- 23.Chu W C, Feiz V, Derrick W B, Horowitz J. J Mol Biol. 1992;227:1164–1172. doi: 10.1016/0022-2836(92)90528-r. [DOI] [PubMed] [Google Scholar]
- 24.Rastinejad F, Lu P. J Mol Biol. 1993;232:105–122. doi: 10.1006/jmbi.1993.1373. [DOI] [PubMed] [Google Scholar]
- 25.Klimasauskas S, Szyperski T, Serva S, Wuthrich K. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold J R P, Fisher J. J Biomol Struct Dyn. 2000;17:843–856. doi: 10.1080/07391102.2000.10506573. [DOI] [PubMed] [Google Scholar]
- 27.Perreault J-P, Wu T, Cousineau B, Ogilvie K K, Cedergren R. Nature (London) 1990;344:565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- 28.Paolella G, Sproat B S, Lamond A I. EMBO J. 1992;11:1913–1919. doi: 10.1002/j.1460-2075.1992.tb05244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J-H, Usman N, Chartrand P, Cedergren R. Biochemistry. 1992;31:5005–5009. doi: 10.1021/bi00136a013. [DOI] [PubMed] [Google Scholar]
- 30.Heidenreich O, Benseler F, Farenholz A, Eckstein F. J Biol Chem. 1994;269:2131–2138. [PubMed] [Google Scholar]
- 31.Menger M, Tuschl T, Eckstein F, Porschke D. Biochemistry. 1996;35:14710–14716. doi: 10.1021/bi960440w. [DOI] [PubMed] [Google Scholar]
- 32.Hansen M R, Simorre J P, Hanson P, Mokler V, Bellon L, Beigelman L, Pardi A. RNA. 1999;5:1099–1104. doi: 10.1017/s1355838299990738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton T E, Clardy D R, DeRose V J. Biochemistry. 1998;37:18094–18101. doi: 10.1021/bi981425p. [DOI] [PubMed] [Google Scholar]
- 34.Hammann C, Cooper A, Lilley D M J. Biochemistry. 2001;40:1423–1429. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]
- 35.Peracchi A, Beigelman L, Scott E C, Uhlenbeck O C, Herschlag D. J Biol Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 37.Amiri K M A, Hagerman P J. Biochemistry. 1994;33:13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- 38.Tuschl T, Gohlke C, Jovin T M, Westhof E, Eckstein F. Science. 1994;266:785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- 39.Hertel K J, Pardi A, Uhlenbeck O C, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach W L, Hodgson R, Symons R H. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]