Abstract
The Group A Streptococcus (GAS) is a strict human pathogen that causes a broad spectrum of illnesses. One of the key regulators of virulence in GAS is the transcriptional activator Mga, which coordinates the early stages of infection. Although the targets of Mga have been well characterized, basic biochemical analyses have been limited due to difficulties in obtaining purified protein. In this study, high-level purification of soluble Mga was achieved, enabling the first detailed characterization of the protein. Fluorescence titrations coupled with filter-binding assays indicate that Mga binds cognate DNA with nanomolar affinity. Gel filtration analyses, analytical ultracentrifugation, and co-immunoprecipitation experiments demonstrate that Mga forms oligomers in solution. Moreover, the ability of the protein to oligomerize in solution was found to correlate with transcriptional activation; DNA binding appears to be necessary but insufficient for full activity. Truncation analyses reveal that the uncharacterized C-terminal region of Mga, possessing similarity to phosphotransferase system EIIB proteins, plays a critical role in oligomerization and in vivo activity. Mga from a divergent serotype was found to behave similarly, suggesting that this study describes a general mechanism for Mga regulation of target virulence genes within GAS and provides insight into related regulators in other Gram-positive pathogens.
INTRODUCTION
Bacterial pathogens must rapidly adapt to changing environments encountered during infection in order to successfully colonize new host tissue sites. This is often mediated by coordinate regulation of virulence gene expression by specific transcription factors in response to signal transduction cascades or via direct interaction with a ligand. The Group A Streptococcus (GAS) or Streptococcus pyogenes is a strict human pathogen that can elicit a broad spectrum of diseases ranging from benign, self-limiting infections (pharyngitis or 'strep throat', impetigo) to immune sequelae (acute rheumatic fever) and life-threatening invasive disorders (necrotizing fasciitis, streptococcal toxic shock syndrome) (Bisno et al., 2003, Cunningham, 2000). Coordinate control of global regulatory networks in response to changing host environments is essential for GAS pathogenesis as evidenced by dramatic changes in the GAS transcriptome upon growth ex vivo (e.g., blood, saliva) as well as growth in vivo (Musser & DeLeo, 2005, Tart et al., 2007).
Although classical two-component signal transduction systems (TCS) are known to contribute to virulence in GAS (Kreikemeyer et al., 2003), these pathogens also rely heavily on so-called ‘stand-alone’ regulators representing transcription factors controlling virulence gene regulons in response to growth phase by sensory components that have yet to be fully defined (McIver, 2009). The multiple virulence gene regulator (Mga) was the first such stand-alone regulatory network described in GAS and allows the pathogen to adapt and flourish in host environments favourable for growth (Hondorp & McIver, 2007).
Over 100 different serotypes of GAS have been identified based upon variability in the cell-surface M protein (emm gene product), an important virulence factor regulated by Mga that helps the bacteria inhibit phagocytosis. The gene encoding Mga (mga) has been found in all sequenced GAS genomes and strains tested, although minor variations exist in the composition of its regulon (Hollingshead et al., 1994). Two different alleles of mga have been described (mga-1 and mga-2) that correlate with emm genomic patterns as well as different tissues sites of infection (Bessen et al., 2005).
Mga strongly activates the transcription of a number of established GAS virulence genes during the exponential phase of growth (carbohydrate rich) in conditions that are conducive to growth (optimal temperature, available iron). These "core" directly Mga-regulated genes encode mostly multifunctional surface molecules important for adherence to host tissues and evasion of the host immune responses, including M protein (emm), M-like proteins (arp), C5a peptidase (scpA), collagen-like protein (scl1, sclA), fibronectin-binding proteins (fba, sof), and the secreted inhibitor of complement (sic) (Hondorp & McIver, 2007). A transcriptome analysis of the Mga regulon from three divergent GAS serotypes (M1, M6, and M4) revealed that Mga regulates greater than 10% of the GAS genome during exponential growth, including apparent indirect repression of operons encoding proteins involved in the transport and utilization of sugar sources (Ribardo & McIver, 2006). Thus, Mga is able to regulate genes important not only for virulence, but also for sugar metabolism.
Mga is a large (62 kDa) DNA-binding protein that interacts with ca. 45–59 bp non-palindromic binding sites within "core" virulence gene promoters in order to activate transcription (Almengor & McIver, 2004, McIver et al., 1995, McIver et al., 1999). An initial consensus Mga binding sequence was proposed (McIver et al., 1995), however, identification of additional binding sites suggests that there is little overall conservation at different promoter sites. The Mga protein possesses two helix-turn-helix motifs (HTH-3, HTH-4) and a conserved region (denoted CMD-1 for conserved Mga domain) in its amino-terminus that are involved in DNA-binding activity, with HTH-4 being essential for binding to all Mga-binding sites tested thus far (McIver & Myles, 2002, Vahling & McIver, 2006). HTH-4 shows homology to the GntR superfamily of winged HTHs (wHTHs) domains (Aravind et al., 2005). Currently, DNA binding is the only functional attribute of Mga known to be necessary for Mga-dependent transcriptional activation.
Despite identification of DNA-binding domains, very little is known about the function of the remaining 380 carboxy-terminal amino acids of Mga and what is required for transcriptional activation at virulence gene promoters. In silico analyses comparing Mga to proteins of known structure identified two potential phosphoenolpyruvate phosphotransferase system (PTS) regulatory domains (PRDs) in the central region of Mga that may allow modulation of activity based on sugar availability (Hondorp & McIver, 2007). Early analysis of the sequence suggested that the C-terminus of Mga might contain a CheY-like receiver domain (Perez-Casal et al., 1991). However, this putative domain includes the extreme C-terminus of the protein where there is little conservation between mga-1 and mga-2 alleles, overall Mga does not resemble typical response regulators, and experimental evidence for a receiver domain is lacking (Hondorp & McIver, 2007). Hence, the role of the carboxy-terminal region of Mga has remained unclear.
Although Mga is the best characterized among a class of homologous regulators found in Gram-positive pathogens (Hondorp & McIver, 2007), only limited biochemical characterization has been performed. Problems obtaining significant amounts of soluble purified protein have long hindered such analyses. Not surprisingly, no structural data are available for Mga or any of its homologs and even the oligomeric status is unknown. Thus there is considerable need for basic biochemical analyses in order to gain insight into the mechanism by which this archetypal protein modulates expression of virulence genes within the pathogen. In this study we present the first high-level purification of soluble Mga, quantification of DNA binding affinity and characterization of oligomerization. Furthermore, we show that DNA binding is insufficient for Mga activity and that the C-terminal region plays a critical role in oligomerization and transcriptional activation of target genes within GAS.
RESULTS
Optimization of Mga Purification
Previous analyses of Mga employed recombinant protein that precluded thorough biochemical characterization due to limited yield and purity (Almengor & McIver, 2004, Almengor et al., 2006, McIver & Myles, 2002). Thus for this study, considerable effort was invested in optimizing expression and purification of Mga from E. coli. We focused our efforts on the M4 Mga (mga-2) from GA40634, a clinically relevant GAS serotype in which the Mga transcriptome has been characterized, since the well-studied M6 JRS4 strain does not appear to provide a general model for Mga regulation (Ribardo & McIver, 2006). Through extensive optimization of conditions (solubility and yield), we were able to purify the His-tagged protein from E. coli lysates to greater than 95% purity (see Supplemental Results and Fig. S1), representing a considerable improvement over previous results.
It is significant to note that while increasing the yield and purity of Mga overcame a major hurdle, considerable precipitation was routinely encountered during purification. After numerous trials to prevent aggregation and resulting loss of Mga, we discovered that the protein is exquisitely sensitive to buffer conditions (see Supplemental Results). Because high concentrations (50 mM) of the metal chelator EDTA were found to abrogate Mga precipitation, we assayed the metal content of the purified protein and found approximately stoichiometric levels of a divalent metal (preliminarily identified as a mixture of Zn2+ and Ni2+, see Supplemental Results and Fig S1). While the physiological identity and role of a metal is unclear, its removal proved critical for manipulating Mga in vitro, and therefore, this study focuses on the apoprotein. The ability to obtain highly purified preparations of homogenous soluble Mga provided a significant advance, allowing for the first extensive biochemical analysis of the protein.
Mga binds DNA with nanomolar affinity in vitro
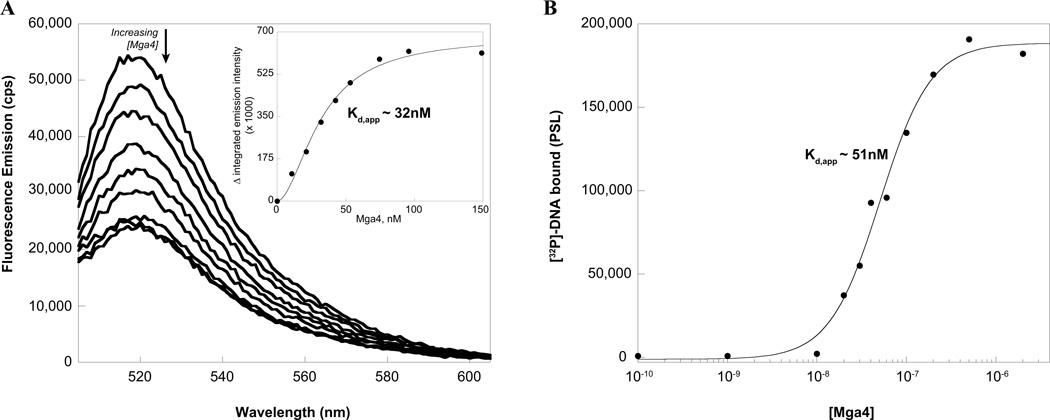
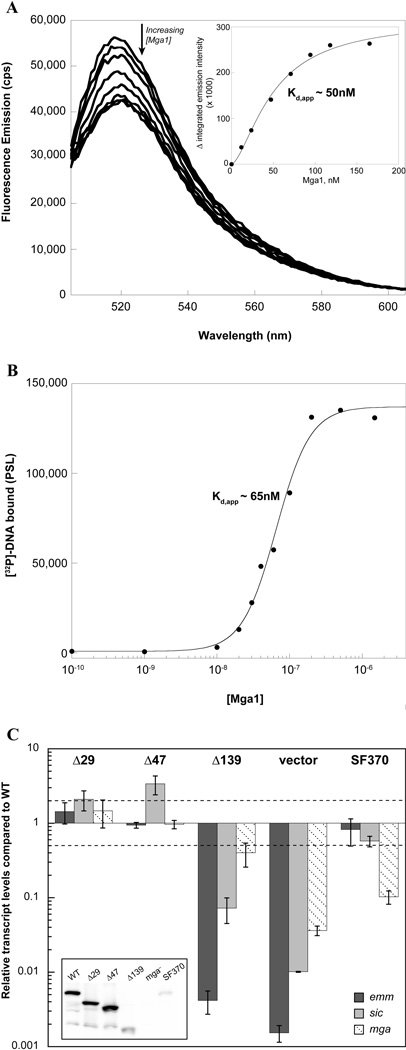
Characterization of the affinity of Mga binding to target DNA is critical to understanding its mechanism of activation. However, the tendency of Mga to aggregate in native gels has hindered quantitative evaluation of DNA binding via traditional gel mobility shift assays in the past. Consequently, binding was assayed by titration of carboxyfluorescein- (FAM-) labeled DNA with Mga4-His6, which was found to quench the fluorophore. The emission spectrum of a FAM-labeled 49 base pair duplex comprising the M4 Mga-binding site of the arp promoter (Parp FAM-49mer) was monitored upon addition of Mga4-His6 (Fig. 1A). Plotting the change in fluorescence versus the concentration of Mga4-His6 indicates that the protein concentration required for half-maximal binding, or the apparent Kd (Kd,app), is approximately 32 nM (Fig. 1A inset).
FIG. 1. Mga4-His6 binds to Parp4 in vitro.
A, Fluorescence titration. The fluorescence spectrum of a FAM-labeled 49mer containing the Mga-binding site of the Parp4 promoter was monitored upon the addition of purified Mga4-His6 to assess binding. The decrease in integrated emission intensity from 515 to 540 nm was plotted versus the concentration of Mga4-His6 to determine the apparent Kd for half maximal binding (inset). B, Filter binding assays. Varying concentrations of Mga4-His6 were incubated with the [32P]-labeled Parp4 49mer and then filtered through nitrocellulose, which retains the protein-bound radiolabel. Phosphorimager analysis followed by densitometry was used to plot the amount of DNA bound as a function of Mga4-His6 concentration and determine Kd,app
Filter-binding assays were employed as an independent means to assay Mga binding to the same DNA sequence. Varying concentrations of Mga4-His6 were incubated with the [32P]-labeled Parp 49mer and then filtered through nitrocellulose. The amount of radiolabeled DNA bound to Mga was quantitated by densitometry and plotted to reveal that half-maximal binding occurs at Mga concentrations of approximately 51 nM (Fig. 1B).
Thus, taken together, these experiments indicate that under these conditions Mga4-His6 binds the Parp 49mer with a Kd,app of approximately 30–50 nM. Interestingly, the data from both methods consistently displayed sigmoidal character, suggesting that binding occurs via a mechanism more complicated than one protein binding to a single site (n = 1.5 and 1.8 for filter binding and fluorescence data, respectively, when fit with the Hill equation).
Mga forms oligomers in solution
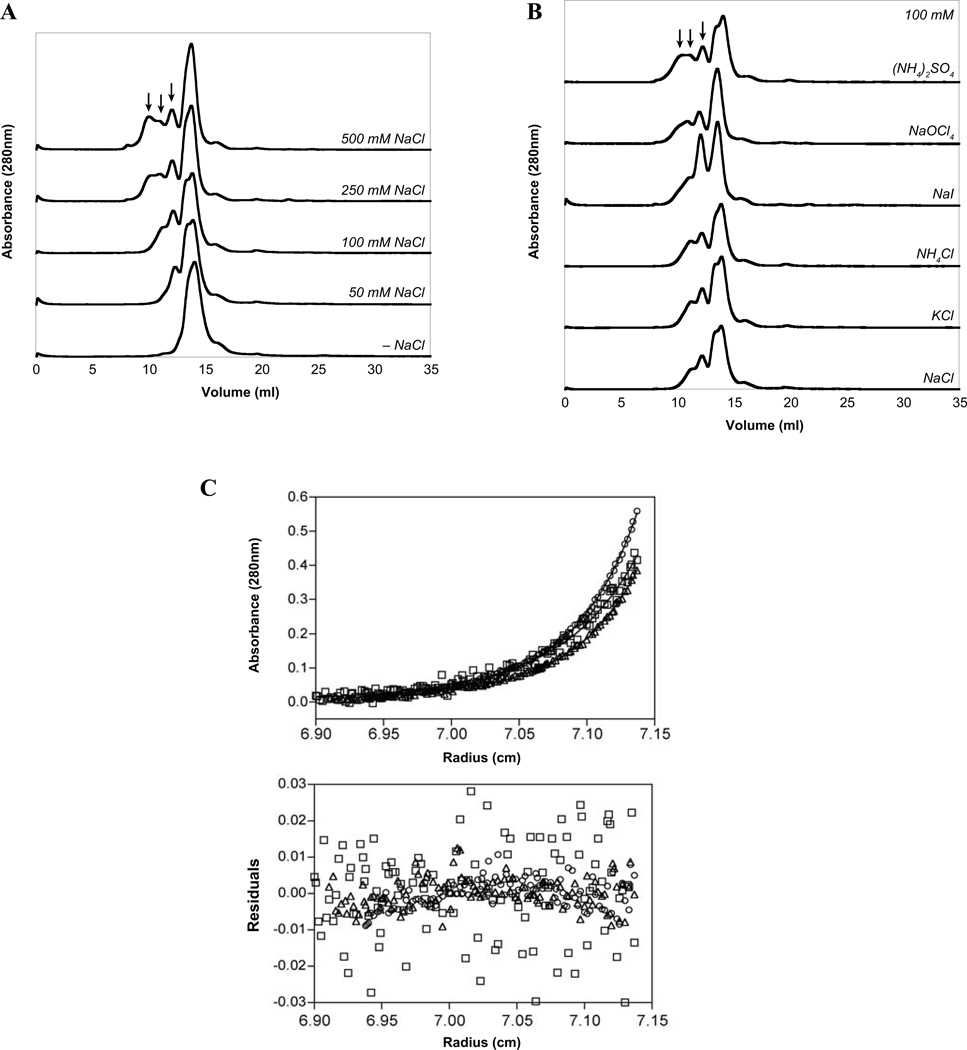
Since transcription factors often function as dimers or higher-order oligomers, it was important to determine the multimeric state of Mga. Attempts to utilize native gels were unsuccessful as the majority of the protein was unable to enter the gel, apparently due to aggregation at the interface. Consequently, Mga was analyzed by size exclusion chromatography in the HEPES/Citrate buffer in which the protein remains soluble. Surprisingly, Mga was found to exist in a dynamic mixture of states that were sensitive to salt. In HEPES/Citrate buffer alone, Mga was almost exclusively a monomer; yet, as the concentration of NaCl increased, multiple higher-order states were observed (Fig. 2A). Based on molecular weight standards, the monomeric species appears to be between 77 and 89 kDa, depending on the salt concentration. This is larger than the calculated analytical molecular weight of 63.2 kDa, which was confirmed by MALDI-MS analysis (64.2 kDa), suggesting that protein conformation causes Mga to exhibit a larger hydrodynamic radius than expected. Furthermore, there appears to be a higher molecular weight shoulder on the monomeric peak (for example, see 50 mM and 100 mM NaCl traces in Fig. 2A) that may indicate conformational heterogeneity, possibly due to multiple monomeric conformations or a monomer-oligomer transition that occurs on the column support. The higher-order species were found to correspond to an average of 2.3, 4.0, and 6.1 times the molecular weight of the monomer peak, suggesting that dimers, tetramers, and hexamers of Mga may be formed with increasing concentrations of NaCl, perhaps mimicking a dimeric array bound to DNA. (Fig. 2A, arrows). Interestingly, the effect of salt on oligomerization of Mga appeared to be directed by the anion, since changing the cation (Na+, K+, or NH4+) did not significantly impact the distribution of oligomers, whereas varying the anion did (Cl−, I−, ClO4−, or SO42−, Fig. 2B). This phenomenon appears to follow the electroselectivity series, where multivalent and more chaotropic anions stabilize multimerization of Mga (SO42− > ClO4− > I− > Cl−).
FIG. 2. Mga forms oligomers in solution.
Mga4-His6 was incubated in a HEPES/Citrate buffer containing varying salts and then subjected to gel filtration analysis in the same buffer. A, Increasing the concentration of NaCl results in the appearance of higher-order multimers (indicated by arrows). B, Maintaining the salt concentration at 100 mM while varying the identity of the anion (but not the cation) also impacts the oligomerization pattern of Mga4-His6. C, Mga4-His6 in the presence of 100 mM NaCl was analyzed by sedimentation equilibrium analytical ultracentrifugation for various protein concentrations (7.5 µM, circles; 20 µM, squares; and 30 µM, triangles) centrifuged at 16,000 rpm. Fifteen datasets were fit to a monomer-dimer model (solid lines) resulting in the residuals depicted.
To further analyze Mga oligomerization, sedimentation equilibrium analytical ultracentrifugation (AUC) experiments were performed on Mga4-His6 in the presence of 100 mM NaCl. Analysis of the data indicates an average molecular weight of 89.9 kDa, significantly larger than the value of 63.2 kDa expected of the monomer. When fit to a monomer-dimer model, the equilibrium dissociation constant for dimerization (Kdim) was found to be 9.41 µM (Fig. 2C). Analysis of the data using monomer-trimer or monomer-tetramer models indicated no improvement in the fit based on the magnitude of the square root of the variance of the fit and the residuals of the fit (data not shown).
The conserved C-terminal region of Mga is critical for its activity in vivo
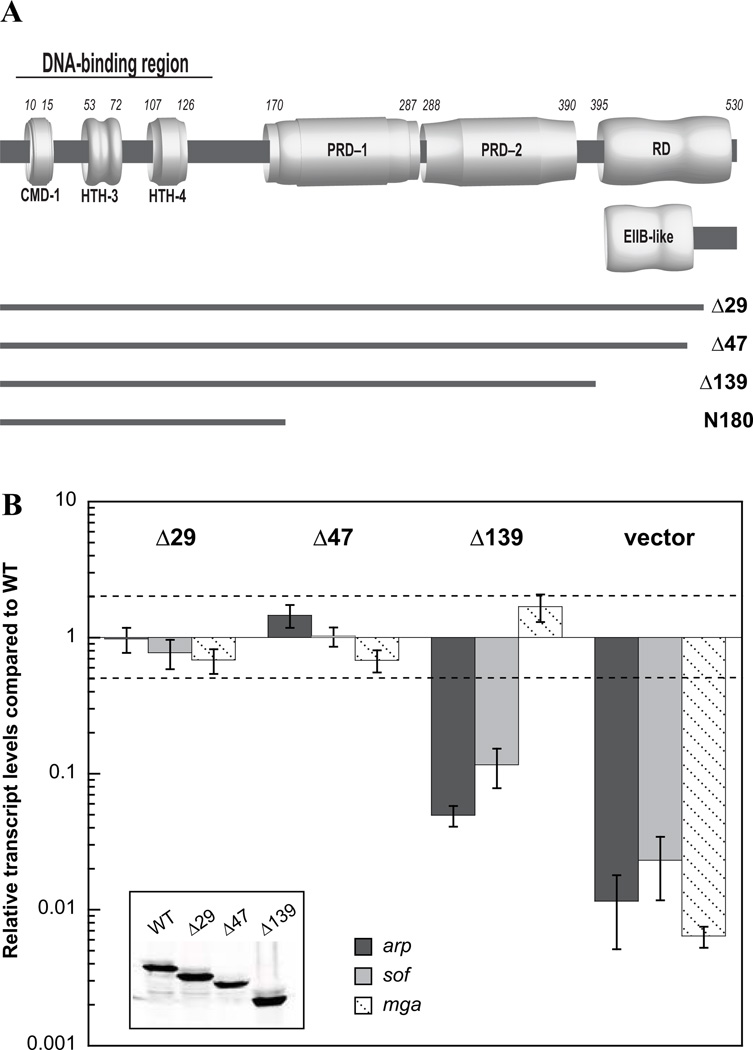
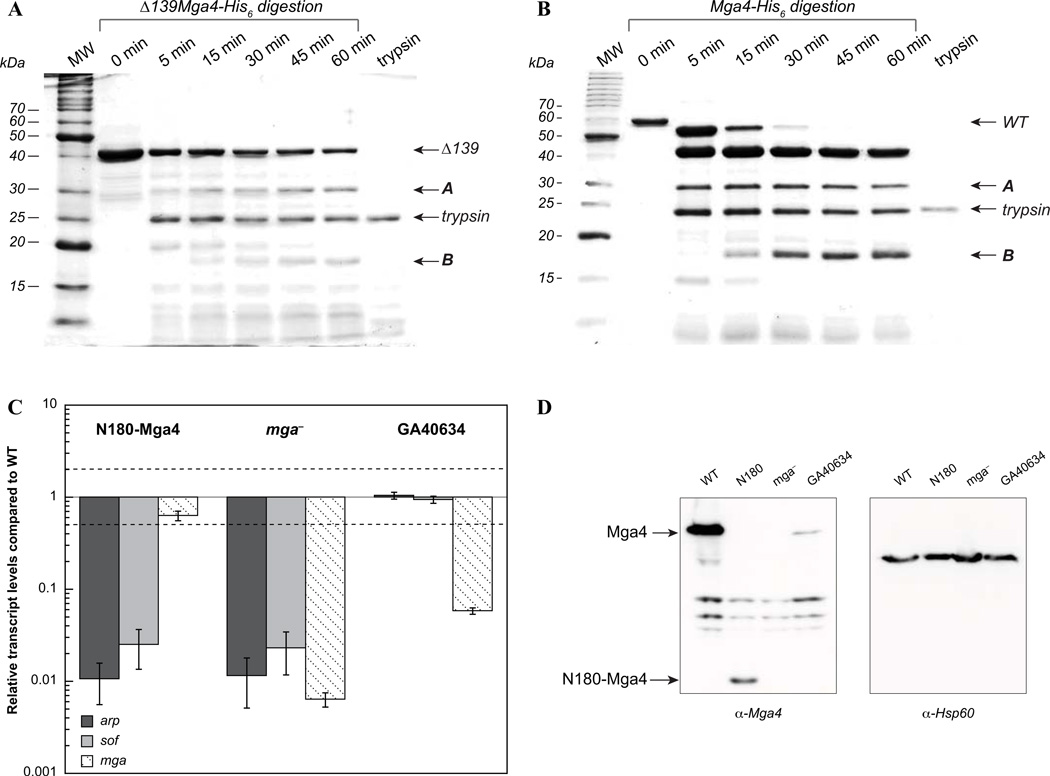
Currently, nothing is known about the role of the carboxy-terminus (aa 390–533) of Mga. In order to gain further insight into the importance of this region, three C-terminal truncation mutants were constructed. First, the C-terminal 29 amino acids that are predicted to be disordered by the program DISOPRED (http://bioinf.cs.ucl.ac.uk/disopred/) were removed to generate Δ29Mga4-His6 (aa 1–504, Fig. 3A). As evidenced from sequence alignment, Mga proteins are highly conserved between GAS serotypes except at the extreme C-terminus (Fig. S2). Therefore, the non-conserved region was similarly eliminated to produce Δ47Mga4-His6 (aa 1–486, Fig. 3A). And finally, the C-terminal 139 amino acids were deleted to generate Δ139Mga4-His6 (aa 1–394), which includes all predicted domains, and terminates after the second putative PTS regulatory domain (PRD, Fig. 3A). The truncation eliminates the region at one time identified by 3D-Jury as having weak homology to a CheY-like receiver domain (RD) (Perez-Casal et al., 1991), which encompasses the extreme C-terminus of the protein where there is limited conservation between mga-1 and mga-2 alleles of Mga (see Fig. 3A and S2). Interestingly, Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/) analysis of Mga detects structural homology of the conserved C-terminal region (aa 407–490) to EIIB domains utilized by the PTS. Thus, while the Δ47 truncation removes the non-conserved region, Δ139 eliminates the putative EIIB-like domain (see Fig. 3A).
FIG. 3. The C-terminal region of Mga4-His6 is required for in vivo activity.
A, Schematic showing N-terminal regions implicated in DNA binding (CMD-1, HTH-3, and HTH-4) and central domains with strong structural homology to PTS regulatory domains (PRD-1 and PRD-2). The C-terminal region at one time predicted to be a CheY-like receiver domain (RD) is indicated. Newly recognized structural homology to an EIIB domain is depicted below the RD. Lines representing the different C-terminal truncation mutants are shown. B, Full-length and truncated Mga4-His6 proteins (Δ29Mga4-His6, Δ47Mga4-His6, and Δ139Mga4-His6) were each expressed on a plasmid under the native Pmga4 promoter in a mga4− strain, KSM547. Mga activity was assayed in vivo by real-time RT-PCR analysis of the Mga-regulated genes, arp (dark gray), sof (light gray), and mga itself (diagonal hashes). Relative transcript levels for the mutant strains compared to the wild type are presented. Error bars represent the standard error from six biological replicates. Differences greater than 2-fold in expression for the mutant strains compared to wild type (denoted by a dashed line) were considered significant. Empty vector mga− results are shown alongside for comparison. The western blot of cell lysates probed with the α-Mga4 antibody (inset) is shown.
To determine the in vivo effects of the truncations on Mga activity, plasmids expressing full-length and truncated Mga proteins from their native promoter (Pmga4) were transformed into an mga4− GAS background, KSM547. All proteins appear to be expressed and stable in GAS, as evidenced by immunoblotting of cell lysates with an 〈-Mga4 antibody (Fig. 3B inset). Mga activity was then assayed by real-time RT-PCR analysis of the M4 Mga-regulated genes that encode an IgA receptor protein (arp) and serum opacity factor (sof), both of which have been used to assess the M4 Mga regulon (Ribardo & McIver, 2006). Changes in transcript levels for the mutant strains that were greater than 2-fold compared to wild type were considered significant (Fig. 3B, dotted line). The Δ29 and Δ47 truncations complement expression of arp and sof indicating that the deleted residues were not required for Mga activity (Fig. 3B). However, deletion of the C-terminal 139 amino acids (Δ139Mga4-His6) causes a dramatic drop in transcript levels for arp and sof compared to full-length Mga4-His6, indicating that the protein is not fully active in vivo. Thus, the C-terminal 139 amino acids of Mga are important for in vivo function. Interestingly, Δ139mga4 expression from its native promoter (Pmga4) appears unaffected despite resulting in an inactive protein. This suggests that either (1) Mga regulation at Pmga is unique and Δ139Mga4-His6 is fully functional at that specific promoter, or (2) mga is not autoregulated under these conditions, which is in contrast to published studies showing Mga autoactivation in the M6 background (McIver et al., 1999, Okada et al., 1993). However we have additional data that suggest a lack of Mga autoregulation in this system (data not shown).
Δ139Mga4 binds DNA but is defective for oligomerization
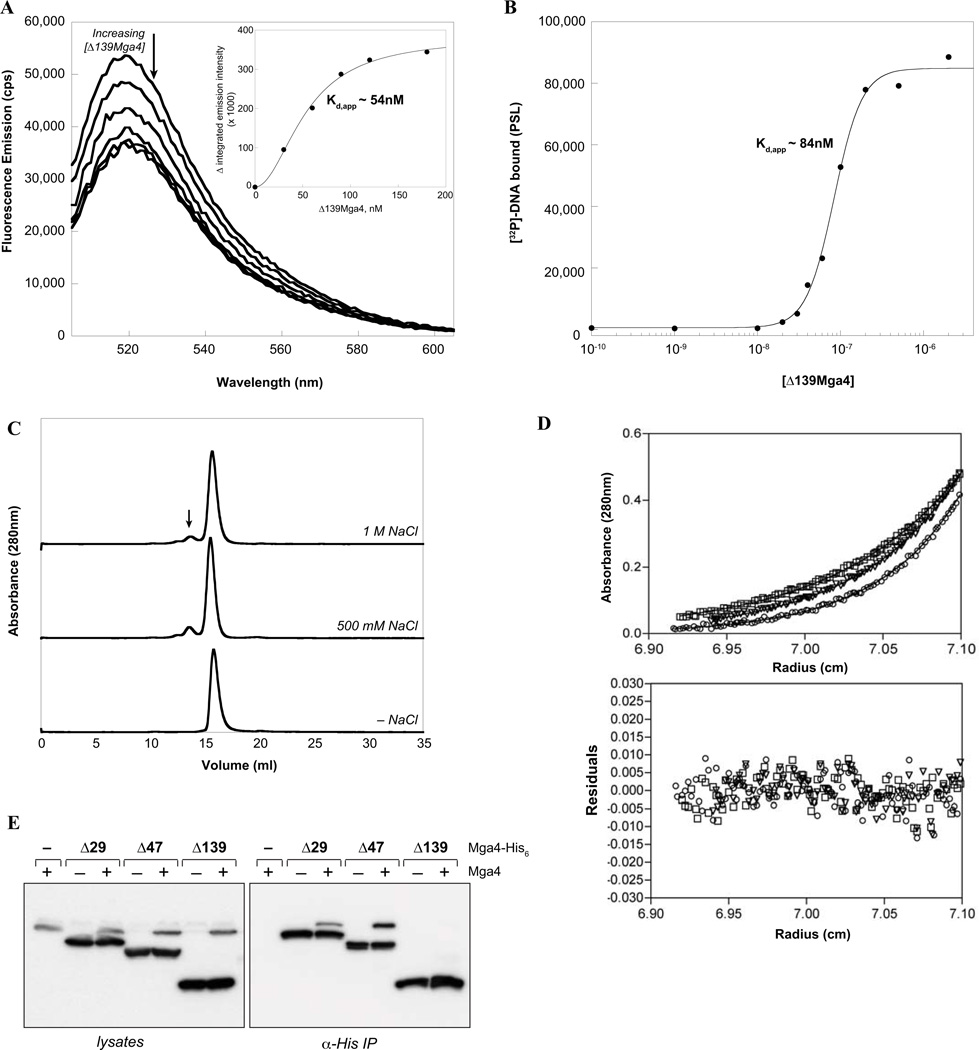
In order to ascertain the means by which the Δ139 truncation hinders Mga activity, it was first important to determine whether Δ139Mga4-His6 maintains the ability to bind to DNA. Therefore, Δ139Mga4-His6 was overexpressed and purified using a similar strategy to wild-type Mga4-His6. It should be noted that Δ139Mga4-His6 was expressed in E. coli at a much higher level compared to the full-length protein, yet also co-purified with a metal that was removed to generate the apoprotein for these studies. Binding of Δ139Mga4-His6 to the Parp 49mer was assayed using both fluorescence and filter-binding studies in the same manner as the wild type. Interestingly, the Δ139 protein was found to bind the Parp 49mer analogously to full-length Mga4-His6 with a Kd,app of 54–84 nM (Fig. 4A,B; n = 2.0 and 2.6 for filter binding and fluorescence data, respectively, when fit with the Hill equation). The similar affinities suggest that DNA binding alone is not sufficient for full activation of transcription.
FIG. 4. Δ139Mga4-His6 binds DNA with an affinity similar to full-length Mga, but is defective for oligomerization.
A, Fluorescence titration. The fluorescence spectrum of a FAM-labeled 49mer containing the Mga-binding site of the Parp4 promoter was monitored upon the addition of purified Δ139Mga4-His6 to assess binding. The decrease in integrated emission intensity from 515 to 540 nm was plotted versus the concentration of Δ139Mga4-His6 to determine the apparent Kd (inset). B, Filter binding assays. Varying concentrations of Δ139Mga4-His6 were incubated with the [32P]-labeled Parp4 49mer. The complex was filtered through nitrocellulose to assess the amount of DNA bound as a function of Δ139Mga4-His6 concentration and determine Kd,app. C, Gel filtration analysis. Δ139Mga4-His6 was incubated in a HEPES/Citrate buffer containing varying concentrations of NaCl and then subjected to size exclusion chromatography. An arrow indicates the position of a higher-order multimer upon increasing salt concentrations. D, Sedimentation equilibrium by analytical ultracentrifugation. Δ139Mga4-His6 (7.5 µM) in the presence of 100 mM NaCl was centrifuged at 18,000 (squares), 20,000 (inverted triangles) and 22,000 (circles) rpm. Six datasets were fit to a model for a single homogenous species (solid lines) resulting in the residuals depicted. E. Co-immunoprecipitation of Mga4 with truncated proteins. Δ29-, Δ47-, and Δ139Mga4-His6 proteins were expressed in wild type (GA40634) and mga4− (KSM547) GAS strains. His-tagged proteins were immunoprecipitated using α-His, and coimmunoprecipitation of endogenous Mga4 was probed by western blot analysis with α-Mga4 following separation of proteins by SDS-PAGE (right). Similar western blots of whole lysates prior to immunoprecipitation are shown to verify protein expression (left).
Since Δ139Mga4-His6 did not appear to be defective in binding DNA, but was inactive in vivo, the ability of the truncated protein to oligomerize was assayed. Gel filtration analysis indicates that, similar to full-length Mga4-His6 (Fig. 2A), Δ139Mga4-His6 is primarily a monomer in solution (Fig. 4C). Interestingly, the average molecular weight was 35.6 kDa based on gel-filtration standards, which was smaller than the expected size of 47.2 kDa (47.18 kDa determined by MS analysis), as opposed to Mga4-His6 which ran larger. Furthermore, in contrast to the full-length protein (Fig. 2A), Δ139Mga4-His6 does not significantly oligomerize upon the addition of high concentrations of NaCl, suggesting a defect in its ability to stabilize a multimeric state (Fig. 4C). These findings were verified by AUC, which indicated a monomeric species (50.7 kDa) in the presence of 100 mM NaCl (Fig. 4D).
To ascertain whether oligomerization of Mga might play a role in its in vivo function, co-immunoprecipitation (co-IP) experiments were performed with all of the truncated proteins. The His-tagged Δ29, Δ47, and Δ139 Mga4 truncations were expressed on a plasmid in wild-type M4 GA40634 GAS, which contains the endogenous untagged Mga4 protein. Immunoprecipitations using an 〈-His monoclonal antibody were then performed with lysates from exponentially growing cells. SDS-PAGE and western blot analysis with the 〈-Mga4 antibody, which recognizes both tagged and native Mga4, indicates that the 29- and 47-residue truncated proteins are able to interact with endogenous full-length Mga4 and coimmunoprecipitate, whereas Δ139Mga4-His6 does not (Fig. 4E). Taken together, the data indicate that while DNA binding is clearly necessary for Mga activity, it is not sufficient to support full Mga-dependent transcription of target genes. However, the ability of Mga to oligomerize in solution correlates with its in vivo activity. Thus, multimerization of Mga may also be required for activation of transcription and the C-terminal region of Mga likely provides important determinants for this interaction. It is also possible that loss of the C-terminus alters the structure of the remaining protein and prevents oligomerization.
The DNA-binding domains of Mga are not sufficient for activity
Based on the putative domain structure of Mga (Fig. 3A), a straightforward mechanism can be envisioned whereby the C-terminal two thirds of the protein serves to regulate the core N-terminal DNA-binding domains. Removal of the C-terminal 139 amino acids could simply result in a defect in the regulatory region, potentially locking the protein in an inactive state. Therefore, it is possible that the isolated N-terminal DNA-binding domains of Mga may retain activity. This is the case for the N-terminal domain of the PRD-containing regulator, LicT, which lacks its C-terminal regulatory PRDs and displays constitutive activity in vivo (Manival et al., 1997).
In order to locate a stable fragment that encompasses the DNA-binding region, the structure of Mga was probed by limited tryptic digestion. Native Δ139Mga4-His6 was incubated with a limiting amount of trypsin in attempts to identify exposed linker regions that might be accessible to proteolysis. SDS-PAGE separation of the digested products indicated that two major fragments are stably formed, which we called A and B (Fig. 5A). A similar pattern was observed with limited proteolysis of the full-length protein (Fig. 5B). The smaller product was isolated (Fig. S3) and mass spectrometry revealed a mass of 20,972 Da, which corresponds to the N-terminal 180 amino acids (20,976 Da expected), indicating that trypsin cleaves at the beginning of the putative PRD-1 domain (Fig. 3A). The mass for the larger ~30 kDa product, A, could not be determined. Nevertheless, the identification of a stable fragment encompassing the N-terminal 180 amino acids served to isolate the DNA-binding region from the putative regulatory region.
FIG. 5. The N-terminal DNA-binding region of Mga is not active in vivo.
A & B, Limited tryptic digestion of native Δ139Mga4-His6 (A) and full-length Mga4-His6 (B) with trypsin (0.12% or 0.08%, respectively) followed over time by SDS-PAGE. The predominant fragments observed over extended digestions (denoted as A and B) are indicated. C, In vivo activity of N180-Mga4. His-tagged wild-type and N180-Mga4 proteins were expressed from the native Pmga4 promoter in a mga4− strain, KSM547. Mga activity was assayed by real-time RT-PCR of the Mga-regulated genes, arp (dark gray), sof (light gray), and mga (diagonal hashes). Relative transcript levels for the mutant strains compared to the wild type are presented. Error bars represent the standard error from three biological replicates. Differences greater than 2-fold (denoted by a dashed line) in expression for the mutant strains compared to wild type were considered significant. The empty vector results in the mga− and wild-type strain are shown for comparison. D, Western blot analyses. Immunoblots of cell lysates probed with α-Mga4 (left) reveal the levels of full-length and N180-Mga4 proteins (indicated by arrows) in a mga4− strain, KSM547 expressing either Mga4-His6 (WT), N180-His6 (N180) or an empty vector (mga −), and an isogenic parental M4 (GA40634) strain. An α-Hsp60 blot (right) was performed to demonstrate equal loading.
To assess whether the N-terminal domain was active, the first 180 amino acids of Mga (Δ353Mga4, here called N180-Mga4) were expressed with a C-terminal His tag, under the Pmga4 promoter in an mga4 − strain of GAS, KSM547. Western blot analysis indicated that N180-Mga4-His6 levels were somewhat reduced compared to full-length Mga4-His6 expressed from a plasmid (Fig. 5D). However, N180-Mga4-His6 levels were still higher than a fully active Mga4 expressed from its native genomic location (Fig. 5C,D). Real-time RT-PCR was employed to assess Mga activity in vivo (Fig. 5C). A complete lack of transcriptional activation of the Mga-regulated genes, arp and sof, was observed for N180-Mga4-His6. This suggests that the remainder of the protein is important for its function and does not simply regulate the activity of the N-terminal DNA-binding domain. Attempts to overexpress and purify recombinant N180-Mga4-His6 have thus far been unsuccessful, as the protein is sequestered in inclusion bodies and is more difficult to manipulate with a near-neutral pI (data not shown).
A divergent M1 Mga behaves similarly to M4 Mga
Finally, in order to ascertain whether the observed results for Mga4 are characteristic of Mga proteins in general, Mga from a divergent mga -1 allele (strain SF370, M1 serotype) was similarly purified. Mga1 is 78% identical to Mga4 at the amino acid sequence level, with the greatest disparity found at the C-terminus (Fig. S2).
Fluorescence and filter binding experiments were performed to assess DNA binding and revealed that Mga1-His6 binds the Parp 49mer with almost the same affinity (50–65 nM Kd,app) as Mga4-His6 (compare Fig. 1 and 6A,B). Binding to Parp, which is exclusive to mga-2 GAS, supports the fact that divergent Mga proteins can function interchangeably as has been previously demonstrated (Andersson et al., 1996, Vahling & McIver, 2006). In fact, DNA-binding experiments with the emm promoter from a M1-type (Pemm1) showed that both M1 and M4 Mga bound with relatively similar affinities (data not shown).
FIG. 6. A mga-1 Mga behaves similarly to a mga-2 Mga.
A, Fluorescence titration. The fluorescence spectrum of a FAM-labeled 49mer containing the Mga-binding site of Parp4 was monitored upon the addition of purified Mga1-His6 to assess binding. The decrease in integrated emission intensity from 515 to 540 nm was plotted versus the concentration of Mga1-His6 to determine the apparent Kd (inset). B, Filter binding assays. Varying concentrations of Mga1-His6 were assayed with the [32P]-labeled Parp4 49mer. The complex was filtered through nitrocellulose to assess the amount of DNA bound as a function of Mga1-His6 concentration and determine Kd,app. C, In vivo activity of M1 truncated proteins. Full-length and truncated Mga1-His6 proteins (Δ29Mga1-His6, Δ47Mga1-His6, and Δ139Mga1-His6) were each expressed from the native Pmga1 promoter in a mga1− strain, KSM1656L. Mga activity was assayed by real-time RT-PCR of the Mga1-regulated genes, emm (dark gray), sic (light gray), and mga (diagonal hashes). Relative transcript levels for the mutant strains compared to the wild type are presented. Error bars represent the standard error from three biological replicates. Differences greater than 2-fold (denoted by a dashed line) in expression for the mutant strains compared to wild type were considered significant. Results for the empty vector in the mga− strain (vector) as well as endogenous Mga1 in the isogenic wild type strain (SF370) are shown alongside for comparison. A western blot of cell lysates probed with α-Mga1 (inset) is shown.
To determine whether the deduced domain structure was similar for the two Mga proteins, the analogous Δ29, Δ47, and Δ139 Mga1-His6 truncated proteins were constructed. In vivo activity was assessed by real-time RT PCR for proteins expressed under the native Pmga1 promoter in an mga1− strain of GAS, KSM165-L. Again, the 29- and 47-residue truncations did not dramatically impact Mga1 activity, whereas the Δ139 protein exhibited a significant loss in its ability to activate transcription of the M1 Mga-regulated genes emm and sic (Fig. 6C). While Δ139Mga1-His6 protein levels appear to be lower than the wild type, they are significantly higher than the fully active endogenous Mga1 (wild-type SF370), suggesting that decreased protein levels do not account for the lack of Δ139Mga1-His6 activity in vivo (Fig. 6C inset). This indicates that the extreme C-terminal 47 amino acids of Mga, where little sequence conservation is apparent between alleles, is indeed dispensable for Mga activity. However, the conserved ~90 residues C-terminal to PRD-2 containing the EIIB-like domain plays an important role in Mga activity (Fig. 3A). Thus, the domain structure and biochemical characterization of M1 and M4 Mga likely describe the general mechanism by which Mga functions to control virulence genes within GAS.
DISCUSSION
Orthologs of Mga have been identified in several Gram-positive pathogens (S. dysgalactiae, S. pneumoniae, S. equi, S. gordonii, S. mitis, S. sanguinis, S. uberis, E. faecalis, L. monocytogenes, and B. anthracis), indicating that it represents an important class of regulators. Yet, while much has been learned concerning the genes regulated by Mga, the means by which the protein controls their transcription has long remained a mystery. Thus, this study provides the first detailed biochemical analysis of Mga and reveals a role for the conserved C-terminal region in oligomerization and transcriptional activation.
Mga-DNA binding and oligomerization
The development of methods to obtain pure soluble Mga overcomes a significant barrier, allowing for analyses that have previously been inaccessible. Consequently, this study provides the first quantification of Mga-DNA binding, which is particularly significant given that DNA binding has not been demonstrated for any other Mga homolog, including the B. anthracis AtxA protein. Both fluorescence titrations and filter-binding assays indicate that Mga binds cognate DNA with nanomolar affinity; an apparent Kd of 30–50 nM was found for binding to a 49-basepair oligonucleotide comprising the Mga-binding site of the arp promoter. The consistency of the equilibrium constants obtained using the two methods, including fluorescence, an equilibrium method and filter binding, a separation method provides confidence in the result. However, given the sigmoidal character of the binding isotherms, the equilibrium constants represent apparent values. Furthermore, this sigmoidal character suggests that Mga binds cognate DNA cooperatively and indicates that determination of the stoichiometry of Mga binding to DNA is important for further clarifying the mechanism of binding and activation.
Transcription factors frequently bind DNA as dimers or higher-order oligomers and the large 45-base-pair binding site of Mga suggests that the protein may activate transcription as a multimer. Initial attempts to determine the stoichiometry of binding suggest that multiple molecules of Mga bind the FAM-labeled 49mer (data not shown). However, stoichiometric titrations require a DNA concentration at least five times the apparent Kd, meaning that micromolar Mga concentrations are needed. The tendency of Mga to form aggregates at increased concentration in the binding buffer complicates the analysis and unfortunately, Mga does not appear to bind DNA in a HEPES/Citrate buffer in which it is fully soluble. Consequently, further studies will be required to determine the binding stoichiometry, although this may prove nontrivial for a large complex, particularly given its sensitivity to buffer composition. It is also possible that protein-DNA stoichiometry is dependent upon Mga association with a metal. For instance, numerous apo-Mga molecules could bind nonspecifically, while metallated Mga binds specifically. Hence, the physiological relevance of metal association to Mga binding will need to be assessed.
Although the stoichiometry of Mga-DNA binding is unclear, coimmunoprecipitation experiments from cell lysates, gel filtration analyses, and AUC demonstrate for the first time that Mga is able to form higher-order oligomers. Moreover, gel filtration experiments found that the multimeric status of Mga in solution was sensitive to the presence of salt. In general, salts can impact protein stability via three different means: (1) Debye-Hückel screening, (2) Hofmeister interactions, or (3) direct binding. Debye-Hückel screening is a nonspecific effect that is solely dependent on the ionic strength of the solution. Since stabilization of Mga oligomerization is sensitive to the identity of the anion, Debye-Hückel screening is probably not responsible. The Hofmeister effect appears to impact protein structure via hydration interactions (Zhang & Cremer, 2006). The Hofmeister series orders anions based on their stabilization properties (sulfate > chloride > iodide > perchlorate), where chaotropic anions (e.g., perchlorate) salt out proteins causing them to precipitate and kosmotropic anions (e.g., sulfate) solubilize proteins by salting-in processes. In contrast, stabilization by direct anion binding is governed by the electroselectivity series, which is based on the affinity of binding to an anion-exchange resin (Gjerde et al., 1980, Gregor et al., 1955). The electroselectivity series may be considered the reverse of the Hofmeister series for monovalent ions, where chaotropic anions stabilize protein structure better than kosmotrophic anions and multivalent interactions are stronger than monovalent (sulfate > perchlorate > iodide > chloride). In this study, increased oligomerization of Mga is clearly stabilized by anions according to the electroselectivity series, indicating that direct anion binding is responsible. Protein stabilization via anion binding is generally attributed to shielding of positively charged residues, which minimizes repulsion between protein molecules. Anion binding has been found to increase the stability of numerous proteins and play roles in folding (Goto et al., 1990, Maity et al., 2005, Muzammil et al., 2000, Ramos & Baldwin, 2002), fibrillation (Jain & Udgaonkar, 2010, Raman et al., 2005), and multimerization (Boffi et al., 1990, Bowie & Sauer, 1989, Gokarn et al., 2009, Sakurai et al., 2001).
Interestingly, AUC experiments performed with Mga4-His6 in the absence of salt indicated a Kdim of 6.13 µM, with confidence intervals overlapping that of the protein analyzed in the presence of 100 mM NaCl (data not shown), suggesting that in this system salt did not have a significant effect. Thus while AUC results confirm Mga oligomerization, they also highlight the complexity of the mechanism and indicate that further studies will be required to fully dissect the nuances of multimerization under various conditions.
Role of the C-terminal region
Previous studies localized DNA-binding determinants to the N-terminal region of the protein, but the function of the rest of the protein has been unclear (McIver & Myles, 2002, Vahling & McIver, 2006). Recent analyses indicate structural homology to phosphotransferase (PTS) regulatory domains (PRDs) by the central ~214 amino acids of Mga, and indeed, we have strong evidence to suggest that the protein is regulated via phosphorylation within these domains (Hondorp et al., manuscript in preparation). Consequently, a simplified view suggests that the N-terminal region is important for DNA binding, while the C-terminal two thirds of the protein functions to regulate Mga activation. If this scheme were correct, removal of the regulatory region could result in constitutive activation of Mga, similarly to other well-characterized PRD-containing proteins, such as LicT (Manival et al., 1997). However, a structurally stable N-terminal domain of Mga composed of amino acids 1–180 was found to be nearly completely inactive in vivo. This suggests that the remainder of the protein is important for function and does not simply regulate the activity of the N-terminal DNA-binding domain.
Truncation analyses revealed that deletion of the nonconserved C-terminal 47 amino acids does not impact activity in vivo. However, removal of 139 amino acids resulted in substantial loss of Mga activity, indicating that the conserved 92 amino acids within the C-terminus are important for transcriptional activation in GAS. Surprisingly, the Δ139 protein bound cognate DNA in vitro with a similar affinity to full-length Mga. However, gel filtration analyses, co-immunoprecipitation experiments, and analytical ultracentrifugation provided evidence that Δ139Mga4-His6 is defective for protein oligomerization in solution. Thus, DNA binding is necessary but not sufficient for full Mga activity; yet the added ability to form oligomers in solution appears to correlate with transcriptional activation, hinting at a more complex mechanism. Moreover, the conserved C-terminal region appears to provide determinants that play an important role in oligomerization and Mga activity in vivo.
Historically, the C-terminal region of Mga has displayed little sequence similarity to other protein domains. However, recent Phyre2 analysis indicates that the conserved region between the Δ47 and Δ139 truncations (amino acids 407–490) is structurally homologous to EIIBGat domains (see Fig. 3A). EIIB proteins are cytoplasmic components of enzyme II (EII) complexes, which are part of the PTS phosphorelay that phosphorylates incoming carbohydrates. Many PRD-containing activators possess EIIB domains at their C-terminus, and recently, the EIIBGat-like domain of the B. subtilis MtlR transcriptional activator was shown to modulate protein activity via phosphorylation of a conserved cysteine that is characteristic of EIIB domains (Greenberg et al., 2002, Joyet et al., 2010). However, a protein alignment reveals that Mga lacks the conserved cysteine and instead possesses a glutamate (Glu 413, Fig. S4). This residue could conceivably serve as a phosphomimetic and play a role in Mga activity, possibly via oligomerization or interaction with other PTS proteins. However, the C-terminal region of Mga likely does not function as a typical EIIB domain that modulates activity via phosphorylation. Nevertheless, the domain structure of Mga appears to be remarkably similar to that of the MtlR sugar regulator, with two PRDs and an EIIBGat-like domain that follow an N-terminal DNA binding domain. Only the C-terminal EIIAMtl domain of MtlR is missing in Mga and its orthologs (Joyet et al., 2010). Interestingly, the E. coli EIIB subunit (GatB) was found to be structurally similar to E. coli CheY (Volpon et al., 2006). Hence, structural analyses coupled with the results of the truncation experiments suggest that the conserved C-terminal region of Mga may possess an EIIBGat-like fold rather than the traditionally held CheY-like receiver domain (Perez-Casal et al., 1991).
The importance of the C-terminal region of Mga is highlighted by newly published studies of the homologous B. anthracis regulator AtxA, in which removal of the C-terminal EIIB-like domain was reported to inactivate the protein and prevent oligomerization (Hammerstrom et al., 2011). Interestingly, AtxA was also found to form oligomers and a role for the C-terminus in multimerization was proposed. Thus, the finding that the EIIB-like domain of Mga impacts the oligomeric status of the GAS protein suggests that the C-terminus of Mga and AtxA have similar functions.
Subsequent to the completion of this study, the crystal structure of a putative Mga family transcriptional regulator from Enterococcus faecalis was deposited in the Protein Data Bank (PDB) of established protein structures (Osipiuk, 2011). The protein was crystallized as a dimer and, as predicted for GAS Mga, the C-terminus folds into an EIIB domain (Fig. S5). Remarkably, the C-termini of the E. faecalis protein are swapped such that each EIIB domain hooks into the opposing monomer to form the dimeric complex. Removal of the EIIB domain would be expected to generate a monomeric protein, precisely as was found with GAS Mga. While EIIB domains are known to interact with EIIA and EIIC proteins, this study suggests an additional role for the EIIB domains of Mga and orthologous regulators in homooligomerization.
It is significant that biochemical characterization and truncation analyses of the divergent M1 Mga (mga-1) gave analogous results to those obtained with the M4 Mga (mga-2), indicating that these are conserved mechanistic features. Prior to this study, the only criteria known to be important for Mga activity in GAS was the ability to bind target promoters. However, our results indicate that oligomerization is a critical facet of a likely complex mechanism extending beyond simple binding and activation. Thus the results of this study provide a critical foundation for further analyses of a novel group of regulators that resemble PTS carbohydrate regulators yet function to control virulence in Gram-positive pathogens.
EXPERIMENTAL PROCEDURES
Detailed additional experimental procedures related to plasmid and strain constructions, protein purifications, metal binding assays, lysate preparations, antibody production, limited tryptic digestion, and primer and strain tables can be found in the supporting material.
Bacterial Strains and Media
GAS strain GA40634 is a clinical isolate of the M4 serotype and KSM547 is the isogenic strain that contains an insertion in the mga gene (Ribardo & McIver, 2006). KSM165L is a similar suicide-based inactivation of mga in the sequenced M1 SF370 (Almengor & McIver, 2004). Escherichia coli strain DH5α was used for cloning, and strains BL21[DE3]-Gold (Stratagene), HMS174[DE3] (Novagen), C41[DE3], and C43[DE3] (Miroux & Walker, 1996) were employed for expression of Mga proteins. GAS cultures were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY) and growth was monitored using a Klett-Summerson colorimeter (A filter). E. coli were grown in Luria-Bertani broth, except where indicated; ZYP-5052 autoinduction medium (Studier & Moffatt, 1986) was used for protein expression. Media was supplemented with the appropriate antibiotics at the following concentrations: 100 µg/ml ampicillin for E. coli, 100 µg/ml spectinomycin for both E. coli and GAS, 50 µg/ml kanamycin for E. coli and 300 µg/ml kanamycin for GAS.
Mass spectrometric analyses of Mga
Matrix-assisted laser desorption/ionization mass spectrometric (MALDI-MS) analysis of Mga4-His6 was performed at the University of Maryland Institute for Bioscience and Biotechnology Research (UM-IBBR) using a sinapinic acid matrix on an ABI Voyager DE instrument with BSA as a calibration reference. Mass spectrometry of Δ139Mga4-His6 and isolated tryptic fragments was performed at the University of Maryland Proteomics Core Facility. Spectra of m/z 400–2000 were acquired with a Thermo LTQ Orbitrap XL mass spectrometer with a resolution of 60,000 at m/z 400. After acquisition, mass spectra over the chromatography peak were averaged and deconvoluted using the program Xtract to obtain the molecular weight.
Western blot analysis
Proteins were separated by SDS-PAGE and then transferred to either nitrocellulose or PVDF. Membranes were blocked with 5% non-fat dry milk in PBS containing 0.5% Tween-20 (PBST). Primary antibody dilutions were as follows: polyclonal rabbit 〈-Mga4 antibody (1:1,000 to 1:2,000); rabbit α-Mga1 conjugated to HRP 1:1,000; mouse α-His monoclonal antibody (Novagen) 1:1,000; mouse α-Hsp60 monoclonal antibody (StressGen Biotechnologies Corp.) 1:2,500. Primary antibodies were incubated for 2 hrs and secondary antibodies were incubated for 1 hr, all at room temperature. Proteins were visualized using SuperSignal West Pico or Femto Chemiluminescent Substrates (Pierce) using a Fuji LAS3000 CCD Scanner.
Real-time RT-PCR
RNA was purified from at least three biological replicates of each strain. GAS cultures were grown to late log phase, cells were pelleted at 4°C, and RNA was isolated using a Triton X-100 method (Sung et al., 2003). Samples (~25 µg) were treated with 10–20 U TURBO DNase (Applied Biosystems) in the presence of 40 U RNase inhibitor (NEB) for 30 min at 37°C, then chloroform extracted and concentrated. The absence of contaminating DNA was verified by PCR analysis using spy1589-1a/spy1589-1b and bgaA-1a/bgaA-1b primers (Table S2) for M4 and M1 serotype, respectively. RNA integrity was analyzed on a formaldehyde-MOPS agarose gel. RNA (25 ng total) samples were added to a SYBR Green Master Mix (Applied Biosystems) containing 250 nM of each primer (Table S2) and 3 U multiscribe reverse transcriptase (Applied Biosystems). Each sample was assessed in triplicate alongside controls lacking reverse transcriptase. Prepared 96-well plates were processed on a Lightcycler 480 real-time PCR system (Roche) using the following cycling parameters: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, then 40 cycles of 95°C for 15 sec and 60°C for 1 min. The Lightcycler 480 software was used to analyze transcript levels in mutant strains compared to the wild type, relative to gyrA expression.
Filter binding assays
A double-stranded (ds) 49 bp oligomer containing the Mga-binding site of the Parp4 promoter was produced by annealing the single-stranded oligomers, Parp MBS 49mer S and Parp MBS 49mer AS (Table S2, Integrated DNA Technologies (IDT); PAGE purified). The two oligonucleotides (30 µM each) in 10 mM Tris, 5 mM MgCl2 were heated at 85°C for 5 min and then slowly cooled to room temperature. The ds 49 bp was 5’-labeled with [32P]-ATP using polynucleotide kinase (NEB) and then purified using G25 sephadex columns (Harvard Apparatus). The DNA concentration was determined spectrophotometrically using an ε260 of 760,136 M−1 cm−1 (calculated at http://biophysics.idtdna.com/UVSpectrum.html). Varying concentrations of Mga proteins were incubated with 0.1 nM [32P]-labeled double-stranded oligonucleotide in 1X BB (50 mM Na-HEPES, 100 mM NaCl, 5 mM MgCl2, pH 7.5) for 15 min at room temperature. Duplicate reactions were filtered through 0.45 µm nitrocellulose (Whatman, pre-equilibrated in 1X BB) using a 24-well slot blot apparatus, washed once with 1X BB, and exposed to a phosphorimager cassette. Bands were visualized using a FLA7000 phosphorimager (Fuji) and intensities were quantified with the MultiGauge software (Fuji). The average intensities were plotted versus the concentration of Mga and then analyzed using Kaleidagraph (Synergy Software) by the Hill equation: I = (Imax [Mga]n) /(Kd,appn + [Mga]n), where I represents the average intensity, Imax the maximum intensity, Kd,app is the apparent dissociation constant for half maximal binding, and n provides an estimate of the Hill coefficient.
Fluorescence titrations
An HPLC-purified 5’ carboxyfluorescein- (FAM-) labeled 49mer (Parp MBS 49 S, Table S2) duplexed to its complementary strand (Parp MBS 49 AS) was prepared by IDT. The oligomer concentration in water was estimated using a calculated ε260 of 788,140 M−1 cm−1 (based on ε260 for the duplex calculated as described above and ε260 of 28,000 M−1 cm−1 for FAM (Favicchio et al., 2009)). Fluorescence emission spectra (505 – 605 nm) were monitored on a QuantaMaster fluorimeter (Photon Technology International) using an excitation wavelength of 494 nm with 4 nm excitation and emission slits. Binding was assessed in a 1 cm quartz cuvette with constant stirring and maintained at 25°C with a Peltier thermal regulator. Increasing concentrations of purified Mga were added to a 2.5 ml solution of 1 nM FAM-labeled 49mer containing the Mga-binding site of the Parp4 promoter (FAM-Parp) in 1X BB. Following Mga addition, the solution was allowed to equilibrate at least 2 min until the emission spectrum stabilized. At each concentration, 3 spectra were obtained and the integrated emission intensity was determined from 515 to 540 nm using Kaleidagraph (Synergy Software). Results at each concentration were averaged (variation was 1% or less) and the buffer data were subtracted. The integrated emission intensity was then plotted versus the concentration of Mga and the data were analyzed as described above.
Gel filtration analysis
Mga proteins in 50 mM HEPES, 50 mM NaCitrate, pH 7.5 (HEPES/Citrate) were diluted 2-fold into HEPES/Citrate containing varying salts, such that the final concentration of salt was as indicated in Figure 3 (with 35 – 40 µM Mga4-His6 or 30 µM Δ139Mga4-His6). Samples were equilibrated on ice for 45 min and then filtered through a 0.22-µm filter unit (Millipore) to remove any particles. The diluted protein was injected onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in HEPES/Citrate and the salt of interest. Gel filtration experiments were performed on an ÄKTA purifier (GE Healthcare) with a flow rate of 0.5 ml/min, and the elution profile was monitored by the absorbance at 280 nm. The molecular weights of eluted proteins were estimated based on a calibration curve standard (high and low molecular weight gel filtration calibration kits, GE Healthcare).
Sedimentation Equilibrium Measurements
Sedimentation measurements were performed in an XLI analytical ultracentrifuge (Beckman Coulter) using cells equipped with 2-hole (3 mm or 1.2 cm path length) charcoal-filled epon centerpieces. Mga4-His6 or Δ139Mga4-His6 was first dialyzed overnight into HEPES/Citrate containing 100 mM NaCl at 4°C. Full-length Mga4-His6 prepared at 7.5, 20 and 30 µM was centrifuged at 14, 16, 18, 20, and 22K rpm and truncated Δ139Mga4-His6 prepared at 7.5 and 30 µM was centrifuged at 18, 20, and 22K rpm. The data were first analyzed using a single species model in WinNonlin (Johnson et al., 1981) to obtain the reduced buoyant molecular mass, σ, from which the molecular weight was calculated using the following equation:
where M is the molecular weight,v̄ is the partial specific volume obtained using SEDNTERP (http://www.rasmb.bbri.org (0.7437 cm3/g for Mga4-His6 and 0.7445 cm3/g for Δ139Mga4-His6), ρ is the buffer density, R is the gas constant, T is the temperature in Kelvin, and ω is the angular velocity. The data for the full-length protein, which yielded a molecular weight higher than that expected for the monomer, was further subjected to analysis using monomer-oligomer models.
Coimmunoprecipitation experiments
GAS strains (GA40634 or KSM547.4 containing a plasmid expressing the His-tagged truncated protein under the constitutive rpsL promoter) were grown to late log phase and 200 µl lysates were prepared using PlyC and DNAse as described in the supplemental material (Preparation of GAS lysates), except that cells were lysed in PBS instead of TN buffer. Lysate samples were diluted in SDS-PAGE loading buffer, and heat denatured. The remainder of the lysates (150 µl) were diluted with 150 µl PBS containing 1X complete EDTA-free protease inhibitors (Roche) and 10 µl mouse α-His monoclonal antibody (Novagen) was added. Samples were incubated 1 hr at 4°C with gentle rocking and then 40 µl Protein A agarose (50% slurry in PBS, CalBiochem) was added. After gently rocking for 1 hr at 4°C, samples were pelleted and the supernatant removed. The resin was washed with 500 µl PBS, resuspended in 40 µl SDS-PAGE loading buffer, and incubated for 10 min at 95°C. Both lysate and coimmunoprecipitation (coIP) samples were analyzed by western blotting as described, using the α-Mga4 polyclonal antibody. Because the efficiency of Δ47Mga4-His6 immunoprecipitation was considerably lower, 100 ml cultures of these GAS strains were grown. Samples were treated in the same manner, except twice the amount of PlyC and DNase were added so that the lysates were 10-fold more concentrated (215 µl total volume); for these samples, only half the volume was analyzed by immunoblotting (compared to other coIP samples).
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Stewart (University of Maryland) for assistance with the fluorescence titrations and helpful comments, O. Herzberg (University of Maryland Institute for Bioscience and Biotechnology Research, UM-IBBR) for assistance with analyzing the crystal structure, Y. Wang (University of Maryland Proteomics Core Facility) for mass spectrometric analyses, and J. Ladner (UM-IBBR) for MALDI-MS results. We are grateful to D. Nelson (UM-IBBR) for supplying the phage lysin PlyC. We thank V. Lee (University of Maryland), R. Blumenthal (University of Toledo), and Z. Kelman (UM-IBBR) for critical review of this manuscript.
REFERENCES
- Almengor AC, McIver KS. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J Bacteriol. 2004;186:7847–7857. doi: 10.1128/JB.186.23.7847-7857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Walters MS, McIver KS. Mga is sufficient to activate transcription in vitro of sof/sfbX and other Mga-regulated virulence genes in the group A streptococcus. J Bacteriol. 2006;188:2038–2047. doi: 10.1128/JB.188.6.2038-2047.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson G, McIver K, Heden LO, Scott JR. Complementation of divergent mga genes in group A streptococcus. Gene. 1996;175:77–81. doi: 10.1016/0378-1119(96)00124-2. [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Bessen DE, Manoharan A, Luo F, Wertz JE, Robinson DA. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J Bacteriol. 2005;187:4163–4172. doi: 10.1128/JB.187.12.4163-4172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- Boffi A, Vecchini P, Chiancone E. Anion-linked polymerization of the tetrameric hemoglobin from Scapharca inaequivalvis Characterization and functional relevance. J Biol Chem. 1990;265:6203–6209. [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry (Mosc) 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favicchio R, Dragan AI, Kneale GG, Read CM. Fluorescence spectroscopy and anisotropy in the analysis of DNA-protein interactions. In: Moss T, Leblanc B, editors. Methods in molecular biology, DNA-protein interactions. New York: Humana Press; 2009. pp. 589–611. [DOI] [PubMed] [Google Scholar]
- Gjerde DT, Schmuckler G, Fritz JS. Anion chromatography with low-conductivity eluents. II. J Chromatogr A. 1980;187:35–45. [Google Scholar]
- Gokarn YR, Fesinmeyer RM, Saluja A, Cao S, Dankberg J, Goetze A, Remmele RL, Jr, Narhi LO, Brems DN. Ion-specific modulation of protein interactions: anion-induced, reversible oligomerization of a fusion protein. Protein Sci. 2009;18:169–179. doi: 10.1002/pro.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Takahashi N, Fink AL. Mechanism of acid-induced folding of proteins. Biochemistry (Mosc) 1990;29:3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- Greenberg DB, Stulke J, Saier MH., Jr Domain analysis of transcriptional regulators bearing PTS regulatory domains. Res Microbiol. 2002;153:519–526. doi: 10.1016/s0923-2508(02)01362-1. [DOI] [PubMed] [Google Scholar]
- Gregor HP, Belle J, Marcus RA. Studies on ion-exchange resins. XIII. Selectivity coefficients of quaternary base anion-exchange resins toward univalent anions. J Am Chem Soc. 1955;77:2713–2719. [Google Scholar]
- Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO(2) -signalling. Mol Microbiol. 2011;82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead SK, Arnold J, Readdy TL, Bessen DE. Molecular evolution of a multigene family in group A streptococci. Mol Biol Evol. 1994;11:208–219. doi: 10.1093/oxfordjournals.molbev.a040103. [DOI] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Jain S, Udgaonkar JB. Salt-induced modulation of the pathway of amyloid fibril formation by the mouse prion protein. Biochemistry (Mosc) 2010;49:7615–7624. doi: 10.1021/bi100745j. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyet P, Derkaoui M, Poncet S, Deutscher J. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Mol Microbiol. 2010;76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Maity H, Mossing MC, Eftink MR. Equilibrium unfolding of dimeric and engineered monomeric forms of lambda Cro (F58W) repressor and the effect of added salts: evidence for the formation of folded monomer induced by sodium perchlorate. Arch Biochem Biophys. 2005;434:93–107. doi: 10.1016/j.abb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Manival X, Yang Y, Strub MP, Kochoyan M, Steinmetz M, Aymerich S. From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins. EMBO J. 1997;16:5019–5029. doi: 10.1093/emboj/16.16.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS. Stand-alone response regulators controlling global virulence networks in streptococcus pyogenes. Contrib Microbiol. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- McIver KS, Heath AS, Green BD, Scott JR. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS, Myles RL. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol Microbiol. 2002;43:1591–1602. doi: 10.1046/j.1365-2958.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- McIver KS, Thurman AS, Scott JR. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J Bacteriol. 1999;181:5373–5383. doi: 10.1128/jb.181.17.5373-5383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Musser JM, DeLeo FR. Toward a genome-wide systems biology analysis of host-pathogen interactions in group A Streptococcus. Am J Pathol. 2005;167:1461–1472. doi: 10.1016/S0002-9440(10)61232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzammil S, Kumar Y, Tayyab S. Anion-induced stabilization of human serum albumin prevents the formation of intermediate during urea denaturation. Proteins. 2000;40:29–38. doi: 10.1002/(sici)1097-0134(20000701)40:1<29::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Okada N, Geist RT, Caparon MG. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Osipiuk J, Wu R, Jedrzejczak R, Moy S, Joachimiak A. In: Structure for the Putative Mga family transcriptional regulator from Enterococcus faecalis. M. C. f. S. G. (MCSG), editor. Protein Data Bank (PDB); 2011. pp. [Google Scholar]
- Perez-Casal J, Caparon MG, Scott JR. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao Ch M, Goto Y. Critical balance of electrostatic and hydrophobic interactions is required for beta 2-microglobulin amyloid fibril growth and stability. Biochemistry (Mosc) 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- Ramos CHI, Baldwin RL. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 2002;11:1771–1778. doi: 10.1110/ps.0205902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Oobatake M, Goto Y. Salt-dependent monomer-dimer equilibrium of bovine beta-lactoglobulin at pH 3. Protein Sci. 2001;10:2325–2335. doi: 10.1110/ps.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Sung K, Khan SA, Nawaz MS, Khan AA. A simple and efficient Triton X-100 boiling and chloroform extraction method of RNA isolation from Gram-positive and Gram-negative bacteria. FEMS Microbiol Lett. 2003;229:97–101. doi: 10.1016/S0378-1097(03)00791-2. [DOI] [PubMed] [Google Scholar]
- Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 2007;15:318–325. doi: 10.1016/j.tim.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Vahling CM, McIver KS. Domains required for transcriptional activation show conservation in the Mga family of virulence gene regulators. J Bacteriol. 2006;188:863–873. doi: 10.1128/JB.188.3.863-873.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Young CR, Matte A, Gehring K. NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA. Protein Sci. 2006;15:2435–2441. doi: 10.1110/ps.062337406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.