Abstract
Background
Network analysis techniques allow a more accurate reflection of underlying systems biology to be realized than traditional unidimensional molecular biology approaches. Here, using gene coexpression network analysis, we define the gene expression network topology of cardiac hypertrophy and failure and the extent of recapitulation of fetal gene expression programs in failing and hypertrophied adult myocardium.
Methods and Results
We assembled all myocardial transcript data in the Gene Expression Omnibus (n = 1617). Since hierarchical analysis revealed species had primacy over disease clustering, we focused this analysis on the most complete (murine) dataset (n = 478). Using gene coexpression network analysis, we derived functional modules, regulatory mediators and higher order topological relationships between genes and identified 50 gene co-expression modules in developing myocardium that were not present in normal adult tissue. We found that known gene expression markers of myocardial adaptation were members of upregulated modules but not hub genes. We identified ZIC2 as a novel transcription factor associated with coexpression modules common to developing and failing myocardium. Of 50 fetal gene co-expression modules, three (6%) were reproduced in hypertrophied myocardium and seven (14%) were reproduced in failing myocardium. One fetal module was common to both failing and hypertrophied myocardium.
Conclusions
Network modeling allows systems analysis of cardiovascular development and disease. While we did not find evidence for a global coordinated program of fetal gene expression in adult myocardial adaptation, our analysis revealed specific gene expression modules active during both development and disease and specific candidates for their regulation.
Keywords: fetal, gene expression, heart failure, hypertrophy, myocardium
Despite advances in diagnosis and management, the clinical syndrome of heart failure remains a leading cause of hospitalization and death in developed countries with a worse five year prognosis than most cancers1. Disparate mechanisms of cardiac stress invoke a common pathway of adaptive myocyte hypertrophy which progresses to impaired systolic function and dilated, thinned myocardium. Expression of several genes is known to differ reproducibly between normal adult myocardium and failing and hypertrophied myocardium. Some of these differentially expressed genes, such as cardiac muscle alpha-myosin heavy chain 6 (MYH6) and cardiac muscle beta-myosin heavy chain 7 (MYH7), comprise the contractile apparatus of the sarcomere. Others, such as sarcoplasmic reticulum calcium ATPase 2 (ATP2A2), phospholamban (PLN), and solute carrier family 2, facilitated glucose transporter member 4 (SLC2A4), are principally involved in myocardial calcium cycling and energetics2–7. It has thus been proposed that myocardial adaptation involves myosin switch and a switch from fatty acid oxidation to glycolysis as the main metabolic pathway of the cardiomyocyte. Similar differential gene expression has been observed in fetal myocardium, leading to a hypothesis that there is a coordinated gene program that overlaps between fetal and failing and hypertrophied myocardium5. It is, however, unclear whether such changes represent a truly coordinated biological response mediated by common regulators.
The incomplete understanding of these shared genomic response pathways is largely due to statistical and computational limitations inherent to traditional differential expression experiments, in which the expression of a pre-defined set of genes is compared between two different phenotypes. These limitations have been compounded by small sample sizes in previous microarray experiments and have precluded prior unbiased genome-wide evaluation of this hypothesis. Network based approaches can be used to identify patterns of shared gene expression, regulation, or protein interaction while limiting the number of statistical comparisons and thus the false discovery rate8, 9. We have previously used literature derived networks to reveal potential targets for abrogation of restenotic atherosclerotic coronary artery disease and in doing so recapitulated targets of known therapeutic agents. Other investigators have used gene coxpression analysis to uncover novel pathways regulated by disease susceptibility genes10. Candidate causal gene relationships have subsequently been validated with high fidelity in transgenic and gene knock-down experiments11, 12.
In this study we used unbiased marginal module analysis of gene coexpression networks13 from all murine myocardial transcript abundance data available in the published literature to formally evaluate the hypotheses that there are gene programs unique to fetal myocardium and these gene programs are re-capitulated in myocardial adaptation.
Methods
Data acquisition and normalization
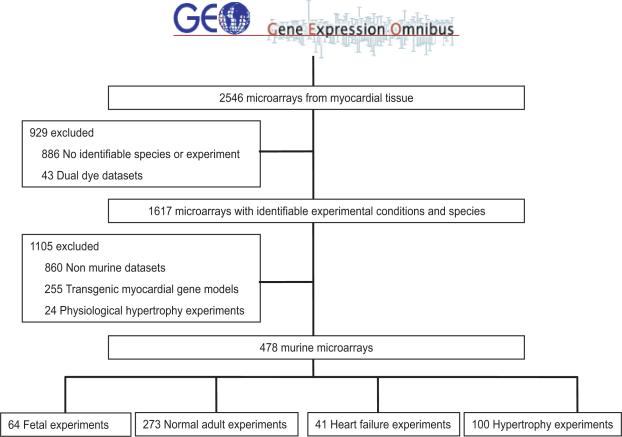
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) is a publicly available repository of all microarray data that are presented in the published literature. This database was queried using the terms heart, myocardium, and cardiovascular to obtain all datasets describing microarray experiments involving myocardial tissue (n = 2546). Because absolute expression data is required for weighted gene coexpression network analysis, log transformed dual dye datasets were excluded (n = 43). Of these datasets, 1617 contained information regarding experimental conditions, species and a phenotype identifiable as developmental (fetal or post natal days 0 to 10 without experimental manipulation of myocardium), normal adult (adult tissue with no experimental manipulation of myocardium), hypertrophy (histologically or anatomically identifiable cardiac hypertrophy), or heart failure (clinical syndrome of heart failure and impaired cardiac systolic and/or diastolic dysfunction). Data combination and normalization was performed as described in the Supplementary methods to allow comparison of gene expression data between different platforms and experimental conditions. Summary statistics showing minimization of bias with this method of normalization are provided in Supplementary Figure 1. As shown in Supplementary Figure 2, gene array experiments clustered by species, suggesting a species dependency for gene coexpression. Therefore, we focused our analysis on the species with the most complete dataset, choosing the 757 arrays performed in mouse myocardium. An additional 255 experiments involved transgenic deletion, insertion, or mutation of genes important to myocardial function and/or myocardial gene expression and were thus excluded. As physiological hypertrophy is a state distinct from pathological hypertrophy that does not progress to heart failure, 24 datasets describing physiological hypertrophy induced by chronic exercise were excluded (Figure 1). The final dataset contained 12,620 genes represented in 478 samples.
Figure 1.
Myocardial gene microarray expression data used in analysis. All murine microarray data from the Gene Expression Omnibus (GEO) database were queried with the search terms heart, myocardium, and cardiovascular.
Weighted gene coexpression network analysis
Weighted gene coexpression network analysis was performed to identify gene coexpression modules, that is, groups of genes with similar patterns of expression across experimental samples14. Gene coexpression networks are defined in this work as a weighted graph in which genes are represented as vertices and connections strengths, or adjacencies, are represented as edges between vertices. We first identified these gene modules in fetal myocardial tissue and then evaluated their reproducibility in normal adult, hypertrophied, and failing myocardial tissue samples. We derived gene modules in a training set of 43 randomly chosen fetal samples (67% of all fetal samples). The remaining 21 fetal samples used for module validation were not used for this initial module identification. To carry out module detection we used agglomerative hierarchical clustering of adjacencies given by the topological overlap measure. The topological overlap measure is a network adjacency based on shared network neighbors, which is in turn based on weighted Pearson correlation between gene expression profiles as described in the Supplement. This network adjacency is robust to noise and amplifies connectivity patterns in sparse networks.15 Separate adjacencies were constructed for each phenotype considered. To cut branches of the tree into gene modules, we used the dynamic tree cut algorithm which iteratively searches for stable branch size and chooses clusters based on the shape of each tree branch16. This algorithm allows manipulation of several parameters controlling resultant cluster size and cohesiveness; sensitivity analysis as described in the Supplement was performed for each of these parameters. The modules from the fetal network were then carried forward for all subsequent analysis. Intramodular connectivity kin was defined for each gene as the sum of adjacencies with all other genes in the module to which it was assigned. A hub gene was then identified for each module as the gene with the single greatest kin. We further assessed module composition according to defined gene ontologies (GO) using the DAVID analysis tool17,18. All p values reported for GO analysis were corrected for multiple comparisons using the Benjamini correction and both corrected and uncorrected p values are reported. Networks were visualized using the visANT network visualization tool19.
Evaluating reproducibility of gene modules
To evaluate reproducibility of modules between networks, we calculated the extent of intramodular connectivity preservation. To this end we calculated for each module the correlation Mpres=cor(kl,km) between intramodular connectivity vectors kl and km. These vectors described intramodular connectivities kin for genes within each module across two networks Nl and Nm The closer Mpres is to 1, the better preserved is the module structure across networks Nl and Nm. An iterative significance test was used to assign a p value to this measure of module reproducibility between networks Nl and Nm. The null hypothesis for this significance test was that derived modules were preserved between networks Nl and Nm no better than modules derived from random clustering. To evaluate this hypothesis, gene module identifiers were randomly permuted such that gene modules of the same size but random gene composition were generated. Ten thousand such permutations were performed and Mrand, the module preservation of each random module assignment, was calculated for each permutation. The probability of the null hypothesis p was then calculated as the proportion of permutations in which the preservation of random modules between networks Nl and Nm was greater than that of derived modules (Mrand > Mpres). This significance test was used for module internal validation and evaluation of fetal module reproducibility in adult normal, failing, and hypertrophied myocardial tissue.
Cluster validation
Clusters were first internally validated in 21 samples not used for module derivation. To prevent spurious inference of association from repeated sampling across the set of gene modules, the Bonferroni correction was employed in each comparison to determine significance of p values derived via permutation derived significance testing described above. Thus, module internal validity was inferred from a p value of less than 5.5 × 10−4 for 90 comparisons.
Conservation of fetal modules in normal adults, heart failure, and hypertrophy
Random permutation was again used to assess the reproducibility of fetal modules in normal adult datasets. Modules with p > 6.9× 10−4 (corrected for 72 comparisons) were defined as specific to fetal tissue. To identify additional modules with conserved topology but significant differential expression between fetal and normal adult myocardium, we used significance analysis of microarrays to evaluate gene-wise differential expression between fetal and normal adult myocardium.20 A d-score was assigned to each gene that quantified the significance of its differential expression between phenotypes. For each module, an average d score was calculated by dividing this aggregate d score by the number of genes in the module. A p value was assigned to this average d score via a permutation-derived significance test. The p value for each average modular d score was given by the proportion of ten thousand permutations in which random gene modules of the same size were associated with a greater average d score than derived clusters. Significant modular differential expression was inferred from p < 0.002 (corrected for 26 comparisons). This set of modules together with the set of modules with topology specific to fetal tissue was thus defined as the “fetal gene program” and evaluated further for reproducibility in hypertrophied and failing myocardial tissue using the iterative module reproducibility algorithm described above. Fetal module reproducibility in heart failure and hypertrophy was inferred from a p value of 0.001 for 50 comparisons.
Transcription factor analysis
We next analyzed gene coexpression modules for over-representation of known transcription factor targets. Each gene g was linked to a transcription factor if it was identified as a target of that transcription factor as described in the Supplementary methods. For each transcription factor t in each co-expression module, we tested the hypothesis that transcription factor t was over-represented in the list of genes comprising the module when compared to what was expected by chance alone. This association was measured using the hypergeometric distribution as the probability of observing k or more genes targeted by t transcription factors in the set of n genes, when there are K genes targeted by transcription factor t in the whole set of N genes. Given that 56 total transcription factors were analyzed in the module shared between fetal and hypertrophied and failing myocardium, significance of association was inferred from a p value less than 9 × 10−24.
Module membership of key genes and higher order topology of fetal gene transcriptome
To summarize the scaled gene expression profiles modules, we used the first singular vector (referred to as the module eigengene) that resulted from singular value decomposition. The module eigengene is equivalent to the first principal component and explains the largest proportion of the variance of the module genes. Based on the correlations between the module eigengenes of different modules, we constructed an eigengene network that can be used to assess higher order topology of gene coexpression21. In this way the gene coexpression network was reduced from thousands of genes to several representative eigengenes that in turn form nodes in a meta-network describing higher-order topology of the transcriptome. Similar eigengenes were calculated for modules in hypertrophied and failing myocardium. To evaluate higher-order reproducibility of fetal gene modules in myocardial adaptation, an adjacency matrix was calculated for eigengene networks in each phenotype considered via the Pearson correlation between eigengenes across all microarrays representing each phenotype. Preservation of inter-modular connections was then assessed via the preservation density, which was calculated as follows:
| (3) |
Where D is the preservation density, and are adjacencies for each phenotype, and n is the total number of genes. The closer the preservation density is to 1, the stronger the evidence that the connectivity pattern is preserved between the two networks.
A measure of module membership kMEi=cor(xi, ME) can be defined as the correlation between expression profile of gene i and the module eigengene. The closer the absolute value of kMEi is to 1, the stronger the evidence that the gene belongs to the module represented by the module eigengene22. Therefore, marker genes not assigned to fetal modules (MYH6, PLN) were re-assigned to the module with highest kME. Module membership of other marker genes (NPPA, MYH7, SLC2A4) was simply the module assignment of each respective gene as given by the algorithm described previously.
Results
Network topology can be defined for large datasets of myocardial gene expression data
We first defined fetal gene programs as modules of gene co-expression specific to fetal myocardial tissue. To this end, we constructed weighted gene co-expression networks for fetal myocardial tissue using every murine transcript abundance dataset available in the Gene Expression Omnibus (GEO) database. Our data search strategy and summary of experimental conditions is presented in Figure 1. Modules were internally validated using a data-splitting technique in which 43 (67%) samples were used as a training set and 21 (33%) samples were used to validate the derived modules. In the training set, hierarchical clustering of the topological overlap matrix identified 90 modules composed of between 25 and 1309 genes. A summary of analyzed genes and modules is presented in the Supplement. The preservation of all gene-gene connections between training and validation datasets as defined by the preservation density was 94%. Of the 90 original modules, 72 had network topology that was significantly preserved in the validation set (p < 5.5 × 10−4 for reproducibility) and were carried forward for all subsequent analysis.
Gene expression in the developing heart involves modules not conserved in normal adults
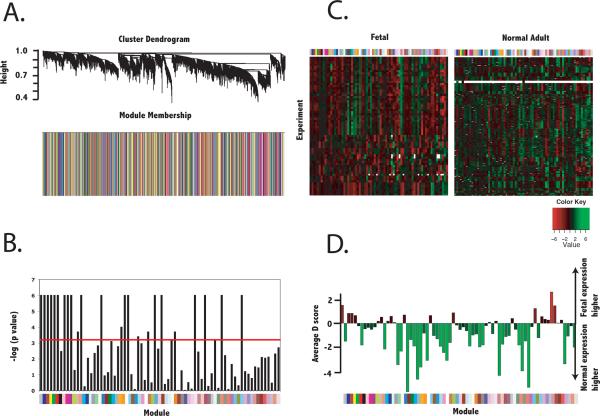
Because there are modules of gene expression that represent constitutive genetic programs of cardiac tissue, we next identified the subset of these validated modules that were not reproduced in normal adult myocardium (n = 273 samples). Of the 72 valid modules identified in fetal myocardium, 26 were also found in normal myocardium (p < 6.9× 10−4 for reproducibility between fetal and normal datasets, Figure 2). Four of these 26 modules were reproduced in adult myocardium but also exhibited significant differential expression (p value for average differential expression < 0.002) as a module. Thus, 50 modules of between 25 and 914 genes were found to comprise the gene expression program of the developing heart.
Figure 2.
Identification of gene coexpression modules specific to fetal tissue. Gene-gene adjacencies were first defined by the topological overlap, which quantifies the degree of shared network neighbors given by gene-wise Pearson correlation. Hierarchical clustering of adjacencies was first performed in the training dataset of 43 fetal samples and resulting coexpression modules were evaluated for significant reproducibility in the validation dataset of 21 fetal samples. The seventy-two valid fetal modules were evaluated for reproducibility in normal adult myocardium as well as differential expression. A.) Cluster dendrogram and barplot describing module membership (according to clusters derived in the training fetal dataset) for normal adult myocardium. B.) Reproducibility of valid fetal modules in normal adult myocardium. Colors below the bargraph correspond to the module membership. Red line indicates cutoff for statistical significance (p < 6.9× 10−4 or − log(p value) > 3.16). Modules below this cutoff were defined as having topology specific to fetal myocardium and defined as fetal modules. C.) Heatmap of average expression of each module in fetal and normal adult myocardium. Colors above the heatmaps correspond to module membership. D.) Average significance level of differential expression (average Significance Analysis of Microarrays d score) for each module. Negative values correspond to lower expression levels in normal than fetal myocardium.
Known marker genes for heart failure are members but not hubs of conserved modules
We next examined the composition of these conserved modules using annotated gene ontologies. Topology of gene modules containing key marker genes is described in Supplementary Figure 7. Matching expected results, MYH7 and NPPA were members of a module up-regulated in fetal tissue compared to normal tissue (156 members, average d score 4.66). SLC2A4 was a member of a module down-regulated in fetal compared to normal tissue (64 members, average d score −4.95). Though not contained in modules specific to fetal cardiac tissue, MYH6 and PLN expression most closely aligned with members of a module down-regulated in fetal compared to normal tissue (90 members, average d score −1.6).
Limited recapitulation of fetal modules in failing myocardium
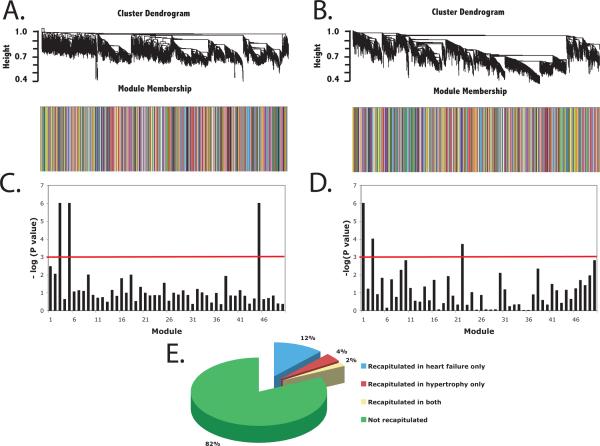
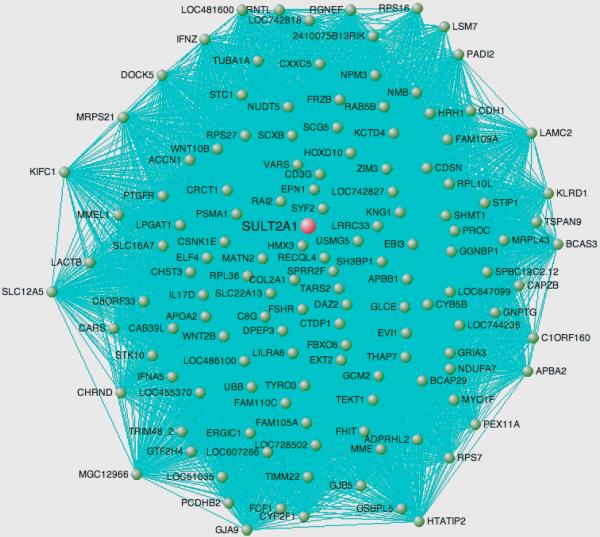
We next evaluated the hypothesis that fetal gene modules are re-capitulated in failing (n=41 samples) and pathologically hypertrophied myocardial tissue (n = 100 samples). These data are summarized in Figure 3 and in tabular form in the Supplement. Seven (14%) of 50 fetal gene modules were reproduced in failing myocardium with p < 0.001. Three (6%) of 50 fetal gene modules were significantly reproduced in pathological hypertrophy. One module (Figure 4) composed of 246 genes was reproduced in both failing and hypertrophied myocardium. The hub gene for this module, dihydroepiandrosterone sulfotransferase (SULT2A1) encodes an enzyme with high levels of expression in heart, adrenal glands, and liver and catalyzes the conjugation of steroids and bile acids23, 24.
Figure 3.
Reproducibility of developmental gene modules in heart failure and cardiac hypertrophy. The modules unique to fetal myocardium were evaluated for reproducibility in heart failure and cardiac hypertrophy. Cluster dendrograms and barplots describe module membership (according to clusters derived in the training fetal dataset) for A.) myocardium in murine heart failure and B.) myocardium in murine cardiac hypertrophy. Bargraphs describe significance of module reproducibility in C.) heart failure and D.) cardiac hypertrophy. Colors below the bargraphs correspond to the module membership. Red line indicates cutoff for statistical significance (p < 0.001 or − log(p value) > 3). E.) Proportion of fetal gene modules recapitulated in heart failure only, cardiac hypertrophy only, both, or neither.
Figure 4.
Topology of gene coexpression module common to developing myocardium and hypertrophied and failing myocardium. The hub gene is colored red and all nodes are colored green. Only edges with adjacency weight > 0.7 are shown for clarity. The proximity of each gene to the center of the figure indicates its connectivity, or sum of connection weights. Genes with connectivity < 50 are omitted for clarity.
Transcription factors are significantly over-represented in discovered modules
We next analyzed over-representation of transcription factor targets in each module. Targets of the transcription factor zinc finger protein 2 (ZIC2, product of gene ZIC2) were significantly over-represented in the fetal module common to both failing and hypertrophied myocardium (p = 3 × 10−4) after correcting for multiple comparisons. Fetal modules reproduced in heart failure were enriched in targets of transcription factor HOXA5 (p = 3 × 10−5) and fetal modules reproduced in hypertrophy were enriched in targets of the transcription factor FOXN1 (p = 4 × 10−4).
Ontology analysis reveals key role of metabolic processes in fetal and failing myocardium
Common biological process pathways were found among fetal and failing myocardium. In the fetal module reproduced in heart failure and cardiac hypertrophy, proteasomal protein catabolic processes (p = 5.9 × 10−5, corrected p = 0.085) and protein catabolic processes (p = 4.2 × 10−4, corrected p = 0.085) were overrepresented. Other biological processes overrepresented in gene modules common to developing and failing myocardium (n = 7) were cytoskeleton organization and biogenesis (corrected p =8.5 × 10−3) and fatty acid oxidation (corrected p =9.2 × 10−3). Fetal modules recapitulated in hypertrophy (n = 3) were enriched in genes involved in membrane lipid metabolic processes (corrected p =0.02). Other fetal modules with significant enrichment for biological processes were involved in stress response, nervous system development, biopolymer metabolism, and innate and humoral immunity (corrected p < 0.05).
Higher order topological overlap is poorly preserved between fetal and failing myocardium
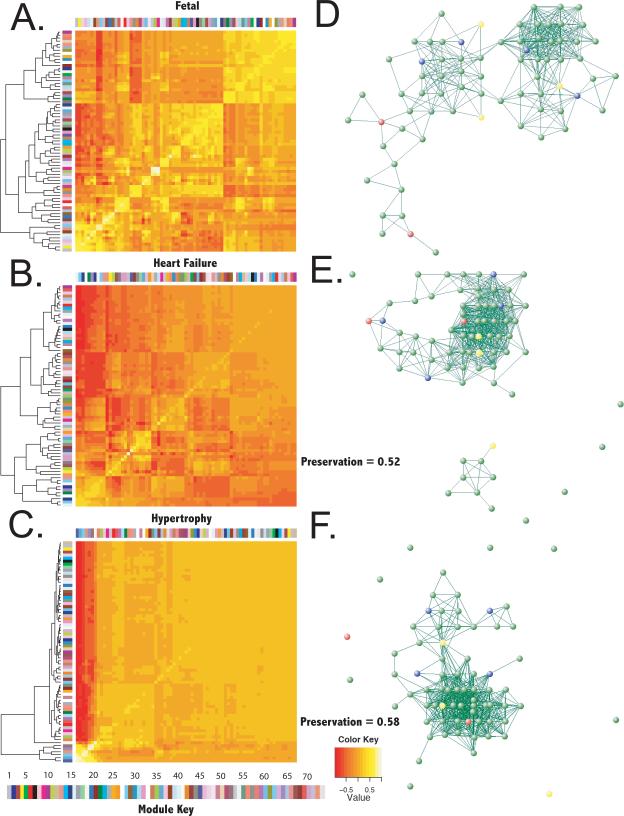
Eigengene networks reveal connectivity patterns between modules, i.e. they allow one to study higher order topology of the transcriptome and relationships between biological pathways represented in each gene module. Derived eigengenes explained between 32% and 67% of the variance in gene expression for each module. Preservation of higher order topology of the module network is presented in Figure 5 in both graphical form and as cluster dendrograms. The preservation of fetal inter-modular connections in failing and hypertrophied myocardial tissue was low (52% and 58%, respectively).
Figure 5.
Reproducibility of higher-order network topology between fetal myocardium and myocardium in murine heart failure and cardiac hypertrophy. Singular value decomposition was used to generate a representative eigengene for each fetal module that described most of the variance in modular gene expression. Hierarchical clustering of eigengenes was then used to identify inter-modular connections. Heatmaps describe the correlation between eigengenes and the dendrograms represent clustering of module eigengenes in A.) fetal myocardium, B.) myocardium in murine heart failure, and C.) myocardium in cardiac hypertrophy. Colors to the left and top of each heatmap correspond to each module and the color of each block represents the pairwise correlation between eigengene expression across all experiments (thus the line of identity has correlation = 1 for all modules). The color bar at bottom maps colors to eigengene identifiers (i.e., 1 = ME1). Graphical representation of eigengene networks is represented for fetal myocardium in D.), heart failure in E.), and cardiac hypertrophy in F.). Nodes colored red were reproduced in both heart failure and cardiac hypertrophy, nodes colored yellow were reproduced in heart failure only and those colored blue were reproduced in hypertrophy only. Values given for preservation refer to preservation of fetal inter-modular connections in heart failure and cardiac hypertrophy.
Discussion
In this study, we applied network analysis tools to the largest dataset of myocardial transcriptomic data assembled to date. We identified and validated modular sets of genes with a high degree of coexpression in developing myocardial tissue and tested their conservation across different pathological states. Specially, we addressed the question of whether a coordinated fetal gene expression program is recapitulated in heart failure and hypertrophy.
In traditional differential gene expression analysis, investigators have applied statistical tools that were developed for evaluation of associations between a small number of predictors (i.e., clinical characteristics or medical interventions) and a large number of outcomes (i.e., mortality). However, these analytical approaches are often inadequate to describe the associations between the large number of gene expression signatures and relatively small number of phenotype experiments in genome-wide expression studies. Identification of a relatively small number of modules and subsequent module-wise, rather than gene-wise, analysis yields a lower false discovery rate than would be expected with more traditional analyses. Modular gene expression analysis also facilitated evaluation of higher-order topology of the transcriptome and associated transcriptional regulators in heart failure and developing myocardium.
Using these analytical tools, we found that a significant proportion of modular gene programs are either unique to fetal cardiac tissue or present but differentially expressed in normal adult myocardium. Analysis of the composition of these fetal gene modules corroborated prior evidence of differences in myocardial energetic pathways and sarcomeric composition between fetal and normal adult myocardium2–5. Furthermore, we identified fetal modules involved in membrane lipid metabolism, nervous system development, and innate and humoral immunity whose importance to developing myocardium, if confirmed in experimental model systems, enriches our understanding of myocardial development.
We also found that key fetal pathways of myocardial energetics previously demonstrated to be recapitulated in myocardial adaptation25 were represented in gene modules that were preserved between fetal and failing and hypertrophied myocardium. Novel pathways that were shared between developing and failing/hypertrophied myocardium were also identified in our analysis and may represent key stress response pathways important to both the fetal circulation and myocardial adaptation. It is yet unclear which of these pathways may represent maladaptive cardiovascular stress responses.
Surprisingly this set of recapitulated modules represented a small proportion of the identified fetal gene program and there was little overlap between fetal modules recapitulated in heart failure and those recapitulated in cardiac hypertrophy. These findings have important implications for the understanding of myocardial adaptation to injury. The hypothesis that a coordinated program of developmental gene expression is recapitulated in myocardial adaptation is widely accepted5, 7, 26. However, our results suggest that this model of myocardial adaptation is inadequate to describe the genomic response to myocardial stress. Instead, the spectrum of myocardial adaptation in which compensatory hypertrophy progresses to myocardial failure appears to involve diverse and plastic pathways of genomic response which are not easily distilled to one coordinated program of fetal gene expression. Furthermore, we found that the higher order organization of genomic response, represented by connections between modules of shared gene expression, was equally disparate between developing, failing and hypertrophied myocardium.
Several regulators of expression, including transcriptional activators and repressors, histone deacetylases, and microRNAs, have been shown to modulate transcription and translation of portions of the gene expression program of the developing heart27–30 and heart failure and hypertrophy31. Here we identified a novel association of the transcription factor ZIC2 with gene coexpression modules common to myocardial development and adult myocardial adapation. Loss of function mutations in this transcription factor are associated with a defect of forebrain development, holoprosencephaly, complex skeletal defects and congenital heart disease32. Modules common to developing and failing myocardium were enriched in targets of HOXA5, a transcription factor with known roles in lung development in mammals33, 34 and apoptotic heart morphogenesis in amphibians35. Targets of the transcription factor FOXN1, which is known to be involved in thymic development and lymphocyte regulation, were found in modules shared between developing and hypertrophied myocardium36. This transcription factor is involved in cardiomyocte-rich embryoid body development in pharyngeal endoderm37 and other members of the forkhead box transcription factor family have been implicated in human and murine heart failure38. In this study we present evidence of the role of these transcription factors in modulation of the gene programs common to developing myocardium and myocardial adaptation. With experimental confirmation of the role of these transcription factors in modulation of gene expression programs of the developing heart and identification of additional common transcription factor and microRNA targets, it may be possible to identify regulators of fetal gene networks that allow for manipulation of maladaptive portions of the program.
Limitations
Our study has limitations. We evaluated myocardial expression data from disparate sources which utilized different experimental model systems and different tissue preparation, transcript isolation, and microarray techniques. However, this is also a strength of our study. It is unlikely that a single model from one laboratory adequately captures the diversity of disease. By concentrating on the findings common to many different models and techniques from many different laboratories, we are able to focus our attention on the most robust changes common to all. Further, normalization minimized between-array differences, while a module identification technique robust to noise was used with an iterative approach to validate modules.
Due to inadequate data from human samples, we concentrated this analysis on murine experimental models. As such, the generalizability of these findings to human samples remains to be demonstrated. Recent data has emphasized the importance of microRNA control of gene expression for heart failure31, 39. Our analysis did not address the potential regulatory role of these RNAs40.
Conclusion
In conclusion, we describe the gene expression topology of the developing and diseased heart. We identify key transcriptional regulators of the fetal gene expression program and apply novel module conservation algorithms to address the question of the recapitulation of fetal gene markers and gene networks in heart failure and hypertrophy. While marker genes are clearly upregulated in gene networks common to developing and diseased myocardium, they are not found to be the most highly connected genes. In addition, limited evidence was found for a coordinated recapitulation of gene expression programs found in the developing heart that is recapitulated in myocardial adaptation.
CLINICAL SUMMARY.
High-throughput techniques allow insight into differential regulation of genes in diverse disease states. Investigation of higher order interactions between genes in networks provides a rich context for exploring fundamental biological questions. Here, using advanced statistical methods for network analysis of the largest collection of myocardial gene expression data assembled to date, we test a fundamental hypothesis in cardiovascular biology, namely, whether a fetal gene expression program is recapitulated in the failing and hypertrophied adult heart. In doing so, we identified networks of genes in developing, failing, and hypertrophied myocardium that may be important to both the developing and failing heart. Limited evidence was found for a single, coordinated program of gene expression shared between the developing and failing heart. However, the definition of higher order organization of the myocardial transcriptome and novel potential transcriptional regulators enriches our understanding of the genomic basis of myocardial development and disease, and may someday lead to therapeutic interventions targeting maladaptive cardiac stress responses.
Supplementary Material
Acknowledgments
Sources of Funding Computational resources were provided by the Stanford BioX2 compute cluster, supported by NSF award CNS-0619926. Dr. Wheeler was supported by an individual National Research Service Award F32 fellowship (NIH award HL097462). Dr. Ashley was supported by NIH/NHLBI KO8 award HL083914 and NIH New Investigator DP2 award OD004613. This work was also partially funded by a grant from the Breetwoor Family Foundation.
Footnotes
Disclosures None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jhund PS, Macintyre K, Simpson CR, Lewsey JD, Stewart S, Redpath A, Chalmers JW, Capewell S, McMurray JJ. Long-term trends in first hospitalization for heart failure and subsequent survival between 1986 and 2003: a population study of 5.1 million people. Circulation. 2009;119:515–523. doi: 10.1161/CIRCULATIONAHA.108.812172. [DOI] [PubMed] [Google Scholar]
- 2.Feldman AM, Weinberg EO, Ray PE, Lorell BH. Selective changes in cardiac gene expression during compensated hypertrophy and the transition to cardiac decompensation in rats with chronic aortic banding. Circ Res. 1993;73:184–192. doi: 10.1161/01.res.73.1.184. [DOI] [PubMed] [Google Scholar]
- 3.Lyn D, Liu X, Bennett NA, Emmett NL. Gene expression profile in mouse myocardium after ischemia. Physiol Genomics. 2000;2:93–100. doi: 10.1152/physiolgenomics.2000.2.3.93. [DOI] [PubMed] [Google Scholar]
- 4.Yue P, Long CS, Austin R, Chang KC, Simpson PC, Massie BM. Post-infarction heart failure in the rat is associated with distinct alterations in cardiac myocyte molecular phenotype. J Mol Cell Cardiol. 1998;30:1615–1630. doi: 10.1006/jmcc.1998.0727. [DOI] [PubMed] [Google Scholar]
- 5.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 6.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 7.Hoshijima M, Chien KR. Mixed signals in heart failure: cancer rules. J Clin Invest. 2002;109:849–855. doi: 10.1172/JCI15380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 9.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, Karlsson J, Castellani LW, Mumick S, Wang K, Xie T, Coon M, Zhang C, Estrada-Smith D, Farber CR, Wang SS, van Nas A, Ghazalpour A, Zhang B, Macneil DJ, Lamb JR, Dipple KM, Reitman ML, Mehrabian M, Lum PY, Schadt EE, Lusis AJ, Drake TA. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 13.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 15.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 19.Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, Delisi C. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–21. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura Y, Xing Y, Sasano H, Rainey WE. The mediator complex subunit 1 enhances transcription of genes needed for adrenal androgen production. Endocrinology. 2009;150:4145–4153. doi: 10.1210/en.2009-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- 25.Hansson A, Hance N, Dufour E, Rantanen A, Hultenby K, Clayton DA, Wibom R, Larsson NG. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci U S A. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W, Geelen P, Verstreken S, Wellens F, Bartunek J. Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol. 2008;51:129–136. doi: 10.1016/j.jacc.2007.07.087. [DOI] [PubMed] [Google Scholar]
- 27.Thattaliyath BD, Livi CB, Steinhelper ME, Toney GM, Firulli AB. HAND1 and HAND2 are expressed in the adult-rodent heart and are modulated during cardiac hypertrophy. Biochem Biophys Res Commun. 2002;297:870–875. doi: 10.1016/s0006-291x(02)02297-0. [DOI] [PubMed] [Google Scholar]
- 28.Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, Usami S, Saito Y, Harada M, Nakao K. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. Embo J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 30.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 31.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grinberg I, Millen KJ. The ZIC gene family in development and disease. Clin Genet. 2005;67:290–296. doi: 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim C, Nielsen HC. Hoxa-5 in mouse developing lung: cell-specific expression and retinoic acid regulation. Am J Physiol Lung Cell Mol Physiol. 2000;279:L863–871. doi: 10.1152/ajplung.2000.279.5.L863. [DOI] [PubMed] [Google Scholar]
- 34.Grier DG, Thompson A, Lappin TR, Halliday HL. Quantification of Hox and surfactant protein-B transcription during murine lung development. Neonatology. 2009;96:50–60. doi: 10.1159/000201739. [DOI] [PubMed] [Google Scholar]
- 35.Gaur A, Bhatia R, Spring-Mills E, Lemanski LF, Dube DK. The heart of metamorphosing Mexican axolotl but not that of the cardiac mutant is associated with the upregulation of Hox A5. Biochem Biophys Res Commun. 1998;245:746–751. doi: 10.1006/bbrc.1998.8514. [DOI] [PubMed] [Google Scholar]
- 36.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 37.Hidaka K, Nitta T, Sugawa R, Schwartz RJ, Amagai T, Nitta S, Takahama Y, Morisaki T. Differentiation of Pharyngeal Endoderm and Derivatives from Mouse Embryonic Stem Cells. Stem Cells Dev. 2010;19:1735–1743. doi: 10.1089/scd.2009.0466. [DOI] [PubMed] [Google Scholar]
- 38.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 39.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.