Abstract
Early diagnosis of lung cancer by low-dose computed tomography is an effective strategy to reduce cancer mortality in high-risk individuals. However, recruitment of at-risk individuals with asymptomatic lung cancer still remains challenging. We developed a minimal invasive serum test, based on the detection of circulating microRNAs, which can identify at-risk individuals with asymptomatic early stage non-small cell lung carcinomas with 80% accuracy.
Background and discussions
Lung cancer is the leading cause of cancer-related deaths in the developed world, accounting for 156,940 estimated deaths in 2011 in the US alone [1]. The lethality of lung cancer is primarily due to the lack of effective strategies for early detection, at a stage when the tumour would still be curable by surgery. However, detection of early stage lung cancer is challenging due to its frequent absence of symptoms [2]. This explains the urgent need for early detection screening programs for lung cancer in high-risk individuals, i.e. current or former heavy smokers (>20 packs/year) aged 50 years or above.
Low-dose spiral computed tomography (LD-CT) is an effective detection method for small lung nodules (even less than 5 mm) that subjects patients to low radiation exposure with no contrast medium, has limited costs and requires only a few seconds of execution [3,4]. The recent report from the large randomised National Lung Screening Trial has demonstrated a mortality reduction of 20% in the arm screened by LD-CT compared with the arm screened by chest x-ray [5]. However, recruitment of at-risk individuals with no symptoms of disease is still demanding.
In such a scenario, the development of blood tests able to detect the presence of lung cancer might form the cornerstone for successful population-based cancer screenings. In recent years there have been many attempts to identify serum/plasma biomarkers for lung cancer detection. Some studies have been based on detection through an enzyme-linked immunosorbent assay (ELISA) of circulating tumour-associated antigens (TAA) such as p53, NY-ESO-1, CAGE, GBU 4-5, annexin, or SOX2, which displayed an overall good specificity and sensitivity (40% sensitivity, 90% specificity [6]). Others have relied on the detection of circulating cancer cells in the blood of patients with metastatic tumours [7]. Such chip-based tests show sensitivity and specificity in detecting cancer cells that is close to 100% [8], warranting further investigations into the applicability of the detection circulating cancer cells at earlier stages of disease.
Recently, we and others have identified a subset of circulating microRNAs (miRNAs) accurate enough to detect symptomatic lung cancer [9–13] and, more importantly, LD-CT-detected asymptomatic lung cancer [14,15]. The detection of circulating miRNAs may be a valid alternative to LD-CT for the early diagnosis of cancer [16], since these tiny molecules are only marginally affected by degradation [17] and can be easily quantified by real-time PCR, a method routinely used in the clinic. In a series of experiments described in more detail elsewhere [14], we used real-time PCR to identify an expression profile of miRNAs extracted from the sera of participants enrolled in a large single-centre observational study (COSMOS study [18]). Among the 147 miRNAs detected by real-time PCR, 34 showed differences in expression in asymptomatic LD-CT-detected lung adenocarcinoma versus normal sera. We developed a multivariate risk-predictor algorithm based on the weighted linear combination of the 34-miRNA expression levels. When the predictor was tested on an independent cohort of patients with asymptomatic LD-CT-detected lung cancer, it displayed an overall accuracy of 80% (sensitivity 71%, specificity 90%; AUC 0.89). Furthermore, through a series of additional experiments, we also showed that the risk-predictor was able to distinguish between LD-CT-detected benign nodules and frankly malignant disease. This very important finding demonstrates the specificity of the test and highlights its utility in the clinic, because of the relative high number of benign lung nodules detected by LD-CT screening [19,20].
The simplicity of the procedure (it is minimally invasive, requiring <1 ml of serum) and its relatively low cost (based on standard real-time PCR) should encourage population compliance to large-scale screening programs, thus accelerating its application in the clinic as a ‘first line screening test’ to identify those high-risk individuals who should undergo further testing, including by LD-CT (Figure 1). However, unresolved questions still remain (as for all newly proposed genomic blood tests) regarding tumour specificity.
Figure 1:
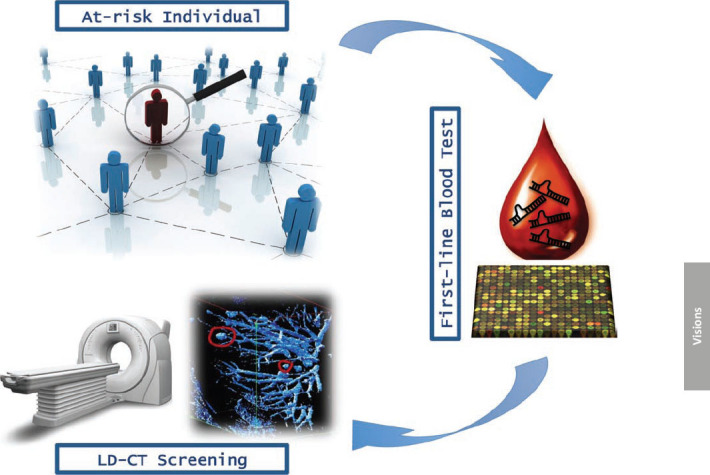
Overview of lung cancer early detection mass population screening.
We tried to initially address this issue by including in our analysis also a group of patients with invasive ductal breast carcinoma or with benign breast fibroadenoma, as a control. The test resulted to be negative in this additional group of patients, thus demonstrating a certain specificity of the 34-miRNA test for lung cancer detection [14].
Lastly, a critical issue relates to the origin of serum circulating miRNAs and their biological function. The current model implies that miRNA are released in membrane-bound vesicles [21–23], which protect them from blood RNAse activity [24–26]. Such kind of vesicles could be either exosomes, 50–90 nm vesicles of endocytic origin arising by inward budding of multivesicular bodies and released by exocytosis [27], or microvesicles, larger (up to 1 um) membrane-bound particles generally derived by membrane shedding of several cell types [24,28].
Strikingly, tumour cells appear to communicate through exosomes with immune cells, leading to immune suppression [29,30]. It is tempting to speculate that lung cancer cells might reprogram the tumour microenvironment, perhaps by altering the expression patterns of surrounding cells, including those of the immune system, through miRNAs contained in exosomes. Undoubtedly, future studies will shed more light on the biological functions of circulating miRNAs and their role, if any, in cancer progression. Such studies will allow researchers to design completely novel strategies for lung cancer therapy.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Diederich S, Wormanns D. Impact of low-dose CT on lung cancer screening. Lung Cancer. 2004;45 Suppl 2:S13–9. doi: 10.1016/j.lungcan.2004.07.997. [DOI] [PubMed] [Google Scholar]
- 3.Mulshine JL, Sullivan DC. Clinical practice. Lung cancer screening. N Engl J Med. 2005;352:2714–20. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 4.Humphrey LL, Teutsch S, Johnson M, U.S. Preventive Services Task Force Lung cancer screening with sputum cytologic examination, chest radiography, and computed tomography: an update for the U.S. Preventive Services Task Force. Ann Intern Med. 2004;140:740–53. doi: 10.7326/0003-4819-140-9-200405040-00015. [DOI] [PubMed] [Google Scholar]
- 5.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD, National Lung, Screening Trial, Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle P, Chapman CJ, Holdenrieder S, Murray A, Robertson C, Wood WC, Maddison P, Healey G, Fairley GH, Barnes AC, Robertson JF. Clinical validation of an autoantibody test for lung cancer. Ann Oncol. 2011;22:383–9. doi: 10.1093/annonc/mdq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–8. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 10.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130:1378–86. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 12.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, Jiang F. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–87. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–86. [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin PY, Yang PC. Circulating miRNA signature for early diagnosis of lung cancer. EMBO Mol Med. 2011;3:436–7. doi: 10.1002/emmm.201100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Veronesi G, Bellomi M, Mulshine JL, Pelosi G, Scanagatta P, Paganelli G, Maisonneuve P, Preda L, Leo F, Bertolotti R, Solli P, Spaggiari L. Lung cancer screening with low-dose computed tomography: a non-invasive diagnostic protocol for baseline lung nodules. Lung Cancer. 2008;61:340–9. doi: 10.1016/j.lungcan.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Bach PB, Jett JR, Pastorino U, Tockman MS, Swensen SJ, Begg CB. Computed tomography screening and lung cancer outcomes. JAMA. 2007;297:953–61. doi: 10.1001/jama.297.9.953. [DOI] [PubMed] [Google Scholar]
- 20.Swensen SJ, Jett JR, Hartman TE, Midthun DE, Sloan JA, Sykes AM, Aughenbaugh GL, Clemens MA. Lung cancer screening with CT: Mayo Clinic experience. Radiology. 2003;226:756–61. doi: 10.1148/radiol.2263020036. [DOI] [PubMed] [Google Scholar]
- 21.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–6. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 27.Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–9. [PubMed] [Google Scholar]
- 29.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang HG. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–75. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 30.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rébé C, Ghiringhelli F. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–71. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]


