Abstract
Efficient cell delivery of antisense oligonucleotides (ONs) is a key issue for their potential therapeutic use. It has been shown recently that some ONs can be delivered into cells without the use of transfection agents (gymnosis), but this generally requires cell incubation over several days and high amounts of ONs (micromolar concentrations). Here we have targeted microRNA 122 (miR-122), a small non-coding RNA involved in regulation of lipid metabolism and in the replication of hepatitis C virus, with ONs of different chemistries (anti-miRs) by gymnotic delivery in cell culture. Using a sensitive dual-luciferase reporter assay, anti-miRs were screened for their ability to enter liver cells gymnotically and inhibit miR-122 activity. Efficient miR-122 inhibition was obtained with cationic PNAs and 2'-O-methyl (OMe) and Locked Nucleic Acids (LNA)/OMe mixmers containing either phosphodiester (PO) or phosphorothioate (PS) linkages at sub-micromolar concentrations when incubated with cells for just 4 hours. Furthermore, PNA and PS-containing anti-miRs were able to sustain miR-122 inhibitory effects for at least 4 days. LNA/OMe PS anti-miRs were the most potent anti-miR chemistry tested in this study, an ON chemistry that has been little exploited so far as anti-miR agents towards therapeutics.
Keywords: 2’-O-Methyl, anti-miR, delivery, Gymnosis, Locked Nucleic Acids, miR-122, miRNA, Peptide Nucleic Acids, phosphorothioate, transfection
Introduction
Antisense oligonucleotides (ONs) have proven to be powerful tools to understand biological processes and more recently as potential therapeutic agents.1-4 However, the use of ONs in vivo has been hampered by their poor cellular uptake. For experiments in cell culture, ONs have generally been delivered by addition of transfection agents, such as lipid-based systems (e.g., Lipofectamine 2000) or by electroporation, but these methods are not very suitable for in vivo or therapeutic applications. Recently ON gapmers containing Locked Nucleic Acids (LNA) flanks and DNA cores and a phosphorothioate (PS) backbone were shown to be able to be delivered into cells without the use of transfection agents.5 In that study, cells were incubated with low-micromolar concentrations of ON for 6–10 d in continuous culture and the phenomenon of unaided delivery was termed ‘gymnosis’ or ‘gymnotic delivery’. Gymnotic delivery and antisense activity of PS/DNA ONs, lipid-conjugated ON phosphoramidates or thio-phosphoramidates had been reported previously involving incubation of ONs in the low-micromolar concentration range with various cell types for several days in continuous culture.6-9
Stein and colleagues showed also that gene silencing of Bcl-2 or Apo-B in melanoma cells by gymnotic delivery of LNA/DNA PS gapmers correlated better with in vivo silencing than ONs delivered by Lipofectamine 2000.5 A similar observation was made by Straarup et al. for short LNA/DNA PS gapmers.10 Furthermore, Zhang et al. showed that gymnotic delivery of LNA/DNA PS gapmer ONs could be used in a wide range of tumor cell lines for gene downregulation.11 Very recently, Koller et al. showed that micromolar quantities of gapmer ONs containing 2’-O-methoxyethyl (MOE) flanks with DNA core and PS linkages could also be delivered without transfection agent and showed RNase-H-dependent antisense activity in MHT liver cells and primary hepatocytes, in this case over a 24–36 h timescale.12 Therefore, it is becoming clear that antisense activity by gymnotic delivery may be a feature of a number of ON chemistries.
We have shown previously that cationic Peptide Nucleic Acid (PNA) ONs can be delivered and are functionally active as steric blocking antisense agents without the use of transfection agents in cells. For inhibition or redirection of nuclear splicing, a cationic cell penetrating peptide is required to be covalently conjugated to the PNA,13,14 whereas for targeting microRNAs we found that only a few Lys residues attached to the PNA are necessary for microRNA inhibition both in cell culture15 and in vivo.16
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression post-transcriptionally and which are involved in complex cellular processes and diseases.17,18 MicroRNA-122 (miR-122) is a 23 nucleotide long, liver-specific miRNA involved in the metabolism of lipids19 and required for hepatitis C virus infection20-22 and is thus an attractive therapeutic target for miRNA inhibition by steric blocking anti-miRs. In addition to Cys-K-PNA-K3 ONs used by us,15 several other anti-miR ON chemistries have been proposed as anti-miR122 agents, including 2’-O-methyl (OMe),20,23-27antagomiRs (OMe-cholesterol conjugated partially or fully modified with PS linkages28,29), 2’-fluoro-2’-deoxy (2’F), MOE and 2’F/MOE mixmers,19,25 tiny all LNA ONs,30 LNA in mixmers with DNA,31-34 and LNA mixmers with OMe15 or with MOE.35 So far only an LNA/DNA anti-miR with PS linkages has moved into clinical trials as reported by Santaris Pharma in 2010 (Santaris Pharma News Release 23 September 2010; www.santaris.com).
As far as we are aware, in all reports of LNA/DNA mixmers targeting miRNAs,30-34,36-38 as well as in the very few studies that have compared side-by-side the potency of anti-miRs of different chemistries,15,25,35,37,39 cell culture experiments were performed using transfection agents. Here we have used a miR-122 sensor dual-luciferase assay to show that PNA, OMe and LNA/OMe containing either phosphodiester (PO) or phosphorothioate (PS) linkages, but not an LNA/DNA PO anti-miR, were able to be internalized in cells by gymnotic delivery and be effective as miR-122 inhibitors at sub-micromolar ON concentrations in liver cells with only 4 h ON incubation times. PNA and the PS ONs thus delivered were found to be capable of sustained anti-miR activity in cells for at least 4 d, while PO ONs showed a peak in activity 3 d after ON incubation, after which activity decreased. The most potent anti-miR was the LNA/OMe PS ON, an anti-miR chemistry that has not been considered as yet sufficiently for therapeutic development.
Results
We and others have shown that quantification of miRNA levels after anti-miR treatment by Real Time quantitative PCR (RT-qPCR) or by use of RNA gel blots can be subject to technical artifacts due to the anti-miR binding strongly to the miRNA and thus preventing its detection.15,25,40 Such strongly binding anti-miRs sequester the miRNA without inducing degradation of the miRNA.25,40 Therefore, evaluation of anti-miR efficiency by quantification of target miRNA upon anti-miR treatment is not a recommended approach. Since target mRNA degradation is a common outcome for miRNA-mediated downregulation of gene expression,41,42 one possible way of studying anti-miR activity is by measurement of mRNA levels of well validated miRNA target mRNAs by RT-qPCR following anti-miR treatment. However, this technique is not sensitive enough to distinguish adequately dose-dependent anti-miR effects, since the corresponding effects on upregulation of mRNA targets are frequently small. We have therefore used a very sensitive miR-122 dual-luciferase reporter assay for comparison of the effectiveness of anti-miRs of different chemistries in unaided cell delivery, an approach initially reported by Vermeulen et al.43 and often now used as a tool for screening of anti-miRs.26,30,32,33,37,38
Briefly, the reporter plasmid contains an exactly complementary miR-122 recognition site cloned into the 3′-untranslated region (UTR) of a Renilla luciferase (RLuc) gene, to bring Rluc under miR-122 control and a non-miR-122 regulated Firefly Luciferase gene (FLuc) that serves as an internal reference. The plasmid was introduced into miR-122 positive Huh7 cells, such that Rluc expression is repressed until anti-miR is added to the cells, whereupon an increase in RLuc expression signifies anti-miR induced derepression. Anti-miR activity is assessed by measurement of the RLuc/FLuc expression ratio, normalized to that of control cells (Relative RLuc/FLuc). Huh7 human hepatoma cells were chosen because they have been shown to express large quantities of miR-122.20,44
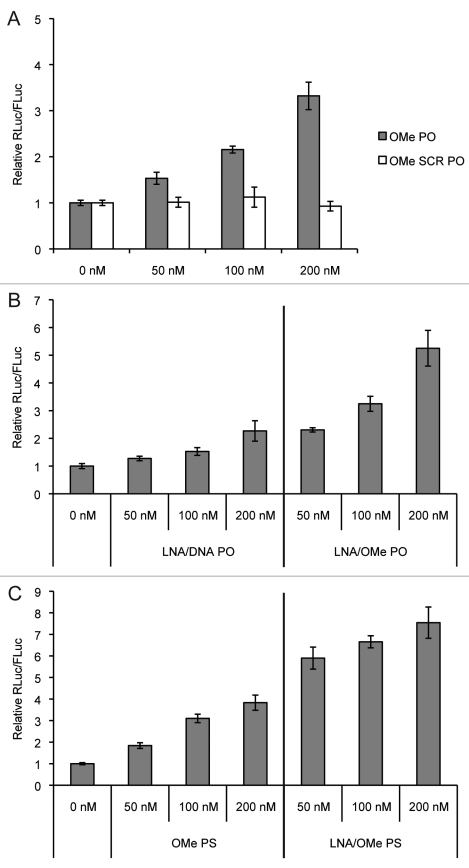
We chose to study the following ONs: a 23mer cationic PNA (Cys-K-PNA-K3) which we have already shown displays anti-miR activity via gymnotic delivery.15 31mer OMe PO (and its scrambled version 31mer OMe SCR PO), 23mer OMe PS, 23mer LNA/OMe PO (43% LNA content), 23mer LNA/OMe PS (17% LNA content) and a commercial 23mer LNA/DNA PO miR-122 knockdown probe (Exiqon, unknown LNA content) anti-miR (Table 1). The chemical structures of the nucleoside and phosphorothioate analogs are shown in Figure 1. Most of these anti-miRs have been previously validated as miR-122 inhibitors.15,20 The anti-miR ONs OMe PO (but not its SCR PO version) (Fig. 2A), LNA/DNA PO and LNA/OMe PO ONs (Fig. 2B) and OMe PS and LNA/OMe PS (Fig. 2C) lipofected at 50, 100 and 200 nM into Huh7 cells are all able to inhibit miR-122 effectively and in a dose-dependent manner, as seen by Rluc/Fluc upregulation. Anti-miRs were lipofected for 4 h (following the manufacturer’s protocol) and luciferase expression was measured 48 h after anti-miR lipofection.
Table 1. Oligonucleotides used in this study.
| Backbone Chemistry | Sequence | Length |
|---|---|---|
| Cys-K-PNA-K3 |
(N-term)-Cys-K-ACAAACACCATTGTCACACTCCA–KKK-(C-term) |
23mer |
| OMe PO |
agacacaaacaccauugucacacuccacagc |
31mer |
| OMe SCR PO |
cacguuaaaaccauacgcacuacgaaacccc |
31mer |
| OMe PS |
acaaacaccauugucacacucca |
23mer |
| LNA/OMe PO |
aCaAaCaCcAuuGuCaCaCuCca |
23mer |
| LNA/OMe PS |
acaaacaccauuguCaCaCuCca |
23mer |
| LNA/DNA PO* | ACAAACACCATTGTCACACTCCA | 23mer |
Unknown LNA content; Lower case = Ome; Underlined = LNA
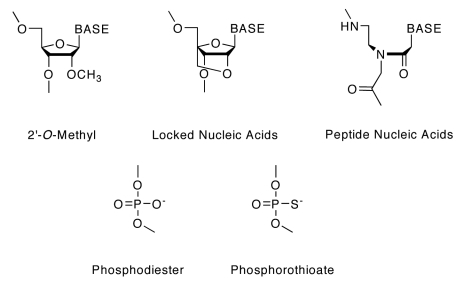
Figure 1.
Chemical structures of nucleoside analogs and nucleoside linkages used in this study. Base: Uracil (OMe only), Thymine (except OMe), Adenine, Guanine or Cytosine (5-methyl-cytosine for LNA bases).
Figure 2.
Dose-dependent miR-122 inhibition as seen by luciferase measurements in Huh7 cells 48 h after lipofection at the indicated concentrations of (A) OMe PO and SCR OMe PO anti-miRs, (B) LNA/DNA PO and LNA/OMe PO anti-miRs and (C) OMe PS and LNA/OMe PS anti-miRs.
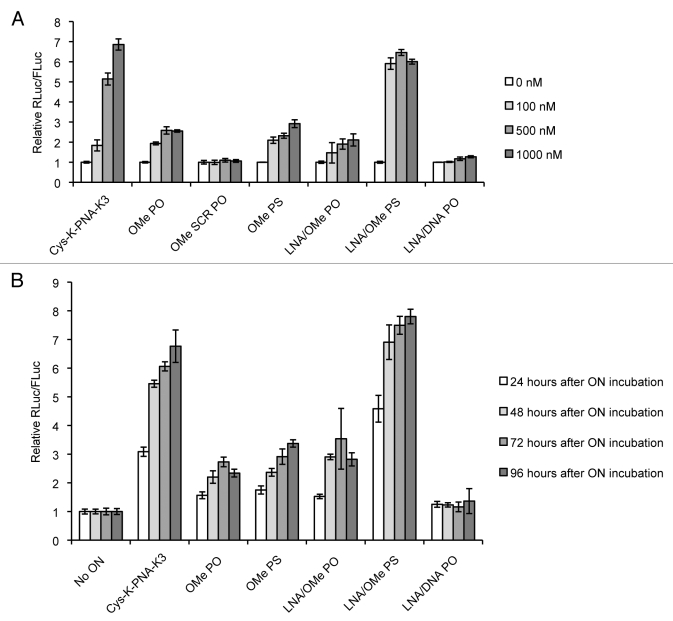
We then examined the miR-122 inhibitory activity of anti-miRs in the absence of transfection agents. Anti-miRs were incubated for only 4 h with Huh7 cells, after which the anti-miR containing media was removed, cells were washed and media was replaced by growing media (Full Media). Luciferase expression was measured after 48 h, just as for the lipofection experiments. Figure 3A shows dose-dependent miR-122 inhibition for Cys-K-PNA-K3, OMe PO (but not its SCR PO version), OMe PS, LNA/OMe PO and LNA/OMe PS anti-miR, while LNA/DNA PO anti-miR did not significantly inhibit miR-122 at the sub-micromolar concentrations tested. It is interesting to note that OMe and LNA/OMe ONs with either PO or PS linkages are capable of miRNA inhibition in the absence of transfection agent, but that by far the most potent anti-miR was the LNA/OMe ON containing PS linkages. The PNA ON was the second most effective. Moreover, at least for miRNA targeting, there was no need for continuous incubation of Huh7 cells with ONs for several days, as seems to be required for gymnotic activity of LNA gapmers against a mRNA target. We have found that activity of Cys-K-PNA-K3 reaches a plateau by 4 h incubation time (Torres et al. manuscript in preparation).
Figure 3.
(A) Dose-dependent miR-122 inhibition as seen by luciferase measurements in Huh7 cells 48 h after gymnotic delivery of anti-miRs at the indicated concentrations. (B) Luciferase measurement in Huh7 cells 24, 48, 72 or 96 h after gymnotic delivery of anti-miRs at 500 nM concentration.
Finally, we decided to determine the sustainability of the observed miR-122 inhibitory effects through unaided delivery. Thus, Huh7 cells were incubated with 500 nM anti-miRs for 4 h in the absence of transfection agents as before and luciferase expression was measured at 24, 48, 72 and 96 h respectively after anti-miR incubation. Figure 3B shows that Cys-K-PNA-K3 and all PS anti-miRs were able to sustain the miR-122 inhibitory effect for at least 96 h (4 d) while for PO anti-miRs (OMe PO and LNA/OMe PO) the anti-miR activity began to decline after day 3. By contrast, the LNA/DNA PO anti-miR did not show any significant activity during the 4 d. Since experiments were performed in 96-well plate high-throughput format, it was not possible to measure luciferase expression beyond 4 d following anti-miR incubation because of high cell confluency. As in the case of Figure 3A, the extended incubation data in Figure 3B revealed that LNA/OMe PS and Cys-K-PNA-K3 anti-miRs were the most potent chemistries of the ones tested in this study.
Discussion
Previous reports have described gymnotic delivery at low-micromolar concentrations of ONs, usually with extended incubation periods, to achieve effective gapmer antisense ON activity in cells.5-9 However, only recently has gymnotic delivery started to be considered as a useful approach for screening of ONs as potential therapeutics, since data obtained by gymnotic delivery of ONs correlated much better with in vivo results as compared with ON cell delivery aided by transfection agents, such as Lipofectamine 2000.5,10,11 Practically all the previous gymnotic delivery experiments have focused on antisense targeting of mRNA, whereas unaided delivery of anti-miRs has not been studied, except for Lys-modified PNA ONs as previously reported by us.15,16,40
The data presented in Figure 3 show that in addition to Cys-K-PNA-K3, OMe PO, OMe PS, LNA/OMe PO and LNA/OMe PS are also all capable of rapidly entering Huh7 liver cells without the use of transfection agents and inhibiting miR-122 at sub-micromolar ON concentrations over a sustained period. Several interesting observations can be made about the rank order of activity of the various ONs. First, the 23-mer LNA/OMe PO was more active than the OMe PO 31-mer with lipofection (Fig. 2). This is consistent with our previous finding that LNA/OMe PO was able to upregulate miR-122 target mRNAs when lipofected into Huh7 cells or primary rat hepatocytes, while the OMe PO anti-miR failed to upregulate such mRNAs in a dose-dependent manner,15 and is validation for the use of the dual-luciferase assay for screening of anti-miR activity. Interestingly, OMe PO and LNA/OMe PO ONs showed similar levels of activity when delivered in the absence of transfection agents (Fig. 3). The OMe PO ON is a 31-mer previously validated against miR-122.20 The 8 additional residues beyond the normal 23-mer anti-miR are complementary to the pre-miR122 sequence, as it was reported that a 23-mer was poorly active.45 Activity of both these all PO ONs, as measured by the RLuc/Fluc ratio, started to decline after 3 d of cell incubation, which can probably be explained by the known instability of phosphodiester linkages to nuclease degradation, whereas the PNA ON and ONs containing PS linkages continued to show an increased RLuc/Fluc ratio on day 4. Although when delivered in the absence of transfection agent, addition of LNA residues did not significantly enhance activity over OMe in the PO context, by contrast significantly increased anti-miR activity was obtained by addition of LNA residues to the OMe ON in the PS context (Fig. 3).
Surprisingly, the commercially obtained LNA/DNA PO anti-miR-122 knockdown probe failed to be internalized and inhibit miR-122 by gymnotic delivery at the tested concentrations (Fig. 3), whereas it was functional when lipofected into cells, albeit with less potency than for the LNA/OMe PO ON (Fig. 2B). We have for several years advocated the use of mixmers of LNA and OMe as steric blocking agents,15,46-48 since the binding strength of OMe residues is higher than that of 2’-deoxynucleotides, because both OMe and LNA residues intrinsically adopt a C-3′-endo, ribose-like conformation. Since the LNA content of the LNA/DNA PO ON is not revealed by the supplier, we cannot rule out the possibility that the increased potency is partly due to a significantly different LNA content between the two ONs, but it seems likely that the commercial LNA/DNA PO probe will have a similar LNA content to the LNA/OMe PO anti-miR (40% LNA). Therefore we believe that the increased potency of LNA/OMe PO over LNA/DNA PO is mostly due to the presence of OMe residues.
The highest potency anti-miR was found to be the LNA/OMe PS ON. Whereas LNA/MOE PS anti-miRs have been described for cellular and in vivo use targeting miR-122,25,37 there is only one previous example of an LNA/OMe PS ON used as an anti-miR, but only with delivery by transfection agent into cells.37 By contrast, LNA/DNA PS ONs targeting miRNAs are well established and have indeed entered clinical trials (Santaris Pharma News Release 23 September 2010; www.santaris.com). Further, LNA/DNA PS gapmers were shown to be subject to gymnosis in mRNA targeting studies.5,10,11 In our study we have used an LNA/OMe PS anti-miR containing only ~15% LNA residues that are all located in the region complementary to the miR-122 seed sequence (Table 1). When lipofected into cells, the LNA/OMe PS anti-miR somewhat outperformed the LNA/OMe PO containing 40% LNA we previously reported (Fig. 2B and 2C). However, the dramatic enhancement in anti-miR activity obtained for the LNA/OMe PS ON over the LNA/OMe PO ON when delivered by gymnosis (Fig. 3) suggests that the presence of the PS linkages enhances productive cellular delivery leading to miRNA inhibition. Interestingly, as seen in Figure 2C and Figure 3A, the observed activity for the LNA/OMe PS ON reached saturation. It is unlikely that this is due to saturation of cellular uptake, since similar saturation was observed for LNA/OMe PS delivered by lipofection. Although we did not correlate precisely anti-miR activity saturation in the dual-luciferase assay with the level of miRNA inhibition, our results indicate that most of the miR-122 is inhibited at the saturating concentrations tested. A significant amount of further work will be necessary to determine the optimum LNA content and placement within an OMe anti-miR sequence, as well as the ON length optimum, and also to compare in greater detail LNA/DNA PS and LNA/OMe PS mixmers, as well as possible mixmers with all three nucleoside types. However, the potent and long-lasting gymnotic anti-miR activity at sub-micromolar concentrations in cells suggests that LNA/OMe PS ONs should be given greater consideration for in vivo use.
While long incubation times (days) with high amounts of ONs (low-micromolar concentrations) in continuous cell culture were necessary for gapmer antisense inhibition of mRNA by gymnotic delivery,5-12 here we showed rapid (only 4 h) gymnotic delivery at sub-micromolar concentrations of Cys-K-PNA-K3, OMe and LNA/OMe PO and PS ONs for miR-122 targeting. Also anti-miR122 activity was sustained for at least 4 d after cell incubation for many of the ONs. Zhang et al. showed that gymnotic delivery of LNA/DNA PS gapmers could be achieved in a wide range of tumor cell lines.11 We previously reported inhibition of miR-155 function in B-cells and in mice by PNA ONs in the absence of transfection agents.16 We have also found recently that at least for PNA ONs, gymnotic uptake does not extend only to Huh7 cells and B-cells but also to HEK293ET cells and H2K mdx mice-derived myoblasts and myotubes (Torres et al., manuscript in preparation). The mechanisms of gymnotic cellular uptake and localization of ONs, such as the highly active Cys-K-PNA-K3 and LNA/OMe PS, and how such anti-miRs find and interact with their miRNA targets still remains to be explored.
Stein and colleagues showed that a 5′-FAM-labeled LNA/DNA PS ON delivered by gymnosis was localized in the cytosol together with GW/P-bodies (site for RNA silencing), while the same ON after lipofection showed bright fluorescence in the nucleus instead.5 By contrast, Zhang et al.11 correlated strong gapmer antisense target down-modulation with nuclear localization following gymnosis of their FAM-labeled LNA/DNA PS ON, although at earlier time-points these ONs localized in the perinuclear space in the cytosol. Koller and colleagues failed to detect MOE/DNA PS gapmer ONs in P-Bodies after gymnotic delivery and instead the bulk of the ON was found in lysosomes after 24 h.12 They also showed that uptake of MOE/DNA PS ON into MHT hepatocarcinoma derived liver cells seemed to be through a clathrin- and caveolae-independent but AP2M1-dependent endocytotic mechanism.12 Since miRNAs have been found within the endosomal pathway49 a very likely possibility is that anti-miR ONs delivered by gymnosis are taken up by cells via a rapid endocytosis mechanism, travel through the whole endocytotic pathway, and are able at some point to encounter the miRNA within one or more of these endosomal compartments. This hypothesis may explain why such short ON incubation times (within 4 h) are effective for miR-122 inhibition in the absence of transfection agents. Our recent investigations into understanding the mechanisms of anti-miR uptake and miRNA targeting will be reported shortly which we hope will provide further insight into improving anti-miR design (Torres et al. manuscript in preparation).
Materials and Methods
Dual-Luciferase miR-122 sensor plasmid. The procedure for plasmid construct is similar to that of Vermeulen et al.43 Insertion of a dsDNA fragment corresponding to perfect matches for hsa-miR-122 was made in the 3′-UTR of Renilla luciferase between the XhoI and NotI restriction sites in the commercially available psiCHECK-2 vector (Promega, Cat Number C8021). The miR-122 dsDNA fragment was made up from the individual strands corresponding to mature miR-122 (miRBase version 14 http://www.mirbase.org/; underlined on ‘miR-122 antisense’ oligonucleotide see below) and its perfect complement. In addition, the oligonucleotides contained necessary sequences for direct cloning into XhoI/NotI-digested psiCHECK-2 plasmid. Oligonucleotide sequences were as follows: miR-122 antisense 5′- GGCCGCAGCTCGAGTGGAGTGTGACAATGGTGTTTGGGTCAT -3′; miR-122 sense 5′- TCGAATGACCCAAACACCATTGTCACACTCCACTCGAGCTGC -3′ and were obtained from Sigma. The identity of the construct was confirmed by sequencing before use.
Anti-miR ONs. Anti-miR sequences are indicated in Table 1. PNA anti-miR15 was synthesized as previously described.16 OMe PO and SCR OMe PO20 were obtained from Dharmacon. LNA/OMe PO15 was synthesized as previously described.50 miRCURY LNA/DNA PO miR-122 knockdown probe was purchased from Exiqon (Cat No 118019–00 hsa-miR-122a).
OMe PS and LNA/OMe PS were synthesized on an Applied Biosystems 394 automated DNA synthesizer using a modified version of the phosphoroamidite method.51 Immediately before use OMe phosphoramidites were dissolved at 0.1 M concentration in anhydrous acetonitrile (OMe), except 5-methyl-C LNA phosphoramidite (Exiqon, Cat Number EQ-0066) which was dissolved in 3:1 anhydrous acetonitrile/anhydrous THF one day before use and left to stand over molecular sieves. Phosphoramidites were coupled for 6 min (OMe) or 75 sec (5-methyl-C LNA) with 0.25 M 5-(bis-3.5-trifluoromethylphenyl)-1H-tetrazole (Activator 42, Sigma-Aldrich, Cat Number L3300002-HH) as activator. ONs were sulfurized for 3 min with 0.2 M phenylacetyl disulfide (Alfa Aesar, Cat Number L19792) in anhydrous acetonitrile containing 10% by volume of N-methylimidazole (Sigma-Aldrich, Cat Number 67560), prepared and left to stand for at least 12 h before use. The terminal 5′-O-DMT group was left on the ONs for purification.
ONs were cleaved and deprotected with 28% aqueous ammonia overnight. After evaporating to dryness ONs were re-dissolved in 0.5 ml 4:1 water/acetonitrile and purified by RP-HPLC on a Zorbax SB-C18 column, 9.4 x 250 mm, 5 µm particle size, (Agilent Technologies, Cat Number 880975–202) with 0.05 M triethylammonium bicarbonate (mobile phase A) and acetonitrile (mobile phase B) at a flow rate of 1.5 mL/min and the following gradient: 0 - 50 min, 0–30% B, 50 – 60 min, 30 – 50% B, 60 – 65 min, 50 – 100% B. The concentrated residues were then treated with 80% acetic acid for 60 min to remove the terminal 5′-O-DMT protecting group. Following evaporation of the deprotection solutions, the ONs were re-dissolved in 0.5 mL 0.05 M triethylammonium bicarbonate, re-applied to the HPLC column, and purified using the same mobile phase gradients.
Cell culture, transfections and gymnotic delivery of anti-miRs. Huh7 cells were maintained at 37°C/5% CO2 in Full Media: Dulbecco’s Modified Eagle Medium (DMEM GlutaMAX, 4.5 g/L D-glucose, pyruvate; Invitrogen Cat Number 31966–021) containing 10% Fetal Bovine Serum (FBS, Perbio/Thermo Scientific, Cat Number CH30160.03) and penicillin/streptomycin antibiotics. Twenty-four hours before transfections, 2x104 cells in 100 μL of DMEM/10% FBS (no antibiotics) per well were plated in 96-well white plates, clear bottom (Corning Costar, Cat Number 3610). The next day, cells were lipofected with 100 ng dual-luciferase miR-122 sensor plasmid per well using Lipofectamine 2000 (Invitrogen, Cat Number 11668–019) following the manufacturer’s protocol in a final transfection volume of 150 μL (100 µL DMEM/10% FBS plus 50 µL plasmid/lipid reaction). Lipofection was performed overnight. For anti-miR lipofection, the following day cells were washed once with 150 μL PBS and lipofected with anti-miRs at the desired concentration following the manufacturer’s protocol in a final lipofection volume of 150 µL (100 µL DMEM/10% FBS plus 50 µL anti-miR/lipid reaction). Four hours later, cells were washed once with 150 μL PBS and were left growing in 150 µL Full Media for 48 h before luciferase measurements. For gymnotic delivery of anti-miRs, 96-well plates containing cells lipofected with luciferase miR-122 sensor plasmid as before were washed twice with 150 µL PBS and incubated at 37°C/5%CO2 in 100 μL Opti-MEM (Invitrogen, Cat Number 31985–047) containing anti-miRs at the desired concentration. Four hours after cell incubation with anti-miRs, cells were washed twice with 150 µL PBS and were grown in 150 μL Full Media until luciferase measurements were performed.
Luciferase measurements. For luciferase measurements, Full media was removed from the cells and replaced with 75 μL PBS at the desired time points (24, 48, 72 or 96 h after anti-miR lipofection/incubation). Luciferase expression was measured using Dual-Glo Luciferase Assay System (Promega, Cat Number E2920) following the manufacturer’s protocol except that Luciferase substrate incubations were performed for 10 min in the dark. Luminescence was measured for 5 sec per well in an Orion Microplate Luminometer (Berthold Detection System, Pforzheim, Germany). All luciferase experiments were performed in triplicates. An RLuc/FLuc ratio was obtained and the data was further normalized to cells containing only dual-luciferase miR-122 sensor plasmid and incubated with Opti-MEM in the absence of ONs (‘0 nM’ for Figure 2 and Figure 3A; ‘No ON’ for Figure 3B). Shown are average values and their standard deviations.
Acknowledgments
We thank Donna Williams for PNA synthesis and Marvin Caruthers (Boulder Colorado, USA) for his support. Special thanks go to Martin Fabani for providing the Luciferase miR-122 sensor plasmid. A.G.T. is funded by a Cesar Milstein Scholarship from the Darwin Trust of Edinburgh, Scotland. The work was supported by the Medical Research Council (MRC Unit program U105178803).
Glossary
Abbreviations:
- ON(s)
Antisense oligonucleotide(s)
- LNA
Locked Nucleic Acids
- OMe
2’-O-Methyl
- PNA
Peptide Nucleic Acids
- 2’F
2’-Fluoro-2’-deoxy
- MOE
2’-O-Methoxyethyl
- PO
phosphodiester linkage
- PS
phosphorothioate linkage
- SCR
Scrambled control ON
- miRNA(s)
microRNA(s)
- miR-122
microRNA-122
- RT-qPCR
Reverse transcription - Real Time quantitative PCR
- RLuc
Renilla Luciferase
- FLuc
Firefly Luciferase
- 3′-UTR
3′-Untranslated Region
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/17731
References
- 1.Persidis A. Antisense therapeutics. Nat Biotechnol. 1999;17:403–4. doi: 10.1038/7973. [DOI] [PubMed] [Google Scholar]
- 2.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr Opin Chem Biol. 2006;10:607–14. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 4.Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers. 2010;7:536–42. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 5.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Høg A, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salas PJ, Rodriguez M, Viciana A. The Apical Submembrane Cytoskeleton Participates in the Organization of the Apical Pole in Epithelial Cells. J Cell Biol. 1997;137:359–75. doi: 10.1083/jcb.137.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dikmen ZG, Wright WE, Shay JW, Gryaznov SM. Telomerase targeted oligonucleotide thio-phosphoramidates in T24-luc bladder cancer cells. J Cell Biochem. 2008;104:444–52. doi: 10.1002/jcb.21635. [DOI] [PubMed] [Google Scholar]
- 8.Abaza M-SI, Al-Saffar A, Al-Sawan S, Al-Attiyah R. c-myc antisense oligonucleotides sensitize human colorectal cancer cells to chemotherapeutic drugs. Tumour Biol. 2008;29:287–303. doi: 10.1159/000156706. [DOI] [PubMed] [Google Scholar]
- 9.Stein CA, Wu S, Voskresenskiy AM, Zhou J-F, Shin J, Miller P, et al. G3139, an anti-Bcl-2 antisense oligomer that binds heparin-binding growth factors and collagen I, alters in vitro endothelial cell growth and tubular morphogenesis. Clin Cancer Res. 2009;15:2797–807. doi: 10.1158/1078-0432.CCR-08-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straarup EM, Fisker N, Hedtjärn M, Lindholm MW, Rosenbohm C, Aarup V, et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–11. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Qu Z, Kim S, Shi V, Liao B, Kraft P, et al. Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther. 2011;18:326–33. doi: 10.1038/gt.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koller E, Vincent TM. Chappell a, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abes S, Williams D, Prevot P, Thierry A, Gait MJ, Lebleu B. Endosome trapping limits the efficiency of splicing correction by PNA-oligolysine conjugates. J Control Release. 2006;110:595–604. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Ivanova GD, Arzumanov A, Abes R, Yin H, Wood MJ, Lebleu B, et al. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res. 2008;36:6418–28. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabani MM, Gait MJ. miR-122 targeting with LNA/2'-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2007;14:336–46. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KGC, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–75. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 18.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, et al. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 21.Triboulet R, Mari B, Lin Y-L, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 22.Chang J, Guo J-T, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215–23. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–7. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson B, Dalby AB, Karpilow J, Khvorova A, Leake D, Vermeulen A. Specificity and functionality of microRNA inhibitors. Silence. 2010;1:10. doi: 10.1186/1758-907X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts APE, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–29. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 29.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–92. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 33.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y-P, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108:4991–6. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–41. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 37.Lennox KA, Behlke MA. A Direct Comparison of Anti-microRNA Oligonucleotide Potency. Pharm Res. 2010;27:1788–99. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 38.Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmén J, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–92. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh SY, Ju Y, Park H. A highly effective and long-lasting inhibition of miRNAs with PNA-based antisense oligonucleotides. Mol Cells. 2009;28:341–5. doi: 10.1007/s10059-009-0134-8. [DOI] [PubMed] [Google Scholar]
- 40.Torres AG, Fabani MM, Vigorito E, Gait MJ. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933. doi: 10.1261/rna.2533811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 42.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, et al. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–30. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu H, He J-H, Xiao Z-D, Zhang Q-Q, Chen Y-Q, Zhou H, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–42. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 45.Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arzumanov A, Walsh AP, Rajwanshi VK, Kumar R, Wengel J, Gait MJ. Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2'-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–54. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]
- 47.Arzumanov A, Stetsenko DA, Malakhov AD, Reichelt S, Sørensen MD, Babu BR, et al. A structure-activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2'-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), alpha-L-LNA, or 2'-thio-LNA residues. Oligonucleotides. 2003;13:435–53. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- 48.Brown D, Arzumanov A, Turner J, Stetsenko D, Lever A, Gait M. Antiviral Activity of Steric-Block Oligonucleotides Targeting the HIV-1 Trans -Activation Response and Packaging Signal Stem-Loop RNAs. Nucleosides Nucleotides Nucleic Acids. 2005;24:393–6. doi: 10.1081/NCN-200059813. [DOI] [PubMed] [Google Scholar]
- 49.Gibbings D, Voinnet O. Control of RNA silencing and localization by endolysosomes. Trends Cell Biol. 2010;20:491–501. doi: 10.1016/j.tcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Turner JJ, Williams D, Owen D, Gait MJ. Disulfide conjugation of peptides to oligonucleotides and their analogs. Curr Protoc Nucleic Acid Chemistry 2006; Chapter 4:Unit 4.28. [DOI] [PubMed] [Google Scholar]
- 51.Beaucage S, Caruthers M. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–62. doi: 10.1016/S0040-4039(01)90461-7. [DOI] [Google Scholar]