Abstract
We have explored the merits of a novel delivery strategy for the antisense oligomers based on cell penetrating peptide (CPP) conjugated to a carrier PNA with sequence complementary to part of the antisense oligomer. The effect of these carrier CPP-PNAs was evaluated by using antisense PNA targeting splicing correction of the mutated luciferase gene in the HeLa pLuc705 cell line, reporting cellular (nuclear) uptake of the antisense PNA via luciferase activity measurement. Carrier CPP-PNA constructs were studied in terms of construct modification (with octaarginine and/or decanoic acid) and carrier PNA length (to adjust binding affinity). In general, the carrier CPP-PNA constructs including the ones with decanoyl modification provided significant increase of the activity of unmodified antisense PNA as well as of antisense octaarginine-PNA conjugates. Antisense activity, and by inference cellular delivery, of unmodified antisense PNA was enhanced at least 20-fold at 6 μM upon the complexation with an equimolar amount of nonamer carrier decanoyl-CPP-PNA (Deca-cPNA1(9)-(D-Arg)8). The antisense activity of a CPP-PNA ((D-Arg)8-asPNA) (at 2 μM) was improved 6-fold and 8-fold by a heptamer carrier CPP-PNA (cPNA1(7)-(D-Arg)8) and hexamer carrier decanoyl-CPP-PNA (Deca-cPNA1(6)-(D-Arg)8), respectively, without showing significant additional cellular toxicity. Most interestingly, the activity reached the same level obtained by enhancement with endosomolytic chloroquine (CQ) treatment, suggesting that the carrier might facilitate endosomal escape. Furthermore, 50% downregulation of luciferase expression at 60 nM siRNA was obtained using this carrier CPP-PNA delivery strategy (with CQ co-treatment) for a single stranded antisense RNA targeting normal luciferase mRNA. These results indicated that CPP-PNA carriers may be used as effective cellular delivery vectors for different types of antisense oligomers and also allows use of combinations of (at least two) different CPP ligands.
Keywords: antisense, carrier, cell penetrating peptide (CPP), cellular delivery, peptide nucleic acid (PNA), siRNA
Introduction
Nucleic acid-based strategies to modulate cellular functions through e.g., RNA interference downregulation of gene expression1 are successfully used both in research and in drug discovery/development, and exploit a variety oligonucleotides, including antisense oligomers, small interfering RNAs (siRNAs), ribozymes, transcription factor-binding decoy and triplex-forming oligonucleotides, but also miRNAs, aptamers and, immunostimulatory CpG oligonucleotides. However, insufficient (cellular) delivery technology has been limiting development in particular for in vivo (therapeutic) applications.
Many methods have been described for the antisense oligonucleotide delivery each having advantages and disadvantages.2,3 In general physical methods (such as microinjection, electroporation or particle bombardment) are efficient and highly selective in terms of cells and tissues but are rather harmful for the target cells and have limited potential for in vivo applications. Viral vector systems are very effective in delivering larger nucleic acids and substantial efforts have been invested in these for gene therapy,4 but they pose challenges in terms of immune responses. Alternatively, non-viral delivery systems (e.g., transfection reagent based on cationic lipids or polymers) may offer advantages. Currently, cationic lipids are the most common carriers for in vitro cellular transfection of nucleic acids (DNA or RNA) as well as for negatively charged DNA/RNA analog oligomers. These methods are rather simple to perform in vitro, but are very difficult to translate to in vivo applications due to substantial in vivo toxicity, although this approach is still being pursued for in vivo delivery of siRNAs using cationic polymers such as cationic lipid,5 atelocollagen or PEI.6,7 A group of cationic peptides, so called cell penetrating peptides (CPPs), are also widely employed as vectors for delivery of large cargo molecules into cells8-10 using either covalent chemical conjugation11-13 or simple complexation via electrostatic (or hydrophobic) interactions. Covalent conjugation of CPP to cargo molecules may be challenging due to tedious chemical synthesis, and will also change the properties of any given cargo molecule by coupling of highly cationic CPPs. Alternatively, the assembly of CPP and cargo molecule can be achieved by electrostatic interactions through the positively charged CPPs and a negatively charged cargo (DNA or siRNAs),14-16 and the assembled complexes are successfully delivered into the cells17-19 both in vitro and in vivo. One drawback of the simple complexation method, is the large excess of CPPs typically required (ratio of 10:118) to obtain an overall positive charge which is essential for the initial interaction (binding) of these complexes to negatively charged (heparansulphate) glycans of the extracellular matrix.20-24 Moreover, complexation based on ionic interaction is not applicable for charge neutral antisense oligomers such as peptide nucleic acid (PNA) or phosphorodiamidate morpholino oligonucleotides (PMO), whereas these synthetic antisense molecules have shown promising biological activity upon covalent conjugation to CPPs.11,25,26
Sequence complementary hybridization is a possible alternative method for non-covalent coupling of CPPs or other targeting ligands (i.e., oligonucleotide aptamer27 or TLR9 agonist28) to antisense oligomers including siRNA. PNA is a promising candidate for the hybridization domain for such carrier CPP as it is charge neutral and forms very stable duplexes with cDNA or RNA.29 For instance Doyle et al. have (in an “inverse” approach) shown that preformed PNA/DNA heteroduplex, using cDNA, are efficiently transfected into cells via cationic lipids exploiting the negative charges of the DNA strand for ionic complexation with cationic lipids.30 In addition, Fischer et al. showed that CPP-PNA conjugates can be used as a delivery vector for a NFκB DNA decoy,31 and Sisido et al. have used PNA conjugates for siRNA delivery.32
Inspired by these findings, we decided to explore in more detail the principle of using CPP-PNA conjugates for cellular delivery of antisense oligomers as illustrated in scheme 1. Employing antisense PNA targeting splicing correction of the mutated luciferase gene in the HeLa pLuc705 cells, we now show that the cellular antisense effect of both unmodified (naked) PNA as well as antisense CPP- PNAs (modified with octaarginine or decanoyl-octaarginine) can be significantly improved by carrier CPP (octaarginine)-PNAs. In addition, we show that effective cellular delivery of single stranded (si)RNA is achieved using a carrier CPP-PNA in combination with the endosomolytic reagent chloroquine.
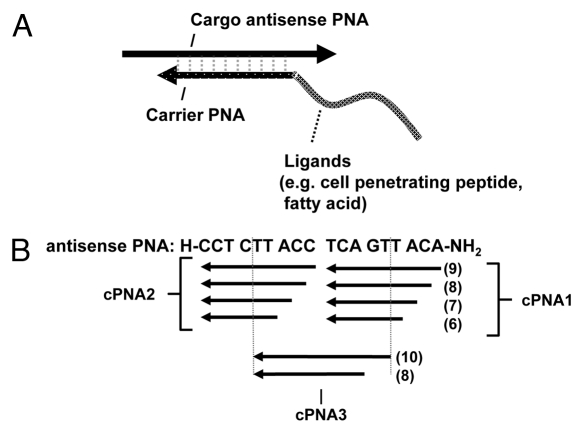
Scheme 1.
Concept of cell delivery carriers for antisense oligomers based on delivery ligands such as cell penetrating peptide (CPP) conjugated to peptide nucleic acids (PNAs). (A) The carrier CPP-PNA is hybridized in an anti-parallel orientation to (part of) the target (antisense) cargo oligomer. (B) Design of carrier PNAs with different length (6–9 nucleobases for both cPNA 1 and cPNA 2 and 8 or 10 or 8 nucleobases for cPNA3) and different target positions within the antisense PNA.
Results and discussion
As an alternative to covalent conjugation or to electrostatic or hydrophobic complexation of CPPs to antisense oligomers hybridization specific assembly using carrier PNAs offers greater flexibility. In order to explore in more detail the possibilities for developing such “carrier CPP-PNAs” for cellular delivery of antisense oligomers, we synthesized a series of carrier PNAs modified with different delivery ligands such as octaarginine and/or decanoic acid (Table 1), complementary to different positions within the antisense PNA oligomer (cPNA1, cPNA2, and cPNA3) (Scheme 1). The well characterized antisense cargo PNA is targeted to an intronic aberrant splice site of the luciferase pre-mRNA in HeLa pLuc705 cells and the antisense activity measured as luciferase activation accomplished as a consequence of splicing correction and interpreted as a measure of (productive) cellular (nuclear) uptake efficiency.33
Table 1. List of PNAs.
| PNAsa | No. | Name | sequenceb |
|---|---|---|---|
| Antisense PNA |
2389 |
Naked asPNA |
H-CCT CTT ACC TCA GTT ACA-NH2 |
| |
2787 |
(D-Arg)8-asPNA |
H-(D-Arg)8-Gly- CCT CTT ACC TCA GTT ACA-NH2 |
| |
2802 |
(D-Arg)8-Deca-asPNA |
H- (D-Arg)8-Lys(Deca)-Gly-CCT CTT ACC TCA GTT ACA-NH2 |
| |
2919 |
Fl-(Arg)8-asPNA |
H-(Arg)8-Flk-CCT CTT ACC TCA GTT ACA-NH2 |
| Carrier PNA1 |
2922 |
Naked-cPNA1(9) |
H- TGT AAC TGA-Gly-NH2 |
| |
2923 |
(D-Arg)8-cPNA1(9) |
H-(D-Arg)8- TGT AAC TGA-Gly-NH2 |
| |
2924 |
(D-Arg)8-Deca-cPNA1(9) |
H-(D-Arg)8- Lys(Deca)- TGT AAC TGA-Gly-NH2 |
| |
2925 |
Deca-cPNA1(9) |
Deca-TGT AAC TGA-Gly-NH2 |
| |
2962 |
cPNA1(9)-(D-Arg) 8 |
H- TGT AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2960 |
cPNA1(8)-(D-Arg) 8 |
H- GT AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2958 |
cPNA1(7)-(D-Arg) 8 |
H- T AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2956 |
cPNA1(6)-(D-Arg) 8 |
H- AAC TGA- (D-Arg)8- Gly-NH2 |
| |
2963 |
Deca-cPNA1(9)-(D-Arg) 8 |
Deca- TGT AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2961 |
Deca-cPNA1(8)-(D-Arg) 8 |
Deca- GT AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2959 |
Deca-cPNA1(7)-(D-Arg) 8 |
Deca- T AAC TGA- (D-Arg)8 Gly-NH2 |
| |
2957 |
Deca-cPNA1(6)-(D-Arg) 8 |
Deca- AAC TGA- (D-Arg)8- Gly-NH2 |
| Carrier PNA2 |
2926 |
Naked-cPNA2(9) |
H-GGT AAG AGG-Gly-NH2 |
| |
2927 |
Deca-cPNA2(9) |
Deca-GGT AAG AGG-Gly-NH2 |
| |
2955 |
Deca-cPNA2(8) |
Deca- GT AAG AGG- Gly-NH2 |
| |
2953 |
Deca-cPNA(7) |
Deca- T AAG AGG- Gly-NH2 |
| |
2951 |
Deca-cPNA(6) |
Deca- AAG AGG- Gly-NH2 |
| Carrier PNA3 |
3229 |
(His)4-cPNA3(8) |
Ac-(His)4 - TGA GGT AA -Lys-NH2 |
| |
3230 |
(His)6-cPNA3(8) |
Ac-(His)6 - TGA GGT AA -Lys-NH2 |
| |
3247 |
(Cholate)3-cPNA3(10) |
Cholic acid - ACTLys(Cholate) GAG GTLys(Cholate)A A -Lys-NH2 |
| |
3202 |
(Lys)3-cPNA3(10) |
H- ACTLys GAG GTLysA A -Lys-NH2 |
| Carrier PNA4 |
3164 |
(D-Arg)8-cPNA4 |
H-(D-Arg)8- AGA CGC CA -Lys-NH2 |
| 3165 | Deca-(D-Arg)8-cPNA4 | Deca-(D-Arg)8- AGA CGC CA -Lys-NH2 |
a PNA types: Antisense PNA, PNAs targeting splicing correction of mutated luciferase gene in the HeLa pLuc705 cells; Carrier PNA1, PNAs complementary to C-terminus end of the antisense PNA; Carrier PNA2, PNAs complementary to the N-terminus end of the antisense PNA; Carrier PNA3, PNAs complementary to the middle of the antisense PNA; Carrier PNA4, PNAs complementary to the antisense strand of siRNA targeting normal luciferase mRNA. bThe sequences of the PNAs are written from N-terminal to C-terminal end. Amino acids, decanoic acid (Deca) or cholic acid (cholate) moiety was covalently linked to PNA at the C-terminal end, N-terminal end, ε-amine group of the lysine side chain, or TLys within the PNA backbone where Tlys is lysine derived backbone unit instead of normal aminoethylglycyl backbone unit. Flk, fluorescein (Fl) attached to theε-amino group of a lysine residue (Lohse, J.et al. (1997) Bioconjugate Chem. 8, 503-509).
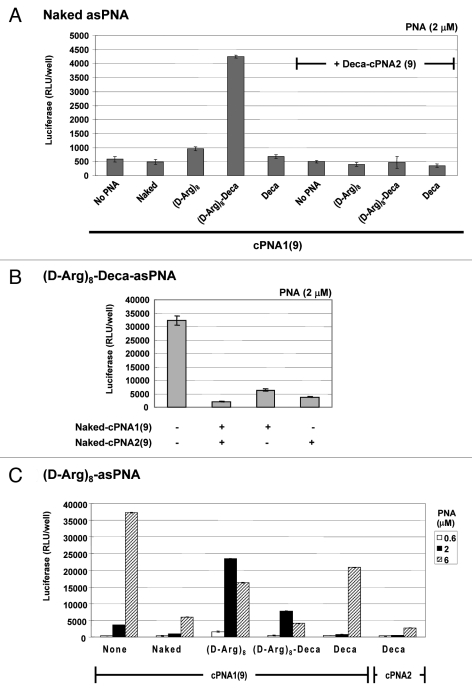
Four carrier PNAs complementary to the C-terminus 9 nucleobases of the antisense PNA and differently modified (Naked-cPNA1(9), (D-Arg)8-cPNA1(9), (D-Arg)8-Deca-cPNA1(9), Deca-cPNA1(9)) were tested for enhancing the cellular antisense effect of unmodified PNA (naked asPNA) (Fig. 1A). Naked antisense PNA itself did not show any antisense activity over background (even at the highest concentration used (6 μM)). However, the activity was increased up to 8-fold at 2 μM in combination with a decanoyl-octaarginine carrier CPP-PNA ((D-Arg)8-Deca-cPNA1(9)), while the analogous decanoyl carrier PNA (deca-cPNA(9)) did not show any significant improvement. These carrier PNAs were also tested in combination with a second decanoyl carrier PNA (Deca-cPNA2(9)) targeting the other half (N-terminus 9 nucleobases) of the antisense PNA, inspired by the previously reported improved cellular uptake by lipidic modification of CPP-PNA.33 However, the additional carrier decanoyl-PNA did not improve the effect of the other carrier PNAs. On the contrary a significant decrease of activity for the active carrier CPP-PNA ((D-Arg)8-Deca-cPNA1(9)) was observed. The reason for the decreased activity with a combination of two carrier PNAs is not entirely clear, but it might be due to steric blocking of the antisense PNA by the bound carrier PNAs or altered uptake (mechanism). To challenge the first hypothesis, we chose an active antisense decanoyl-octaarginine-PNA ((D-Arg)8-Deca-asPNA) and transfected this in complex with one or two unmodified carrier PNAs ((Naked cPNA1(9)) and (Naked-cPNA2(9)) at 2 μM (Fig. 1B). These two carrier PNAs both dramatically inhibited the antisense effect around 80% and in combination the antisense activity was reduced to background level. This clearly suggests that dissociation of the carrier PNA(s) from the antisense PNA strand is a rate limiting step for the antisense activity. Nonetheless, the results also indicate that carrier CPP-PNAs may improve the (spontaneous) cellular uptake of unmodified antisense PNA without direct chemical conjugation to the CPP. In order to further explore the carrier PNA aided transfection strategy, these carrier PNAs were also tested for effects on antisense octaarginine CPP-PNA conjugates ((D-Arg)8-asPNA)(Fig. 1C). The antisense activity of the (D-Arg)8-asPNA (at 2 μM) was improved up to 6-fold and 2-fold by (D-Arg)8-cPNA1(9) and (D-Arg)8-Deca-cPNA1(9), respectively, while the decanoic acid carrier PNA did not show any improvement, but rather showed inhibitory effects. Especially, decanoic acid PNA2 (Deca-cPNA2(9)) showed the most significant inhibition among the carrier PNAs. These results indicate that carrier CPP-PNAs can improve the cellular uptake of antisense CPP-PNA. Based on these findings, we decided to optimize the binding strength (affinity) between the antisense PNA and the carrier CPP-PNA by changing a sequence length of carrier PNAs.
Figure 1.
Carrier PNA effect on different antisense CPP-PNAs. Relative antisense activity in HeLa pLuc705 cells of antisense PNAs hybridized to a carrier PNA1 (Naked-cPNA1(9), (D-Arg)8-cPNA1(9), (D-Arg)8-Deca-cPNA1(9), Deca-cPNA1(9)) and/or a carrier PNA2 (Naked cPNA2(9), Deca-cPNA2(9)). Antisense PNA was hybridized to a carrier PNA(s) at 1:1 molar ratio and used for transfection at the indicated concentrations. After 24 h transfection, cells were subjected to the luciferase analysis. Each data set represents the mean ± SD of three independent experiments Antisense PNAs used: (A) Unmodified PNA (Naked asPNA (PNA2389)) at 2 μM, (B) Decanoyl-octaarginine PNA ((D-Arg)8-Deca-asPNA (PNA2802)) at 2 μM or (C) octaarginine PNA ((D-Arg)8-asPNA (PNA2787)).
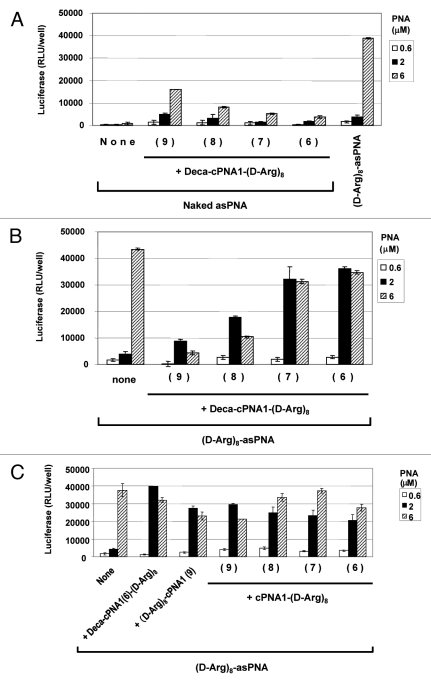
We synthesized a new series of carrier CPP-PNAs of different PNA length (9–6 nucleobases) with octaarginine-decanoic acid modification (Deca-cPNA1(9–6)-(D-Arg)8) and tested their effect on PNA antisense activity. In combination with unmodified antisense PNA (Fig. 2A), the longest carrier CPP-PNA with 9 nucleobases showed the highest antisense activity improvement (at least 20-fold at 6 μM), and the activity at 2 μM equals antisense the effect of (D-Arg)8-asPNA at 2 μM. However, this activity improvement was decreased with shorter carrier CPP-PNAs. This could suggest that a certain length of carrier PNA (hence certain binding strength) is required to form a sufficiently stable complex during cellular delivery. Then, we tested the effect of these carrier CPP-PNAs on the antisense activity of octaarginine (D-Arg)8-asPNA (Fig. 2B). In contrast to the results with the unmodified antisense PNA, the antisense CPP-PNA showed higher antisense activity in combination with the shorter carrier CPP-PNAs. The shortest carrier CPP-PNA of 6 nucleobases ((Deca-cPNA1(6)-D-Arg)8) showed the highest improvement of the activity yielding almost 10-fold activation at 2 μM whereas no activation was seen at 6 μM. The reason for the opposite length dependency for unmodified antisense PNA and CPP-PNA is not clear at this stage, but most likely it is reflecting a delicate balance of sufficient stability of the cPNA-asPNA complexes (which exhibit thermal stabilities (Tm) ranging from 45–64°C (hexa- to nonamers) for delivery and adequate lability for dissociation inside the cell.
Figure 2.
Effect of carrier PNA length. Relative antisense activity in HeLa pLuc705 cells of antisense PNAs hybridized to carrier CPP-PNA of different PNA length (9–6 nucleobases) modified with decanoyl-octaarginine (Deca-cPNA1-(D-Arg)8 (PNA2963, 2961, 2959, 2957)) (A, B) or octaarginine (cPNA1(9–6 nucleobases) -(D-Arg)8 (PNA2962, 2960, 2958, 2956)) (C) (see Scheme 1). Antisense PNA was hybridized to the carrier CPP-PNA at 1:1 molar ratio and used for transfection at the indicated concentrations. After 24 h transfection, cells were subjected to the luciferase analysis. Each data set represents the mean ± SD of three independent experiments. Antisense PNAs used: (A) Unmodified PNA (Naked asPNA (PNA2389)) ((D-Arg)8-asPNA (PNA2787) as control) (B) and (C) (D-Arg)8-asPNA (PNA2787).
In order to explore further the structure activity relations of CPP carrier PNAs, we next studied a series of similar cPNAs modified only with octaarginine. In this case no clear length dependence was found and as before activation was seen at 2 μM (Fig. 2C). Likewise moving the peptide to the N-terminal instead of the C-terminal of the PNA did not affect the enhancement activity.
Although the results so far (Figs. 1A,C) would indicate negative effects on antisense activity of a lipophilic carrier PNA, despite the clear positive effect on antisense activity upon covalent conjugation to CPPs in the CatLip approach,33 we decided to more systematically test the effect on the antisense activity of (D-Arg)8-asPNA of a series Deca-cPNA2 (9–6 nucleobases)) complementary to the N-terminus of the antisense PNA. However, these carrier PNAs all showed inhibitory effects on the antisense activity of the (especially at high PNA concentration (6 μM) (Fig. S1). The longest nonamer carrier PNA (Deca-cPNA2(9)) showed the highest inhibition, and the inhibition remained even with the shortest hexamer carrier PNA (Deca-cPNA2(6)). The reason for this inhibitory effect when supplied in trans (hybridization) rather than in cis (direct conjugation) is not clear, but because of this (unexpected) inhibitory effect, these carrier PNAs were not studied further.
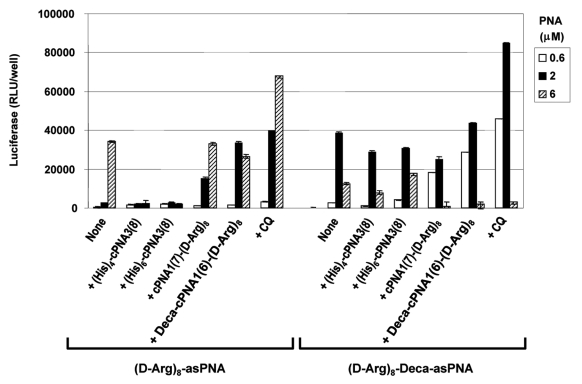
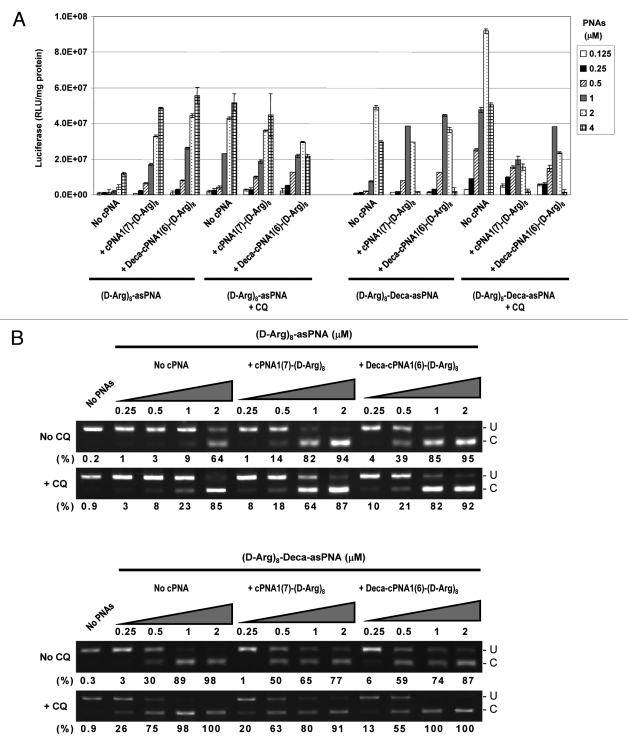
Although CPP conjugation can very significantly improve PNA antisense activity, the efficacy of CPP-PNA conjugates may be further enhanced (up to 100-fold depending on CPPs and formulation) by endosomolytic agents such as chloroquine (CQ).34 In order to study this approach in relation to carrier CPP-PNAs, two of the most active carrier CPP-PNA constructs, cPNA1(7)-(D-Arg)8 and Deca-cPNA1(6)-(D-Arg)8 were chosen. The results (Figs. 3 and 4) show that the antisense activity of the CPP-asPNAs (D-Arg)8-asPNA and (D-Arg)8-Deca-asPNA were improved (up to 15-fold) by CQ treatment and the activation is significantly more pronounced at lower PNA concentrations (e.g., 2 μM for (D-Arg)8-asPNA and 0.5 μM for (D-Arg)8-Deca-asPNA). Interestingly, employment of carrier CPP-PNAs Deca-cPNA1(6)-(D-Arg)8 and cPNA1(7)-(D-Arg)8 enhanced the antisense activity to virtually the same level as did CQ treated CPP-PNA at 0.5 μM and 1 μM, respectively. The decrease of activity at higher PNA concentrations (6 μM for (D-Arg)8-Deca-asPNA) is ascribed to cellular toxicity of PNA conjugates33 (Fig. S2). In addition to the two promising carrier CPP-PNAs, we also tested two carrier PNA constructs conjugated to oligo-histidine residues, the tetrahistidine (His)4-cPNA3(8) and hexahistidine (His)6-cPNA3(8) carrier PNAs, which could potentially provide endosomal disruption by the “proton sponge effect” of histidine residues in the acidic endosomal compartment as previously reported.35 However, no improvement was observed (in fact activity decreased) in contrast to apparent successes reported in the literature.36,37 Finally, we studied the effect of CQ treatment on carrier CPP-PNA enhanced PNA antisense activity. The results presented in Figure 4 show that additional CQ treatment does not further enhance the activity obtained with the antisense PNA alone in combination with CQ. In fact some inhibition is observed, especially when using CatLip type PNAs and at least some of this effect could be due to cellular toxicity under these conditions (Fig. S2).
Figure 3.
Oligohistidine carriers. Relative antisense activitiy in HeLa pLuc705 cells of antisense CPP-PNAs ((D-Arg)8-asPNA (PNA2787) and (D-Arg)8-Deca-as PNA (PNA2802)) hybridized to carrier CPP-PNA. Two carrier PNA constructs with oligohistidine ((His)4-cPNA3(8) (PNA3229) and (His)6-cPNA3(8) (PNA3230)) were compared with two octaarginine conjugated carrier CPP-PNA constructs (cPNA1(7)-(D-Arg)8 (PNA2958) and Deca-cPNA1(6)-(D-Arg)8 (PNA2957)). For the chloroquine (CQ) treatment, 100 μM CQ was added to the medium for PNA transfection. Antisense CPP-PNA was hybridized to the carrier PNA at 1:1 molar ratio and used for transfection at the indicated concentrations. After 24 h transfection, cells were subjected to the luciferase analysis. Each data set represents the mean ± SD of three independent experiments.
Figure 4.
Effect of chloroquine. Relative antisense activity in HeLa pLuc705 cells of two antisense CPP-PNAs ((D-Arg)8-asPNA (PNA2787) and (D-Arg)8-Deca-asPNA (PNA2802)) hybridized to a carrier CPP-PNA. Antisense PNA was hybridized with one of the carrier CPP-PNAs (cPNA1(7)-(D-Arg)8 (PNA2958) or Deca-cPNA1(6)-(D-Arg)8 (PNA2957)) at 1:1 molar ratio and transfected to the cells at the indicated concentrations in the absence or the presence of 100 μM chloroquine (CQ). After 24 h transfection, cells were subjected to further analysis. (A) Luciferase activity was measured and normalized by the protein concentration (presented as RLU/mg protein). Each data set represents the mean ± SD of three independent experiments. (B) RT-PCR analysis of the mis-splicing correction of luciferase pre-mRNAs by antisense PNA. The numbers under the figure indicate the relative fraction (%) of the corrected form. U: PCR product without correction (268 bp). C: PCR product with mis-splicing correction (142 bp).
The luciferase data (Fig. 4A) were fully corroborated by mRNA splice correction analyses using RT-PCR (Fig. 5B), and again these results demonstrate that the effect of carrier CPP-PNA delivery is comparable to or even higher than that obtained using CQ with the antisense PNA alone (compare for instance “no cPNA/+CQ with + cPNA1(7)-(D-Arg)8/no CQ and Deca-cPNA1(6)-(D-Arg)8/no CQ in Figure 4B). These results suggest that carrier CPP-PNA-mediated enhancement of antisense CPP-PNA delivery could be an attractive alternative to auxiliary CQ treatment since this method gives comparable or even higher activity than that achieved by CQ treatment, without significantly increased cell toxicity (Fig. S2), despite the cellular toxicity of (especially the CatLip) antisense CPP-PNAs observed at higher PNA concentrations (2-4 μM).
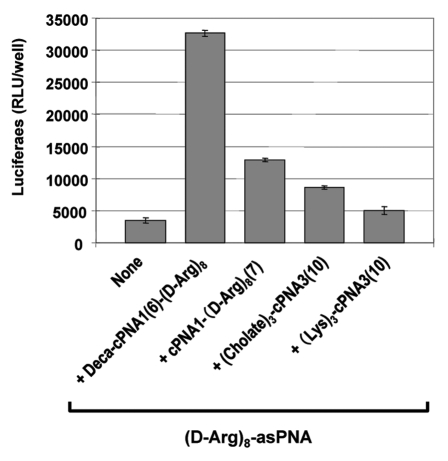
Figure 5.
Effect of cholate carrier PNA. Relative antisense activity in HeLa pLuc705 cells of octaarginine conjugated antisense PNA ((D-Arg)8-asPNA (PNA2787)) hybridized to carrier PNA. Two carrier PNAs with backbone modification by Lys-derived backbone units ((Lys)3-cPNA3 (PNA3202)) or Lys residues with cholic acid modification ((Cholate)3-cPNA3 (PNA3247)) were compared with two carrier CPP-PNAs (Deca-cPNA1(6)-(D-Arg)8 (PNA2957) and cPNA1(7)-(D-Arg)8 (PNA2958)). Antisense PNA was hybridized to the carrier PNA at 1:1 molar ratio and transfected to the cells at 2 μM. After 24 h transfection, cells were subjected to the luciferase analysis. Each data set represents the mean ± SD of three independent experiments.
Lipophilic ligands such as cholesterol and cholic acid2,38 have been found to increase cellular uptake and bioavailability of oligonuceotides, and especially cholic acid moieties were used to improve a lipid bilayer penetration of rather large hydrophilic compounds including 16 mer oligonucleotide,39 presumably by shielding of the hydrophilic cargo by a cholic acid “molecular umbrella.” Thus we synthesized a 10-mer carrier PNA containing three cholic acid moieties in the backbone of the PNA ((Cholate)3-cPNA3(10), Figure 5). The activity of the antisense CPP-PNA ((D-Arg)8-asPNA) together with (Cholate)3-cPNA3(10) carrier at 2 μM was slightly higher (ca. 2-fold) than the antisense CPP-PNA itself while only a negligible increase was seen with a control carrier PNA construct without cholic acid moieties ((Lys)3-cPNA3(10)). Thus the cholic acid modified carrier PNA did indeed enhance the antisense PNA activity, but to a significantly lesser degree than the carrier CPP-PNAs (up to 6-fold). The mechanism of the enhancement (improved membrane passage?) is not clear at this point.
In order to study the effect of carrier CPP-PNA on the cellular localization of the antisense CPP-PNAs by fluorescence microscopy, we synthesized fluorescein-labeled octaarginine antisense PNA (Fl-(Arg)8-asPNA (PNA2919)) and used this for transfection of the cells in combination with carrier CPP-PNAs (cPNA1(7)-(D-Arg)8 or Deca-cPNA1(6)-(D-Arg)8). Fluorescence microscopy indicated a slightly higher intensity of fluorescence with more even distribution in both cytoplasm and nucleus upon delivery with carrier PNAs or upon CQ co-treatment as compared with treatment with the antisense PNA alone (Fig. S3). The changes of the cellular localization of the cargo Fl-CPP-PNA by carrier CPP-PNAs were not as evident as suggested from the luciferase experiments. However, this preliminary experiment together with the luciferase experiments do suggest that carrier CPP-PNAs (like chloroquine) facilitate endosome disruption.
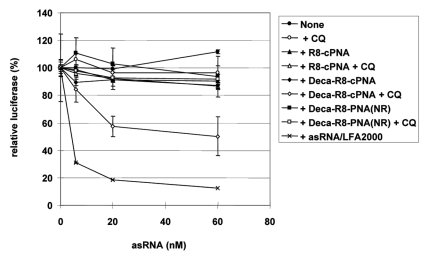
To further explore the carrier CPP-PNA delivery method for other (antisense) oligomers, we chose a single stranded short RNA originating from the antisense strand of an siRNA targeting normal luciferase mRNA. This single stranded antisense siRNA showed 70% downregulation of luciferase activity at 80 nM in p53R cells (expressing normal luciferase) when delivered by cationic lipid transfection (LFA2000) under conditions where the complete double stranded siRNA showed up to 90% downregulation (Fig. S4). Two carrier CPP-PNAs targeting the PNA to the 3′-end 8 nt of the antisense siRNA strand and modified with either octaarginine alone ((D-Arg)8-cPNA4) or with decanoyl-octaarginine ((D-Arg)8-Deca-cPNA4) were synthesized. The antisense siRNA strand was hybridized to the carrier CPP-PNA at equimolar ratio and used for transfection in the absence or presence of CQ (Fig. 6). In the absence of CQ, the antisense RNA - alone or combined with either of the carrier CPP-PNA - did not show any significant luciferase downregulation even at the highest concentration (60 nM), while upon cationic lipid transfection very siginificant downregulation (90%) was seen. However, in combination with CQ treatment the decanoyl-octaarginine carrier CPP-PNA ((D-Arg)8-Deca-cPNA4) very significant downregulation (59%) at 60 nM was obtained. This result emphasizes the general possibilities of exploiting carrier CPP-PNA based delivery formulation (via base-pair recognition) at low non-cytotoxic concentrations as it does not require a (large) molar excess of the cationic carrier, but also strongly suggests that delivery in this case occurs via endosomal pathways.
Figure 6.
Downregulation of luciferase in p53R cells by a single stranded antisense siRNA (asRNA) delivered by carrier CPP-PNAs. Two octaarginine conjugated carrier CPP-PNAs (R8-cPNA, (PNA3164) and Deca-R8-cPNA (PNA3165)) with a complementary sequence to the asRNA and one PNA with non-related (NR) sequence but with analogous modifications (Deca-R8-PNA(NR), PNA2961)) as a control, were tested. The asRNA was hybridized to the carrier CPP-PNA at the molar ratio 1:1 and transfected to the p53R cells at the indicated concentrations in the absence or presence of 100 μM chloroquine (CQ). After 48 h transfection, luciferase activity was measured and is shown as relative luciferase activity (%) after normalization by protein concentration (non-siRNA treated sample set as 100%). Each data point represents the mean ± SD of three independent experiments.
Conclusion
In conclusion we have found that sequence complementary carrier CPP-PNAs can significantly (more than one order of magnitude in some cases) enhance the cellular antisense effects of both unmodified PNA as well as of PNA-CPP conjugates. The effect depends on the length and modification of the carrier PNA, and is most pronounced at lower PNA concentrations, and thus requires extensive optimization. Furthermore, the method allows use of combinations of at least two different CPP ligands. It is noteworthy that the effect is not significantly enhanced by chloroquine, but reached activities comparable to those obtained without the carrier but in the presence of chloroquine, thereby suggesting that (at least part of) the effect of the carrier CPP-PNA may be ascribed to endosome disruption, and thus may substitute chloroquine enhancement. We are confident that these findings can be extended to other uncharged antisense oligomers such as phosphorodiamidate morpholino oligonucleotides (PMOs). Finally, this strategy may also be exploited (with CQ co-treatment) for negatively charged nucleic acids and their analogs such as locked nucleic acid (LNA) and RNA based molecules (short hairpin RNA, miRNA) without using molar excess of the carrier CPP for complexation.
Materials and methods
Synthesis of PNAs and CPP-PNA conjugates
The sequences of the PNAs are listed in Table 1. PNA synthesis was performed using boc-chemistry as reported elsewhere.40 Peptides or amino acids were linked to the PNA either at the C-terminal or N-terminal via continuous solid phase synthesis. The fatty acid was conjugated to the PNA N-terminus directly or to the ε-amino group of a lysine-moiety during synthesis using orthogonal Fmoc protection of the amine. The PNA conjugates were purified by HPLC and characterized by MALDI-TOF mass spectrometry. The synthesized PNAs were lyophilized and stored at 4°C until use.
Cell culture
HeLa pLuc705 cells and p53R cells (expressing normal luciferase) were purchased from GENE TOOLS (USA) and ATCC (USA) respectively. Cells were grown in RPMI1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma), 1% glutamax (Gibco), 100 U/ml penicillin (Gibco) and 100 μg/ml streptomycin (Gibco) at 37°C in humidified air with 5% CO2. For the studies in 96 well or 24 well plate format, cells were trypsinized and seeded at 1.2 x 104 or 7.2 x 104 cells/well, respectively, 16 -24 h before transfection.
Antisense PNA transfection
Antisense PNA was hybridized with complementary carrier PNA(s) at the molar ratio 1:1 and this pre-formed duplex was used for transfection. Hybridization was performed in a thermal cycler with the following temperature reduction profile: 95°C, 5 min; 85°C, 1 min; 75°C, 1min; 65°C, 5 min; 55°C, 1 min; 45°C, 1 min; 35°C, 5 min. For the transfection, cells replated in the multiple well plates were transfected with PNA (0.1 ml or 0.3 ml for 96 well and 24 well plate, respectively) by incubating in OPTI-MEM (Gibco) containing the PNAs for 4 h, and incubated further for 20 h after supplementing with the same volume of growth medium containing 20% FBS and 1% glutamax. For endosome disruption treatment by chloroquine (CQ), 100 μM CQ was added to the OPTI-MEM medium.
siRNA transfection
Double strand siRNA (dsRNA) was prepared by annealing sense strand RNA (ssRNA, 5′-ACGCCAAAAACAUAAAGAAAG-3′) and antisense strand RNA (asRNA, 5′-UUCUUUAUGUUUUUGGCGUCU-3′) in 10 mM TRIS-HCl (pH 7.5) with 20 mM NaCl. The mixture was heated at 95°C for 10 min, then gradually cooled down to room temperature, and incubated for 16–20 h at room temperature. For the transfection by cationic lipid LipofectAMINE2000 (LFA2000, Invitrogen), RNAs (ssRNA, asRNA, dsRNA) were complexed with LFA2000 and transfected to the cells according to the manufacturer’s instruction. For transfection by carrier CPP-PNA, single strands of siRNA (asRNA) was annealed with CPP-PNA at the molar ratio 1:1 and delivered as described above. RNA oligonucleotides were obtained from Eurofin (Germany).
Luciferase assay
Luciferase activity of the cells or cell lysates was measured by using the Bright-Glo Luciferase assay system (Promega) according to the manufacturer’s instructions. Luminescent readings from 96 well plate formats (with background subtracted) are presented as relative light units (RLU/well), or (as indicated) after normalization with protein concentration obtained with BCA protein assay kit (Pierce) shown as RLU/mg protein.
Cytotoxicity test
Cells in 96-well plates were subjected to the MTS-assay for cell viability testing by using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer’s instructions. The absorbance is presented as relative cellular viability (absorbance from non-PNA treated cells was set as 100%).
RT-PCR
Total RNA was extracted from the cells by using RNeasy Mini kit (Qiagen) and subjected to RT-PCR analysis following company instructions. Three ng of total RNA was used for each RT-PCR in 20 μl reaction. RT-PCR was performed by using OneStep RT-PCR kit (Qiagen) following company instructions. Primers for the RT-PCR were as follows: forward primer, 5′-TTGATATGTGGATTTCGAGTCGTC-3′; reverse primer, 5′-TGTCAATCAGAGTGCTTTTGG-CG-3′. The RT-PCR program was as follows: {(55°C, 35 min) x 1cycle, (95°C, 15 min) x 1cycle, (94°C, 0.5 min; 55°C, 0.5 min; 72°C, 0.5 min) x 29 cycles}. RT-PCR products were analyzed on 2% agarose gel with 1x TBE buffer and visualized by ethidium bromide staining. Gel images were captured by ImageMaster (Pharmacia Biotech) and analyzed by UN-SCAN-IT software (Silk Scientific Corporation).
Supplementary Material
Supplementary PDF file supplied by authors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Lundbeck Foundation and The Danish Council for Independent Research/Medical Sciences
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/18739
References
- 1.Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nat Rev Drug Discov. 2002;1:503–14. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- 2.Manoharan M. Oligonucleotide conjugates as potential antisense drugs with improved uptake, biodistribution, targeted delivery, and mechanism of action. Antisense Nucleic Acid Drug Dev. 2002;12:103–28. doi: 10.1089/108729002760070849. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh C, Iversen PL. Intracellular delivery strategies for antisense phosphorodiamidate morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2000;10:263–74. doi: 10.1089/108729000421448. [DOI] [PubMed] [Google Scholar]
- 4.Verma IM, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–38. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–6. doi: 10.1016/S0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 6.Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–82. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grzelinski M, Urban-Klein B, Martens T, Lamszus K, Bakowsky U, Hobel S, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17:751–66. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- 8.Ezzat K, El Andaloussi S, Abdo R, Langel U. Peptide-based matrices as drug delivery vehicles. Curr Pharm Des. 2010;16:1167–78. doi: 10.2174/138161210790963832. [DOI] [PubMed] [Google Scholar]
- 9.Dietz GP, Bahr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Fischer R, Fotin-Mleczek M, Hufnagel H, Brock R. Break on through to the other side-biophysics and cell biology shed light on cell-penetrating peptides. ChemBioChem. 2005;6:2126–42. doi: 10.1002/cbic.200500044. [DOI] [PubMed] [Google Scholar]
- 11.Zatsepin TS, Turner JJ, Oretskaya TS, Gait MJ. Conjugates of oligonucleotides and analogues with cell penetrating peptides as gene silencing agents. Curr Pharm Des. 2005;11:3639–54. doi: 10.2174/138161205774580769. [DOI] [PubMed] [Google Scholar]
- 12.Cesarone G, Edupuganti OP, Chen CP, Wickstrom E. Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjug Chem. 2007;18:1831–40. doi: 10.1021/bc070135v. [DOI] [PubMed] [Google Scholar]
- 13.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–8. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 14.Deshayes S, Morris M, Heitz F, Divita G. Delivery of proteins and nucleic acids using a non-covalent peptide-based strategy. Adv Drug Deliv Rev. 2008;60:537–47. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Meade BR, Dowdy SF. Enhancing the cellular uptake of siRNA duplexes following noncovalent packaging with protein transduction domain peptides. Adv Drug Deliv Rev. 2008;60:530–6. doi: 10.1016/j.addr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59:134–40. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Unnamalai N, Kang BG, Lee WS. Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells. FEBS Lett. 2004;566:307–10. doi: 10.1016/j.febslet.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–24. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veldhoen S, Laufer SD, Trampe A, Restle T. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: quantitative analysis of uptake and biological effect. Nucleic Acids Res. 2006;34:6561–73. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi M, Rusnati M, Presta M, Giacca M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem. 2001;276:3254–61. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J Biol Chem. 2002;277:2437–43. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 22.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) "protein transduction domains" promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J Biol Chem. 2003;278:35109–14. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 23.Foerg C, Ziegler U, Fernandez-Carneado J, Giralt E, Rennert R, Beck-Sickinger AG, et al. Decoding the entry of two novel cell-penetrating peptides in HeLa cells: lipid raft-mediated endocytosis and endosomal escape. Biochemistry. 2005;44:72–81. doi: 10.1021/bi048330+. [DOI] [PubMed] [Google Scholar]
- 24.Mano M, Teodosio C, Paiva A, Simoes S, Pedroso de Lima MC. On the mechanisms of the internalization of S4(13)-PV cell-penetrating peptide. Biochem J. 2005;390:603–12. doi: 10.1042/BJ20050577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–45. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Turner JJ, Jones S, Fabani MM, Ivanova G, Arzumanov AA, Gait MJ. RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol Dis. 2007;38:1–7. doi: 10.1016/j.bcmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–49. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–32. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 30.Doyle DF, Braasch DA, Simmons CG, Janowski BA, Corey DR. Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry. 2001;40:53–64. doi: 10.1021/bi0020630. [DOI] [PubMed] [Google Scholar]
- 31.Fisher L, Soomets U, Cortes Toro V, Chilton L, Jiang Y, Langel U, et al. Cellular delivery of a double-stranded oligonucleotide NFkappaB decoy by hybridization to complementary PNA linked to a cell-penetrating peptide. Gene Ther. 2004;11:1264–72. doi: 10.1038/sj.gt.3302291. [DOI] [PubMed] [Google Scholar]
- 32.Kitamatsu M, Kubo T, Matsuzaki R, Endoh T, Ohtsuki T, Sisido M. Carrier PNA for shRNA delivery into cells. Bioorg Med Chem Lett. 2009;19:3410–3. doi: 10.1016/j.bmcl.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 33.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, et al. Solid-phase synthesis of peptide nucleic acids. J Pept Sci. 1995;1:175–83. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 34.Koppelhus U, Shiraishi T, Zachar V, Pankratova S, Nielsen PE. Improved cellular activity of antisense peptide nucleic acids by conjugation to a cationic peptide-lipid (CatLip) domain. Bioconjug Chem. 2008;19:1526–34. doi: 10.1021/bc800068h. [DOI] [PubMed] [Google Scholar]
- 35.Shiraishi T, Pankratova S, Nielsen PE. Calcium Ions Effectively Enhance the Effect of Antisense Peptide Nucleic Acids Conjugated to Cationic Tat and Oligoarginine Peptides. Chem Biol. 2005;12:923–9. doi: 10.1016/j.chembiol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Midoux P, Kichler A, Boutin V, Maurizot JC, Monsigny M. Membrane permeabilization and efficient gene transfer by a peptide containing several histidines. Bioconjug Chem. 1998;9:260–7. doi: 10.1021/bc9701611. [DOI] [PubMed] [Google Scholar]
- 37.Midoux P, LeCam E, Coulaud D, Delain E, Pichon C. Histidine containing peptides and polypeptides as nucleic acid vectors. Somat Cell Mol Genet. 2002;27:27–47. doi: 10.1023/A:1022931923153. [DOI] [PubMed] [Google Scholar]
- 38.Pichon C, Goncalves C, Midoux P. Histidine-rich peptides and polymers for nucleic acids delivery. Adv Drug Deliv Rev. 2001;53:75–94. doi: 10.1016/S0169-409X(01)00221-6. [DOI] [PubMed] [Google Scholar]
- 39.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 40.Janout V, Jing B, Regen SL. Molecular umbrella-assisted transport of an oligonucleotide across cholesterol-rich phospholipid bilayers. J Am Chem Soc. 2005;127:15862–70. doi: 10.1021/ja053930x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.