Abstract
How various epigenetic mechanisms restrict chromatin plasticity to determine the stability of repressed genes is poorly understood. Nuclear transfer to Xenopus oocytes induces the transcriptional reactivation of previously silenced genes. Recent work suggests that it can be used to analyze the epigenetic stability of repressed states. The notion that the epigenetic state of genes is an important determinant of the efficiency of nuclear reprogramming is supported by the differential reprogramming of given genes from different starting epigenetic configurations. After nuclear transfer, transcription from the inactive X chromosome of post-implantation-derived epiblast stem cells is reactivated. However, the same chromosome is resistant to reactivation when embryonic fibroblasts are used. Here, we discuss different kinds of evidence that link the histone variant macroH2A to the increased stability of repressed states. We focus on developmentally regulated X chromosome inactivation and repression of autosomal pluripotency genes, where macroH2A may help maintain the long-term stability of the differentiated state of somatic cells.
Keywords: epigenetic reprogramming, macroH2A, nuclear reprogramming, nuclear transfer, somatic stability, X chromosome inactivation, Xenopus oocytes
Introduction
Cellular differentiation is a unidirectional process starting from uncommitted, undifferentiated cells and ending with differentiated, stable somatic cells. During and after embryonic development, multiple mechanisms are used to ensure that cells do not go backward along their differentiation pathway, or change from one kind to another. Instead, they are stabilized within specific developmental paths, and, once differentiated, cells remain extremely stable; only in very rare cases do they fail to do so. Since the genetic information carried over from the undifferentiated to the differentiated state is maintained during differentiation, it is the epigenetic state of cells that determines their differentiated state and its stability. It is thought that differentiation entails a progressive epigenetic restriction of developmental potency, because the efficiency of nuclear reprogramming decreases as the differentiated state of somatic cells increases.1,2 How various epigenetic mechanisms contribute to the remarkable stability of the differentiated state is a fundamental but poorly understood question.
Resistance to Nuclear Reprogramming Revealed by Nuclear Transfer to Xenopus Oocytes
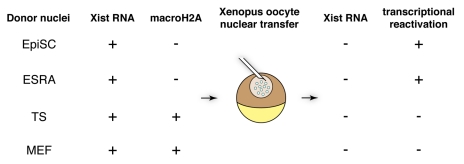
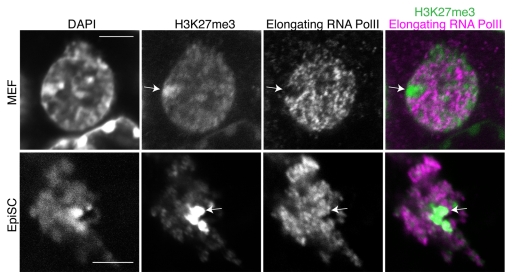
The Xenopus oocyte nuclear transfer system is highly suited to probe the stability of repressed states.3,4 Up to several hundred mammalian somatic nuclei can be directly transplanted into the oocyte’s own nucleus,4 the germinal vesicle (GV) (Fig. 1). Under these conditions, the oocyte imposes a new transcriptional state onto the incoming somatic chromatin. Transcription of previously silenced genes in somatic nuclei is directly induced, after transplantation to an oocyte, in the absence of cell division or DNA synthesis. The notion that the epigenetic state of genes in somatic nuclei is a critical determinant of reprogramming efficiency is directly supported by Xenopus oocyte nuclear transfer studies.3,4 A given gene, in distinct starting epigenetic configurations (i.e., different cell types or different alleles) is reprogrammed with different efficiencies. For example, pluripotency genes are reactivated by Xenopus oocytes with drastically different efficiencies depending on the cell type used for donor nuclei in nuclear transfer experiments.4 This is well exemplified by the mouse X chromosome, for which dosage compensation results in inactivation of one of the two X chromosomes in every differentiated female cell.5 After nuclear transfer of mouse embryonic fibroblast (MEF) or trophoblast stem (TS) cell nuclei, active transcription of the active X chromosome (Xa) was detected while the inactive X chromosome (Xi) was fully resistant to transcriptional reactivation by Xenopus oocytes.3 Although gene expression analysis was limited to a X-linked GFP transgene (Xi-GFP) and a single X-linked gene, the failure to reactivate genes from the Xi of MEFs was also associated with reduced RNA Pol II on the Xi (Fig. 2). Conversely, the transplantation of post-implantation derived epiblast stem cells (EpiSCs) or retinoic-acid differentiated ES cell (ESRA) nuclei led to transcriptional reactivation of the Xi, consistent with the reacquisition of elongating RNA Pol II signals. This does not correspond to a full reactivation of the X chromosome, first because histone modifications such as H3K27me3 were not reversed on the Xi after nuclear transfer (Fig. 2) and second because this dosage compensation mechanism does not exist in Xenopus laevis. However, the transcriptional induction of Xi-linked genes critically depended on the cell type used as a donor. Nuclear transfer also induced a loss of the non-coding RNA Xist from the Xi territory. Xist splicing was not found to be defective and absolute Xist transcript levels increased after nuclear transfer.3 Loss of Xist RNA from the Xi territory may result from the loss of an important factor required for Xist localization to chromatin. In addition to SafA and SATB1, this could be the recently identified YY1, because its knock-down in MEFs results in partial delocalization of Xist RNA from the Xi.6-8 Changes in non-coding RNAs association with chromatin may be an important fundamental mechanism by which oocytes induce nuclear reprogramming. In summary, nuclear transplantation to Xenopus oocytes induces transcriptional reprogramming of silenced genes, and conveniently reveals the epigenetic stability of repressed states.
Figure 1. Nuclear transfer to Xenopus oocyte reveals the stability of repressed states. The nuclei of differentiated cells can be transplanted into the germinal vesicle (GV) of first meiotic prophase oocytes. This induces reactivation of previously silenced genes. A repressed transgene on the inactive X (Xi-GFP) is reactivated when EpiSC are used as donors. Reactivation of Xist-induced repression is also seen when ES cells differentiated for 4 d with retinoic acid (ESRA) are used as donors, correlating with the absence of macroH2A on the Xi. However, when TS or MEF nuclei are used as donors, the Xi-GFP fails to reactivate, correlating with the presence of the histone variant macroH2A. Xist RNA is lost from the Xi following nuclear transfer.
Figure 2. Elongating RNA Pol II exclusion from the Xi is maintained in transplanted MEF nuclei, but not in transplanted EpiSC nuclei. The elongating form of RNA Pol II (H5 antibody) remains excluded from the Xi in transplanted female MEF nuclei (arrows), but not in transplanted EpiSC nuclei. Xenopus oocyte GVs containing transplanted nuclei were fixed 48 h post transplantation and immunostained as described.3 The antibodies used were a rabbit IgG anti-H3K27me3 (1/200, Upstate 07–449) and a mouse IgM anti-Serine2 phosphorylated RNA Pol II (1/200, H5 Covance MMS-129R). The secondary antibodies used were: Alexa 488 goat anti-mouse IgG (1/200, Invitrogen), Alexa 647 goat anti-mouse IgM (1/200, Invitrogen). Confocal sections were projected and merged using ImageJ. Number of transplanted nuclei showing RNA Pol II exclusion: MEF 81.15% (n = 16), EpiSC 0% (n = 15). Scale bars = 5 μm.
Resistance to Reprogramming Correlates with the Incorporation of the Histone Variant MacroH2A in the Presence of DNA Methylation
The sequence of epigenetic events leading to the stable inactivation of the Xi has been particularly well studied.5,9 This includes the formation of a nuclear compartment devoid of transcriptional machinery, into which genes are recruited upon their silencing.10,11 Initial silencing of genes is followed by other epigenetic changes such as the acquisition of histone marks associated with gene repression, including those deposited by Polycomb group proteins.5,12-14 Similarly, on an autosome, Oct4 silencing during differentiation also occurs in several steps, whereby initial repression is followed by deacetylation and H3K9 methylation, followed by DNA methylation.15 During X chromosome inactivation (XCI), one of the last events, occurring after silencing is induced, is DNA methylation and the incorporation of the histone variant macroH2A.16-18 On the Xi, a transition occurs about 2 d after ESRA induction in which Xist-induced silencing switches from a Xist-dependent repressed state to an epigenetically stable, Xist-independent state.19 This transition to stable Xist-independent silencing is not well understood. Surprisingly, we found that in the absence of macroH2A, both Xist-dependent and independent silencing can be reactivated following Xenopus oocyte nuclear transfer.3 Therefore, resistance to reactivation is acquired late, after initiation of XCI and after the transition to Xist-independent silencing. This suggests that a single epigenetic event during cell differentiation is not sufficient to induce resistance toward gene reactivation by Xenopus oocytes.
DNA methylation of Xi genes is a late event of XCI and is known to stabilize the Xi in the somatic cells of developing mouse embryos, because Xi reactivation is seen in DNA methylation-deficient Dnmt1 or Smchd1 mutant embryos.18,20 However, Xi reactivation in Dnmt1 mutant embryos is not seen in extraembryonic lineages, where macroH2A is associated with the Xi of extraembryonic cells under normal conditions, reflecting differential regulation of XCI between these lineages.20,21 Treatment of MEFs carrying an X-GFP transgene on the Xi (Xi-GFP) with the 5-methylcytosine analog 5-azacytidine (5-Aza) leads to a 10-fold increase in transgene reactivation.22 However, this concerns a small proportion of the cells (0.25%), suggesting that additional mechanisms contribute to the stability of the repressed Xi.22 Indeed, a study showed that 5-Aza treatment in combination with Xist deletion or TSA treatment has a synergistic effect on Xi-GFP transgene reactivation.22,23 Overall, these results support the view that DNA methylation is required for stable repression of the Xi in somatic cells, together with other mechanisms. Furthermore, the Xi failed to reactivate from MEFs after nuclear transfer to Xenopus oocytes, while several genes, methylated at their promoters and including lineage specific genes such as MyoD, were reactivated following nuclear transfer.3,24-26 The extreme stability of the Xi after nuclear transfer to Xenopus oocytes further suggests that several mechanisms may be used to confer resistance to nuclear reprogramming.
Resistance toward Xi reactivation could not be explained by differences in DNA methylation before nuclear transfer.3 Instead, resistance correlated with the presence of the histone variant macroH2A.3 macroH2A is incorporated on the Xi of MEF and TS cells, but not on the Xi of EpiSCs and ESRA cells, thereby correlating with the observed resistance toward reprogramming.
Previous work has shown that the loss of Xist RNA on the Xi results in the disappearance of macroH2A enrichment on the Xi.27 To determine the localization of macroH2A in transplanted nuclei, we used a macroH2A-GFP expressing C2C12 cell line, because antibodies failed to work in Xenopus oocytes and a stable macroH2A-GFP expressing MEF cell line was not available. Intriguingly, when macroH2A-GFP expressing C2C12 nuclei were transplanted into Xenopus oocytes, macroH2A-GFP remained associated with the Xi, despite delocalization of Xist RNA from the Xi.3 Furthermore, macroH2A-GFP time-lapse imaging revealed a major reorganization of chromatin within transplanted nuclei (Fig. 3). This reorganization strikingly resembles that occurring upon deletion of Dnmt1 in MEFs.28 In addition to the Xi, macroH2A-GFP seems to become enriched at heterochromatic foci, unlike what is seen in the starting nuclei, which show a more uniform nuclear staining (Fig. 3). We think that these heterochromatic foci are pericentric heterochromatin. Pericentric association of macroH2A has been previously reported.21 It could also be that Xenopus oocyte nuclear transfer conditions reveal differential association and turnover of macroH2A within different regions of the transplanted nuclei. Altogether, the continuous association of macroH2A with heterochromatin after nuclear transfer raises the possibility that it may contribute to resistance toward transcriptional reprogramming.
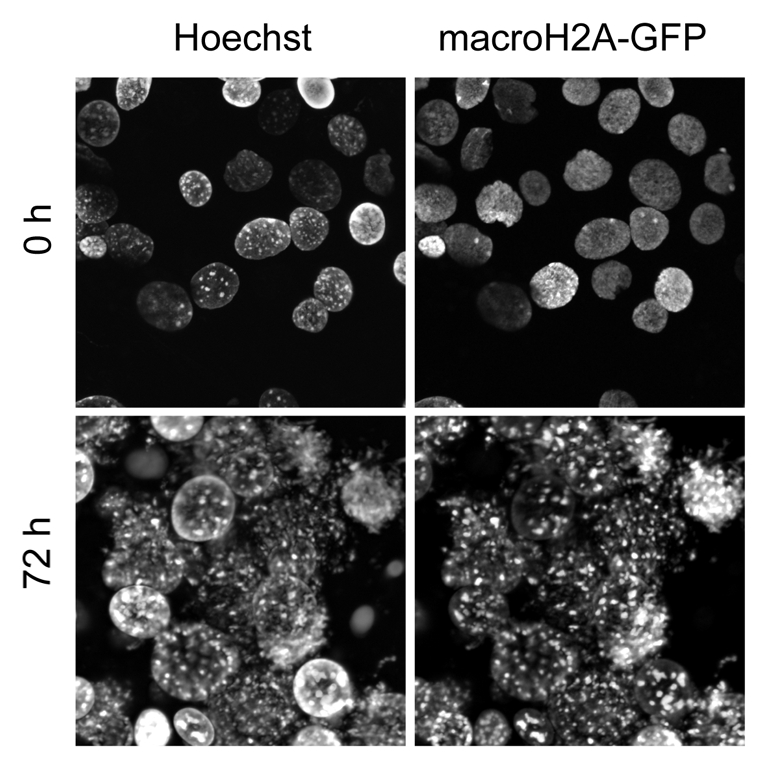
Figure 3. macroH2A-GFP reveals a major reorganization of chromatin following transplantation of nuclei into Xenopus oocytes. The nuclei of C2C12 cells expressing macroH2A-GFP were transplanted into Xenopus oocyte GVs and imaged immediately (0 h) or 3 d after nuclear transfer (72 h).
macroH2A Contributes to the Stability of Repressed States
It is important to stress that the histone variant macroH2A is not required for silencing, nor required for the initiation of XCI and furthermore, in the mouse, mutant embryos are viable, fertile, and their Xi is maintained in a repressed state.29,30 This does not support the view that macroH2A is critical for the establishment of stable Xi repression. Moreover, macroH2A removal is not sufficient to induce Xi-GFP reactivation.3,31 However, it was demonstrated previously that depletion of macroH2A by RNAi together with 5-Aza and TSA treatment has a synergistic effect on Xi-GFP reactivation from MEFs, indicating a role of macroH2A in the maintenance of the silenced X chromosome.22,23 macroH2A seems to also confer resistance toward transcriptional reprogramming, because removal of macroH2A1 and/or macroH2A2 by RNAi in MEFs relieved, partially, the resistance of Xi-GFP to transcriptional reprogramming by Xenopus oocytes.3 Interestingly, HDAC inhibition under nuclear transfer conditions relieved resistance to the same extent, and also had an additive effect when combined with macroH2A depletion.3 The overall picture that emerges from these studies is that macroH2A is not required to induce Xi repression, but instead adds another layer of epigenetic repression on top of other silencing mechanisms already in place, in order to ensure the long-term maintenance of silenced states, which restricts nuclear reprogramming.
Implications for the Stability of X Chromosome Inactivation During Development
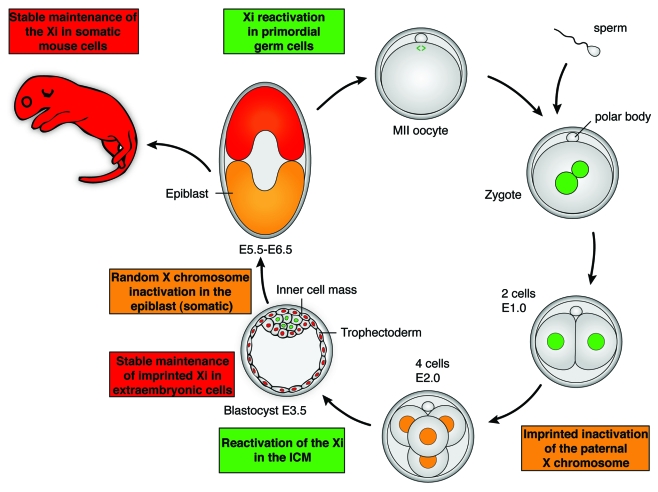
XCI is a developmentally regulated process and is tightly coupled to the loss of pluripotency.32,33 The X chromosomes inherited by the gametes are initially active, but the paternal X chromosome quickly becomes inactivated during the first few cell divisions of female mouse embryos (Fig. 4).10 This imprinted XCI is maintained in the extra-embryonic lineage, where macroH2A associates early with the Xi.21 In the developing inner cell mass, induction of pluripotency is associated with X reactivation.34 Hence, ES cells, derived from the inner cell mass (ICM), have two active X chromosomes (Xa). A second round of XCI takes place in the post-implantation epiblast at around stage E5.5. This round of XCI is random; each X chromosome has a 50% chance of becoming inactivated. Importantly, EpiSCs, derived from E5.5-E6.5 epiblasts, express pluripotency genes, have an Xi but do not show macroH2A enrichment on the Xi.3 This may reflect the in vivo situation in the post-implantation epiblast. We speculate that macroH2A levels increase upon differentiation of epiblast cells after E6.5, because ESRA induces a 4-fold induction of macroH2A1 protein levels.35 In addition, high induction of macroH2A and its incorporation into the Xi was seen in differentiated EpiSCs.3 We propose that the Xi of the post-implantation epiblast is inactivated but that its repressed state is not fully stabilized. This may be because the Xi is reactivated in developing primordial germ cells (PGCs) specified from post-implantation epiblast. Hence, long-term, stable inactivation of the Xi may only occur after germ cell lineage specification from the epiblast, when epiblast cells further differentiate. In addition, it has been reported that macroH2A is removed from PGCs during their development.36 Interestingly, macroH2A is also removed (perhaps actively) from chromatin just after fertilization, during reprogramming in the zygote, but reappears by the morula stage, and this occurs both in fertilized embryos and nuclear transfer embryos.37,38 Altogether, association of macroH2A with the Xi correlates with its stable and irreversible inactivation during development. Conversely, the removal of macroH2A also correlates with epigenetic reprogramming during embryonic development.
Figure 4. Developmental dynamics of X chromosome inactivation and reactivation during female mouse embryogenesis. One active X chromosome (green) is contributed by both egg and sperm to give zygotes in which the two X chromosomes are active (green). Due to maternal imprints, the paternal X chromosome is always inactivated during early mouse development (E2.0, orange). The inactivation of the paternal X is then stably maintained in the extra-embryonic lineage (red), and this is correlated with the incorporation of macroH2A on the inactive X chromosome. In the inner cell mass of the E3.5 blastocyst, induction of pluripotency is linked to the reactivation of the X chromosome (green). Hence, female ES cells, derived from the ICM at E3.5, possess two active X chromosomes. Random X chromosome inactivation is initiated during differentiation of the epiblast from ICM cells (orange). A subset of the epiblast cells is induced to form primordial germ cells, in which X chromosome reactivation is induced (green). Stable maintenance of Xi in the embryo and in the adult is also associated with the incorporation of macroH2A (red). Green: active or reactivation of the X chromosome. Orange: X chromosome inactivation. Red: maintenance of X chromosome inactivation. Adapted with permission, from reference 5 (2011) Macmillan Publishers' Ltd.
Implications for the Repression of Autosomal Pluripotency Genes
Importantly, macroH2A incorporation in somatic cells is not limited to the Xi. Somatic levels of macroH2A do not differ between male and female cells.39 macroH2A evolved before XCI and macroH2A variants have been conserved in non-mammalian vertebrates that do not use XCI as a dosage compensation mechanism.40 As noted above, the levels of macroH2A1 increase upon cellular differentiation and the different macroH2A variants show diverse tissue-specific expression patterns during development.41 Notably, macroH2A RNAi in MEFs led to enhanced reprogramming efficiencies following nuclear tansfer.3 Despite the lack of obvious defects in developing macroH2A mouse mutants, it may be that, in a similar manner as for the Xi, macroH2A backs-up other epigenetic repression mechanisms already in place to ensure the remarkable stability of repressed states. Hence, loss of macroH2A might be, at least in part, compensated for by other mechanisms. Combined HDAC inhibition and macroH2A depletion also had an additive effect on reprogramming by Xenopus oocytes, indicating that macroH2A may also restrict reprogramming through non-HDAC related pathways. Future areas of research include testing the efficiency of nuclear reprogramming in the absence of macroH2A by other procedures, such as cell fusion, nuclear transfer to eggs and induced pluripotency. We conclude that macroH2A shapes chromatin to confer stability to transcriptional states in somatic cells.
Concluding Remarks
Histone variants, and especially macroH2A variants, offer unparalleled means to alter chromatin plasticity and structure.42 While many epigenetic mechanisms are used to induce and maintain the repressed state of genes, histone variants such as macroH2A may help to reinforce the mechanisms already in place to confer increased long-term stability of repressed states. If this were the case, one would expect the loss of macroH2A to be associated with decreased stability of the differentiated state, for which evidence is accumulating.43,44 The Xenopus oocyte system is uniquely suited to address the mechanisms regulating epigenetic memory and in particular those that restrict reprogramming and confer stability to repressed states.45
Acknowledgments
We would like to thank our colleagues who made this research possible. We apologize to those authors for which we could not cite their work due to space contraint. V.P. and A.G were supported by Wellcome Trust PhD Scholarships [081277 and RG44593], RPH-S by the MRC. V.P. was also supported by a Wallonia-Brussels International Excellence Grant. This work was also supported by The Wellcome Trust [RG54943].
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/17799
References
- 1.Gurdon JB, Melton D. Nuclear Reprogramming in Cells. Science. 2008;322:1811–5. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 2.Pasque V, Miyamoto K, Gurdon JB. Efficiencies and Mechanisms of Nuclear Reprogramming. Cold Spring Harb Symp Quant Biol. 2010;75:189–200. doi: 10.1101/sqb.2010.75.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30:2373–87. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halley-Stott RP, Pasque V, Astrand C, Miyamoto K, Simeoni I, Jullien J, et al. Mammalian Nuclear Transplantation to Germinal Vesicle stage Xenopus Oocytes - A method for Quantitative Transcriptional Reprogramming. Methods. 2010;51:56–65. doi: 10.1016/j.ymeth.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–53. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–76. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, et al. SATB1 Defines the Developmental Context for Gene Silencing by Xist in Lymphoma and Embryonic Cells. Dev Cell. 2009;16:507–16. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon Y, Lee JT. YY1 Tethers Xist RNA to the Inactive X Nucleation Center. Cell. 2011;146:119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–67. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 11.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–38. doi: 10.1016/S0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 13.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 14.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 16.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen TP, Wutz AP, Pehrson JR, Jaenisch RR. Expression of Xist RNA is sufficient to initiate macrochromatin body formation. Chromosoma. 2001;110:411–20. doi: 10.1007/s004120100158. [DOI] [PubMed] [Google Scholar]
- 18.Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–9. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 19.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/S1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 20.Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- 21.Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–9. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- 22.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–84. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herńndez-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–40. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biddle A, Simeoni I, Gurdon JB. Xenopus oocytes reactivate muscle gene transcription in transplanted somatic nuclei independently of myogenic factors. Development. 2009;136:2695–703. doi: 10.1242/dev.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–90. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 26.Byrne JA, Simonsson S, Western PS, Gurdon JB. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr Biol. 2003;13:1206–13. doi: 10.1016/S0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- 27.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–4. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 28.Ma Y, Jacobs SB, Jackson-Grusby L, Mastrangelo MA, Torres-Betancourt JA, Jaenisch R, et al. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J Cell Sci. 2005;118:1607–16. doi: 10.1242/jcs.02291. [DOI] [PubMed] [Google Scholar]
- 29.Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol. 2007;27:2758–64. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanasijevic B, Rasmussen TP. X Chromosome Inactivation and Differentiation Occur Readily in ES Cells Doubly-Deficient for MacroH2A1 and MacroH2A2. PLoS ONE. 2011;6:e21512. doi: 10.1371/journal.pone.0021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herńndez-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–40. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 33.Deuve JL, Avner P. The Coupling of X-Chromosome Inactivation to Pluripotency. Annu Rev Cell Dev Biol 2011. [DOI] [PubMed]
- 34.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–74. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 36.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CC, Gao S, Sung LY, Corry GN, Ma Y, Nagy ZP, Tian XC, Rasmussen TP. Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cellular reprogramming 2010; 12:43-53. [DOI] [PMC free article] [PubMed]
- 38.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137:3785–94. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen TP, Huang T, Mastrangelo MA, Loring J, Panning B, Jaenisch R. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 1999;27:3685–9. doi: 10.1093/nar/27.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pehrson JR, Fuji RN. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–42. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pehrson JR, Costanzi C, Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J Cell Biochem. 1997;65:107–13. doi: 10.1002/(SICI)1097-4644(199704)65:1<107::AID-JCB11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 42.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662–74. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–8. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- 44.Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–9. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wutz A. X inactivation: a histone protects from reprogramming by the frog. EMBO J. 2011;30(12):2310–1. doi: 10.1038/emboj.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]