Abstract
Transcription factors that regulate monocyte subset differentiation in bone marrow have not yet been identified. Here we show that the orphan nuclear receptor NR4A1 controls Ly6C− monocyte differentiation. Ly6C− monocytes , which function in a surveillance role in circulation, were absent in Nr4a1−/− mice. Normal numbers of myeloid progenitor cells were present in Nr4a1−/− mice, indicating that the defect occurs during later stages of monocyte development. The defect is cell-intrinsic, as wild-type mice receiving bone marrow from Nr4a1−/− mice developed reduced numbers of patrolling monocytes. Ly6C− monocytes remaining in the bone marrow of Nr4a1−/− mice were arrested in the S phase of the cell cycle and underwent apoptosis. Thus, NR4A1 functions as a master regulator of differentiation and survival of ‘patrolling’ Ly6C− monocytes.
Introduction
NR4A1 (also known as Nur77; mouse gene name Nr4a1), along with NR4A2 (Nurr1) and NR4A3 (NOR-1), constitute the NR4A subfamily of orphan nuclear receptors in the steroid thyroid receptor family1. NR4A1 was originally identified as a growth factor inducible gene, and is often overexpressed in a variety of cancer cells including lung, prostate, breast and colon cancers2–5. A number of growth factors and mitogens rapidly induce the expression of NR4A1, suggesting a survival role for this gene in mediating growth of cancer cells. Conversely, Nur77 has been implicated in the programmed cell death of T and B lymphocytes6, 7. NR4A1 is rapidly induced by T cell receptor signals, and dominant-negative NR4A1 inhibits clonal deletion of developing T cells, indicating a role for Nur77 and its family members in thymocyte negative selection8–10. NR4A1 mRNA is also rapidly (<1 h) and potently induced in macrophages in response to a variety of inflammatory stimuli, including lipopolysaccharide (LPS), cytokines and oxidized lipids11.
Structural and functional studies of Nur77 and other NR4A family members suggest that these nuclear receptor family members do not need to bind small-molecule ligands to be activated12, 13. Instead they appear to be primarily regulated by post-translational modifications, such as phosphorylation14. Acting as a transcription factor, Nur77 can directly bind specific DNA response elements alone or can heterodimerize with the retinoid X receptor (RXR)15, 16. In response to apoptotic stimuli, Nur77 may dimerize with RXR and translocate from the nucleus to the cytoplasm where it can target mitochondria to induce cytochrome c release and apoptosis17. Consistent with its role as an apoptotic mediator in lymphocytes, Nur77 triggers mitochondrial apoptosis though its interaction with Bcl-2, converting Bcl-2 from an anti-apoptotic to a proapoptotic molecule18, 19.
Little is known about the exact functions of Nur77 in monocyte biology. Of clinical relevance, Nur77 is localized within macrophages in human atherosclerotic lesions and reduces macrophage lipid loading and inflammatory responses20, 21. Nur77 also inhibits macrophage accumulation and vascular remodeling in mice22. Nr4a3−/−Nr4a1−/− double knockout mice develop acute myeloid leukemia with abnormal expansion of myeloid progenitor cells23. Subsequent studies of NOR-1 and Nur77 in hypoallelic (Nr4a3+/− Nr4a1−/− and Nr4a3−/− Nr4a1+/−) mice recapitulates the pathological features of myelodysplastic–myeloproliferative neoplasms, with enhanced proliferation and excessive apoptosis of hematopoietic stem cells and myeloid progenitors24. However, mice deficient in either Nur77 or NOR-1 showed relatively subtle abnormalities and lacked overt defects in general physiology, consistent with the idea that NR4A family members have some functional redundancy25. Interestingly, Nurr1, a closely related NR4A family member, was recently demonstrated to play a regulatory role in maintaining hematopoietic stem cell quiescence26. Such work illustrates an important role for members of the NR4A nuclear receptor family in myeloid differentiation and inflammatory diseases.
In mice and humans, at least two distinct CD11b+ CD115+ blood monocyte subsets exist. In mice, Ly6C+CCR2+CX3CR1loCD62L+ monocytes are recruited to inflamed or infected tissues and lymph nodes27–29, and can differentiate into antigen-presenting cells that produce tumor necrosis factor (TNF), nitric oxide and reactive oxygen species (ROS)30, 31. Ly6C−CCR2−CX3CR1hiCD62L− monocytes patrol the resting vasculature and can populate inflamed sites28, 32, 33. Murine Ly6C+ monocytes are analogous to CD14+ monocytes in humans which exhibit a strong inflammatory response to LPS, and Ly6C− mouse monocytes are most likely analogous to CD14dimCD16+ human monocytes, which patrol blood vessels, and exhibit a TNF and interleukin 1β (IL-1β) proinflammatory response induced via a TLR7-MyD88-MEK pathway34. Several groups have demonstrated that both populations of monocytes are recruited to inflammation or injury sites32, 35, 36. Both Ly6C+ monocytes37 and Ly6C− monocytes35 can also participate in the resolution of inflammation and tissue repair.
Monocyte subsets arise from a common macrophage dendritic precursor (MDP) in the bone marrow38. However, the details of the differentiation steps and intermediaries between MDPs and mature monocyte subsets in the bone marrow are unclear. Adoptive transfer experiments demonstrate that Ly6C+ monocytes can down-regulate Ly6C expression, and move between blood and bone marrow39–41, but whether the loss of Ly6C expression corresponds to a conversion of one monocyte subset into another has been questioned42. A number of transcription factors including PU.1, JunB, CEBPα/β, and IRF8 have important roles in myeloid lineage differentiation, but the specific factors that drive differentiation of Ly6C− monocytes are unknown38. In the current study, we sought to identify novel transcription factors that may control the differentiation of monocyte subsets in vivo. We identified an absence of Ly6C− monocytes arising from Nur77-deficient MDP progenitors in the bone marrow. Ly6C− monocytes remaining in the bone marrow of Nur77-deficient mice underwent apoptosis due to abnormal cell cycle progression, implying a critical role for Nur77 in Ly6C− monocyte differentiation and survival.
RESULTS
NR4A expression in monocytes and progenitors
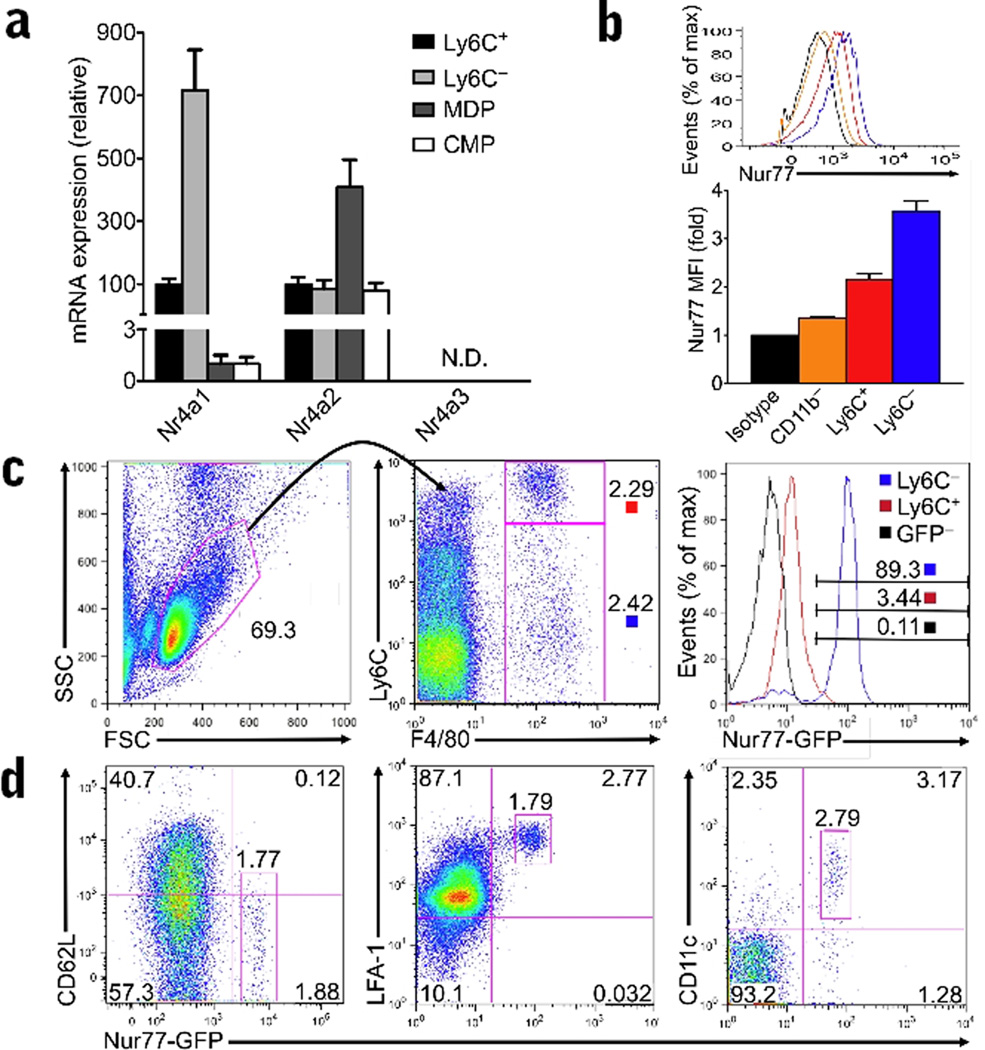
We first examined the relative expression of NR4A1 and other NR4A family members in Ly6C+ and Ly6C− monocytes and myeloid progenitor populations in wild-type (C57BL/6J) bone marrow (Fig. 1a). Ly6C− monocytes expressed on average 7-fold more Nr4a1 mRNA than Ly6C+ monocytes. Similarly, Nur77 protein expression was highest in Ly6C− monocyte populations (Fig. 1b). Nur77 protein expression was also much higher in monocyte populations compared to non-myeloid (CD11b−) cells (Fig. 1b), or MDP or common myeloid precursors (CMP) (Fig. 1a). The NR4A family member, Nr4a2, was also expressed in monocytes and myeloid progenitor cells, while expression of Nr4a3 was undetectable (Fig. 1a). Interestingly, Nr4a2 expression was highest in MDP cells, the precursor cell to monocytes, but was less abundantly expressed in differentiated monocyte populations (Fig. 1a). We confirmed that expression of Nur77 distinguished Ly6C− “patrolling” monocytes from Ly6C+ “inflammatory” monocytes using a new reporter transgenic mouse model where induction of the Nr4a1 promoter drives green fluorescent protein (GFP) expression (Nur77-GFP). The majority (over 85%) of CD11b+F4/80+Ly6C−/lo monocytes circulating in the blood (Fig. 1c) and spleen (data not shown) of these reporter mice expressed abundant Nur77-GFP; the majority of Ly6C+ monocytes (over 95%) expressed low, although not negligible amounts of Nur77-GFP (Fig. 1c). Interestingly, as Ly6C expression decreased, Nur77-GFP expression increased among CD11b+F4/80+ monocytes (Supplementary Fig. 1). GFPhi monocytes in Nur77-GFP mice exhibited other defining phenotypic features of patrolling monocytes, including abundant LFA-1, low amounts of CD62L, intermediate CD11c expression, and, by qPCR, increased expression of CX3CR1 and less CCR2 than their Ly6C+ counterparts (Fig 1d, and data not shown). All GFPhi cells circulating in the blood of Nur77-GFP mice were F4/80+CD11b+ monocytes, and exhibited the characteristic horseshoe nuclear morphology (data not shown). Thus, Nur77 is highly expressed in Ly6C− monocytes compared to Ly6C+ monocytes or monocyte progenitors, suggesting that Nur77 may have a unique function in this particular monocyte subset.
Figure 1. Nur77 is expressed in Ly6C− monocytes.
(a) Relative mRNA expression of NR4A1 family members Nr4a1, Nr4a2, and Nr4a3 in wild-type bone marrow of FACS sorted Ly6C+, Ly6C−, MDP and CMP populations analyzed by qRT-PCR (n = 6 mice; expressed as a percentage of Ly6C+ monocyte transcript). (b) Nur77 protein expression in Ly6C+ and Ly6C− monocyte populations from wild-type bone marrow, and CD11b− non-myeloid cells measured by flow cytometric intracellular staining with a Nur77-specific antibody. For A and B, isolated monocyte populations were determined to be over 95% pure by Cytospin preps of sorted cells stained with HEMA3 dye. (c) Live F4/80+ CD11b+ monocytes from the peripheral blood of a Nur77-GFP transgenic reporter mouse and Nur77-GFP negative (GFP-) littermate control were assessed for Ly6C and GFP expression by flow cytometry. Left and center, representative gating strategy to identify Ly6C+ and Ly6C− monocytes in the blood. Right, representative histogram of Ly6C+ and Ly6C− monocytes, and cells from Nur77-GFP negative (GFP−) littermate control in the blood with a gate defining Nur77-GFPhigh expression. (d) Circulating GFPhi cells in Nur77-GFP reporter mice express other features of patrolling monocytes. Flow cytometry was performed on Nur77-GFP and control peripheral blood cells stained with antibodies indicated. Data from Nur77-GFP mice are representative of 4 experiments.
Nur77-deficient mice lack Ly6C− monocytes
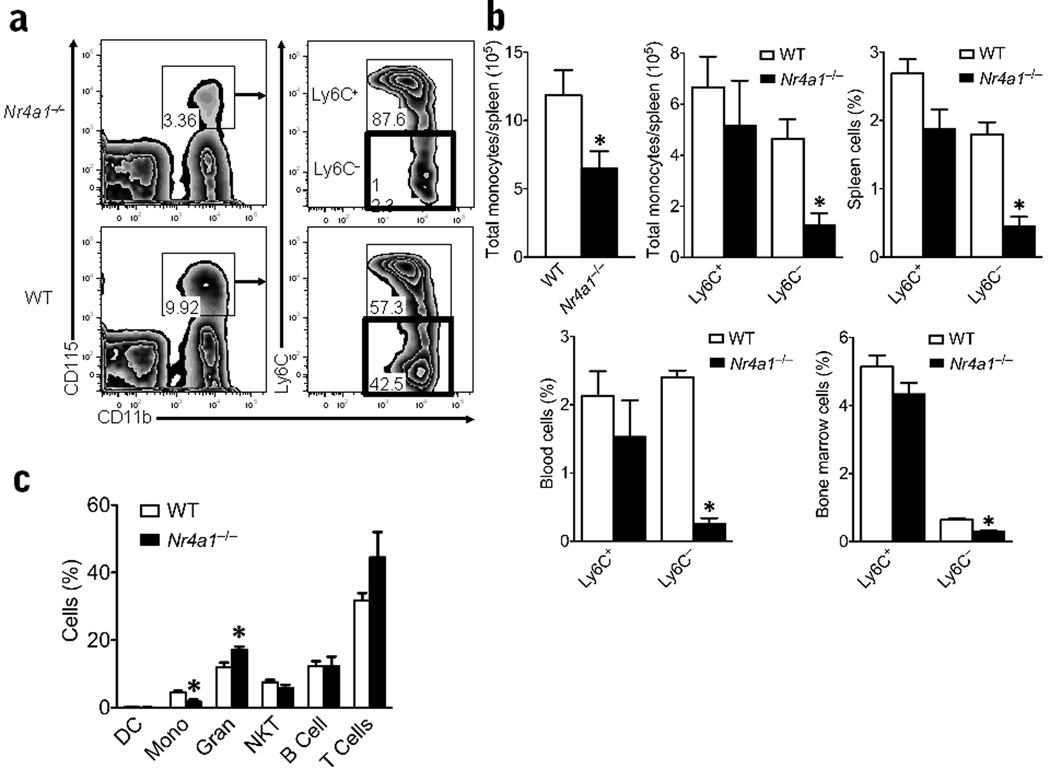
Nur77-deficient (Nr4a1−/−) mice presented with significantly lower numbers of monocytes in bone marrow, blood and spleen in comparison to wild-type, Nr4a1+/+ mice (Fig. 2a,b). Gating strategies for monocyte subsets in bone marrow are shown in Supplementary Fig. 2 and for spleen and blood are shown in Supplementary Fig. 3. Monocytes in bone marrow, spleen and blood were identified as cells with low side scatter, Lin− (CD19, CD49b, CD3ε, Ly6G) and positive for CD115 and CD11b. This monocyte population is distinct from CD11chi DCs identified as CD11chiCD11b+CD115− (CD3e−CD19−CD49b−Ly6G−) (Supplementary Fig. 4a,b). Furthermore, we observed no differences in numbers of CD11chi dendritic cells in Nr4a1−/− mice (Supplementary Fig 4a).
Figure 2. Absence of Ly6C− monocytes in Nr4a1−/− mice.
(a) Live cells with low side scatter and Lin− (CD3ε, CD19, CD49b, Ly6G) were plotted for CD115+ and CD11b+ expression and then gated for Ly6C+ and - expression. Representative flow cytometric scatter plot of CD11b+CD115+ monocyte populations further gated on Ly6C + or - expression in spleens of Nr4a1−/− mice or wild-type (WT) control mice (percentage of total population displayed on plot; red box highlights Ly6C− monocyte population). (b) Quantification of the number of total monocytes/spleen (left panel), the number of Ly6C + and – monocytes/spleen (center panel), and the percent of Ly6C + and – monocytes/spleen of all live cells (right panel) analyzed by flow cytometry (*P < 0.001, n = 10 mice/group). Quantification of Ly6C + and – monocyte populations in blood and bone marrow of global Nr4a1−/− mice and wild-type control mice analyzed by flow cytometry (bottom panel). (*P < 0.001, n = 10 mice/group) (c) Quantification of other major hematopoietic cell populations found in the blood of global Nr4a1−/− mice and wild-type control mice analyzed by flow cytometry (*P <0.01, n = 10 mice/group). Results are expressed as % of live cells unless otherwise noted.
We refined our assessment of the CD115+CD11b+ monocyte population to distinguish between Ly6C+ and Ly6C− subsets (Fig. 2a,b) and found that the reduction in the monocyte population in Nr4a1−/− mice was almost entirely due to a loss of the Ly6C− monocyte population (Fig. 2a,b). Ly6C− monocytes were missing from bone marrow, blood, and spleen of Nr4a1−/− mice (Fig. 2b). Ly6C+ monocyte numbers were also slightly reduced in the blood of Nr4a1−/− mice, although this observation was somewhat variable and not statistically significant. Except for a slight increase in granulocytes (<10%), we found little variation in other immune cell subsets examined in blood (Fig. 2c) and in spleen (Supplementary Fig. 4a).
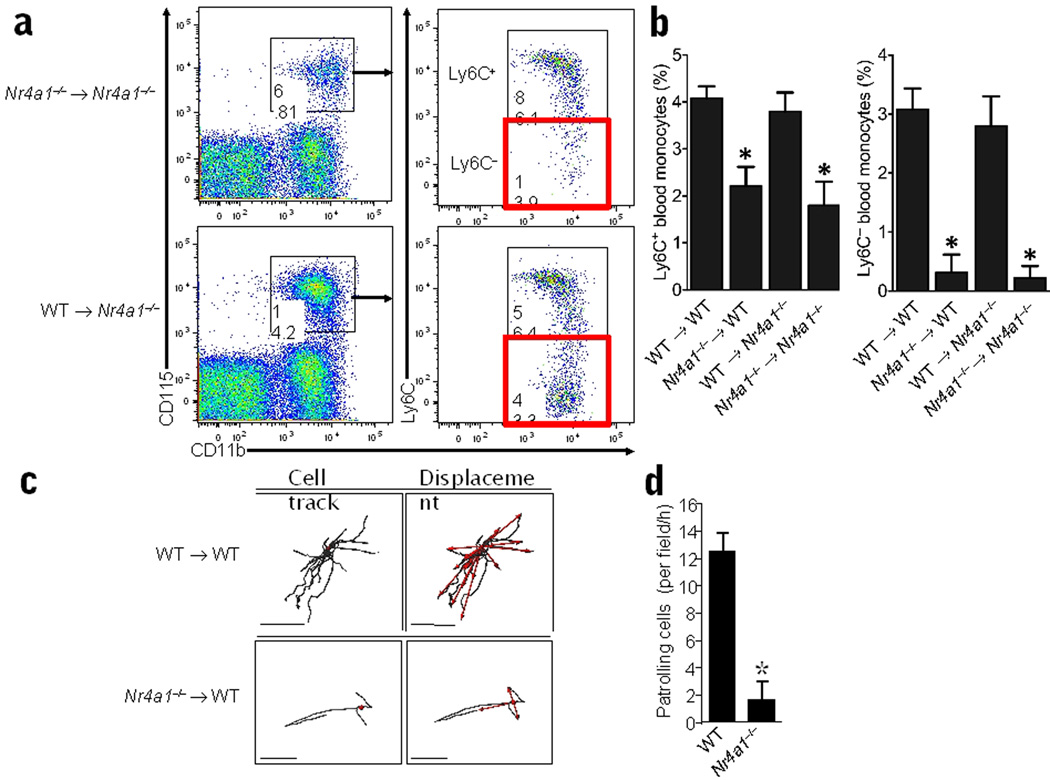
To determine if the defect in Ly6C− monocyte differentiation in Nur77-deficient mice is bone marrow-derived and cell-intrinsic, we grafted Nr4a1−/− and wild-type irradiated recipients with either Nr4a1−/− or control wild-type bone marrow (Fig. 3a,b). Grafts of wild-type bone marrow into Nur77-deficient recipients restored the population of Ly6C− monocytes to normal percentages (Fig. 3b). However, six weeks after reconstitution of wild-type mice with Nr4a1−/− bone marrow, we observed a 90% reduction in Ly6C− monocytes in blood, and a more modest 40% reduction in Ly6C+ monocytes (Fig. 3b).
Figure 3. Cell-intrinsic defect in monocyte development and lack of patrolling ability in monocytes derived from Nr4a1−/− bone marrow.
(a) Representative analysis of Ly6C+ and Ly6C− monocyte populations in the blood of recipients of Nr4a1−/− or wild-type (WT) control transplanted whole bone marrow. (b) Quantification of monocyte populations in blood from whole bone marrow transplants of either wild-type (WT) or Nr4a1−/− donor bone marrow into lethally irradiated wild-type or Nr4a1−/− recipient mice. For whole bone marrow transplants, mice were irradiated with 2 doses of 600 RAD, reconstituted with a total of 5×106 bone marrow cells from donors, and allowed to reconstitute 6 weeks before analysis (*P <0.001, n = 7 mice per group; results expressed as percentage of live cells). (c) CD45.1 recipient mice were irradiated with 9.5 Gy and reconstituted with bone marrow from either CD45.2 wild-type or CD45.2 Nr4a1−/− donors. After 6 weeks the mice were imaged and representative CD11b+ tracking data for wild-type recipients (upper 2 panels), and Nr4a1−/− BM recipients (lower 2 panels) are shown. Anesthetized mice were injected i.v. with 10 µg PE conjugated anti-mouse CD11b (clone M1/70) and cell tracks (left) and displacement vectors of individual cells (red arrows, right) were plotted. (d) Mean number (±SD) of patrolling CD11b+ cells/field/h in wild-type and Nr4a1−/− bone marrow recipients. Scale bars represent 60 um. * P <0.01 Mean ±SD, patrolling cells per field/h wild-type (3 fields/h and 2 animals) vs. Nr4a1−/− BM recipients (7 fields/h and 4 animals).
Ly6C− monocytes have been shown to patrol the endothelium of blood vessels28, 32. Therefore we investigated the patrolling behaviors of Nur77-deficient monocytes. Bone marrow from either CD45.2 wild-type or Nr4a1−/− mice was transplanted into irradiated CD45.1 wild-type recipients (Supplementary Fig. 5). Recipients of wild-type bone marrow had normal percentages of Ly6C− and Ly6C+ monocyte subsets in the blood, whereas recipients of Nr4a1−/− bone marrow showed an 8-fold reduction in the Ly6C− subset (33±1.4% to 4±1.4%; Supplementary Fig. 5). The number of patrolling cells in the blood vessels of the ear of Nr4a1−/− bone marrow recipients was also reduced (by 6-fold) in comparison with mice receiving wild-type bone marrow (1.7± 1.3/cells/field/h for Nr4a1−/− bone marrow recipients vs 12 ±1.5 cells/field/h for wild-type bone marrow recipients; Fig. 3c,d). Taken together, these data indicate that Nur77 selectively regulates the production of patrolling Ly6C− monocytes in vivo.
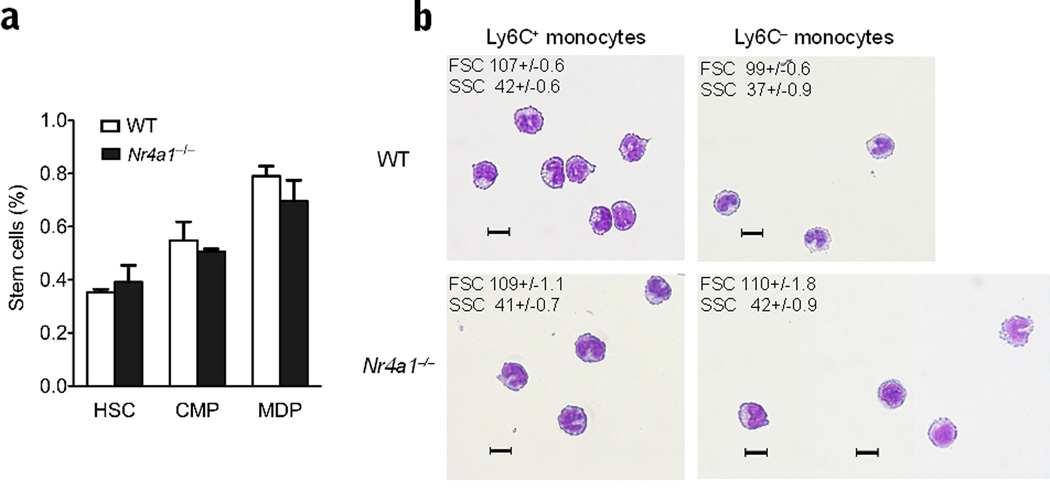
Nur77-deficient MDP fail to differentiate into Ly6C− monocytes
Numbers of myeloid progenitor cells, including MDP, were unaltered in Nr4a1−/− bone marrow (Fig. 4a). The Ly6C− monocytes that did appear in the bone marrow of Nur77-deficient mice were morphologically larger and appeared less differentiated, with enlarged non-horseshoe shaped nuclei and increased granularity compared to their counterparts in wild-type mice (Fig. 4b). It has been hypothesized that both Ly6C+ and Ly6C− monocytes arise from MDP in the bone marrow, but the cellular factors that mediate their specific differentiation are currently unknown43. Our findings suggested that Nur77 is a factor that regulates the differentiation of Ly6C− monocytes from MDP in bone marrow.
Figure 4. Normal stem cell populations and abnormal Ly6C− monocytes in Nr4a1−/− mice.
(a) Quantification of hematopoietic stem cells (HSC), common myeloid precursors (CMP) and macrophage dendritic precursor (MDP) cell population in the bone marrow of Nr4a1−/− mice and wild-type (WT) control mice analyzed by flow cytometry, (n = 10 mice per group) (b) CD115+ CD11b+ Ly6C + and – monocytes were isolated from Nr4a1−/− and wild-type bone marrow by cell sorting. Forward and side scatter (FSC and SSC) values are displayed in the upper left corner as mean+/−SE. (scale bar=10µm). Cells were stained with HEMA3.
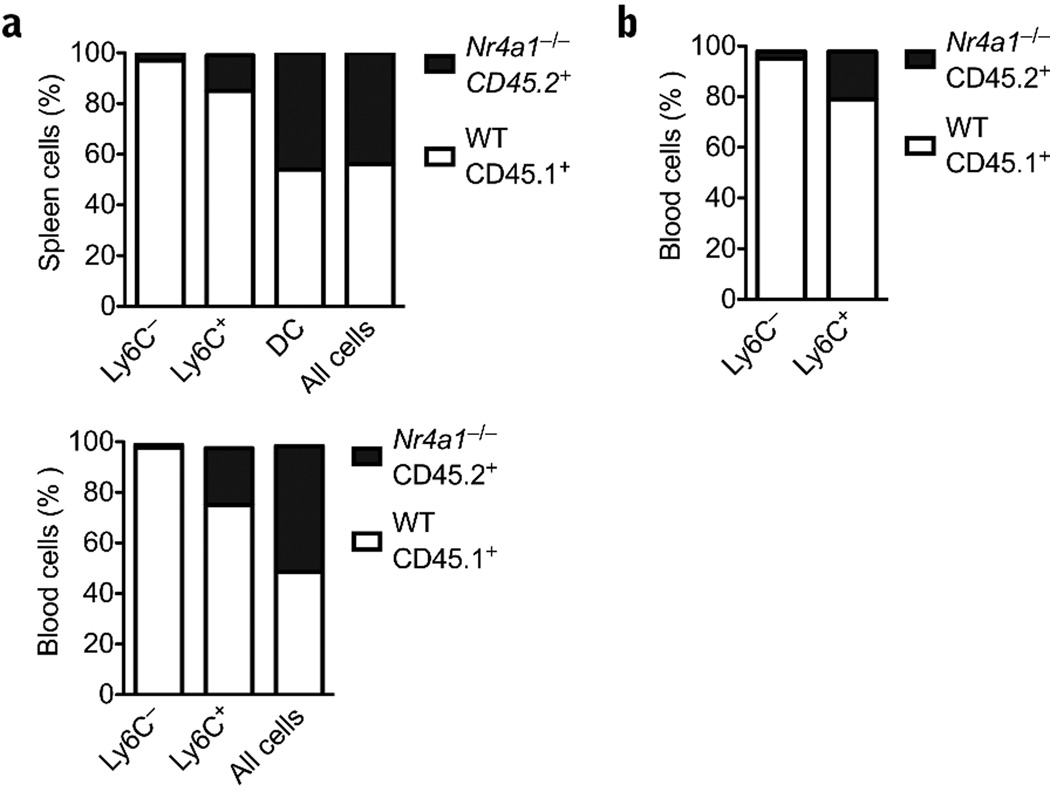
Competitive reconstitution experiments were performed to further examine the role of Nur77 in monocyte development. Bone marrow from CD45.2 Nr4a1−/− mice and CD45.1 wild-type control mice were mixed 1:1 and transplanted into CD45.1 recipients. Six weeks after bone marrow transfer, only 25% of Ly6C+ monocytes and less than 5% Ly6C− monocytes in spleen (Fig. 5a) and blood (Fig. 5a) were derived from Nr4a1−/− bone marrow. In contrast, about equal numbers of CD11chi DCs and all other CD45+ cells were derived from CD45.2 and CD45.1 donors (Fig. 5a). Therefore, Nur77-deficient progenitor cells were specifically inefficient at generating monocytes in general and Ly6C− monocytes in particular.
Figure 5. Specific defect in differentiation of Ly6C− monocytes from myeloid dendritic precursors (MDP) in the bone marrow.
(a) Mixed chimera transplants of whole bone marrow 1:1 mixed donors (Nr4a1−/− CD45.2: wild-type CD45.1) into wild-type CD45.1 recipients (b) MDP cells were isolated by cell sorting from Nr4a1−/− or wild-type control bone marrow and mixed 1:1 (Nr4a1−/− CD45.2: wild-type CD45.1), and then reconstituted for seven days into lethally irradiated mice before analysis. For the whole bone marrow mixed chimera, 2.5×106 cells from each donor were reconstituted into each recipient (P < 0.005, n = 4; results expressed as percent of cell population). For the MDP transfer, approximately 1×104 cells from each donor were transferred into mice. (P < 0.005, n = 6 mice per group; results expressed as percent of cell population).
To determine if the expression of Nur77 in MDP was required for monocyte differentiation, mixed chimeric experiments were conducted with MDP cells purified by fluorescent activated cell sorting. Isolated MDP cells (Lin−CD115+CD117int) from CD45.2 Nr4a1−/− mice and CD45.1 wild-type control mice were mixed 1:1 and transplanted into irradiated CD45.2 recipients. Seven days after bone marrow transfer, recipients had less than 10% Ly6C− and 30% Ly6C+ monocytes derived from Nr4a1−/− donors, while the vast majority of monocytes were derived from wild-type donors (Fig. 5b). These results were similar to the mixed chimera reconstitution using whole bone marrow and, together, indicate that Nur77 expression is required for optimal differentiation of Ly6C− monocytes. These data also suggest that Nur77 deficiency influences the competitive advantages of Ly6C+ monocytes or their precursors, although to a lesser extent.
Nur77-deficient Ly6C− monocytes accumulate in S phase
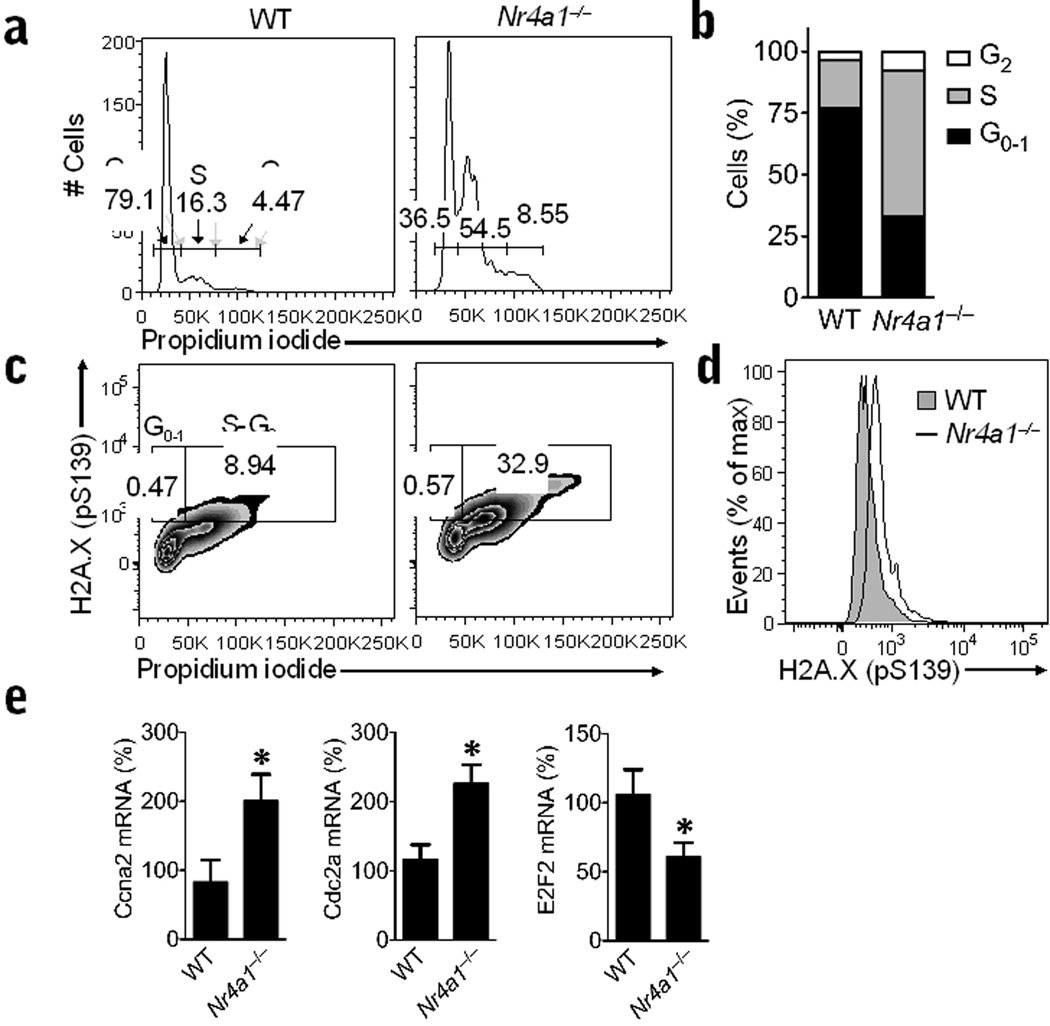
Given that other NR4A family members have been associated with cell cycle regulation, we next examined the progression of monocytes through the cell cycle. Surprisingly, we found that Nr4a1−/− mice had approximately 3-fold more Ly6C− monocytes present in S phase and 2-fold more Ly6C− monocytes present in the G2 phase of the cell cycle compared to Ly6C− monocytes in wild-type control mice in bone marrow (Fig. 6a,b). To assess potential DNA damage in the abnormally dividing Ly6C− monocytes, we examined phosphorylation of H2A.X (Ser139), a marker of double-stranded DNA breaks, during cell cycle progression. We found evidence of increased DNA damage in Ly6C− monocytes in S and G2 phases from Nr4a1−/− mice compared to wild-type mice (Fig. 6c,d). Transcripts of the S phase mediators Cyclin A2 (Ccna2) and Cdk1 (Cdc2a) were both about 2-fold higher in Ly6C− monocytes from bone marrow of Nr4a1−/− compared to wild-type mice implying an acceleration of cell cycle entry (Fig. 6e). Consistently, expression of the transcription factor E2F2, which represses S-phase entry, was reduced by about 45% in Ly6C− monocytes from Nr4a1−/− mice (Fig. 6e). No differences in cell cycle progression were observed in Ly6C+ monocytes or other cells found in the bone marrow of Nr4a1−/− mice (data not shown). Thus in Ly6C− monocytes, Nur77 is required for cell cycle progression during bone marrow differentiation.
Figure 6. Abnormal cell cycle and DNA damage in Ly6C− monocytes from Nr4a1−/− mice.
(a) Representative flow cytometry analysis of cell cycle progression in bone marrow Ly6C− monocytes from wild-type (WT) control or Nr4a1−/− mice stained with propidium iodide. Gates show percentage of cells in G1-0 phase (left), S phase (middle), and G2 phase (right) of the cell cycle. (b) Quantification of data in panel A expressed as an average percentage of cells in each phase of cell cycle (*P < 0.009, n = 6 mice/group). (c) Representative flow analysis of DNA damage during cell cycle progression in bone marrow Ly6C− monocytes from wild-type control or Nr4a1−/− mice as measured by H2A.X phospho-serine139 and propidium iodide staining. Gates show percentage of cells in G1-0 phase (left) and S-G2 phase (right) of the cell cycle. (d) Representative histogram showing H2A.X phospho-serine139 measurement of DNA damage in bone marrow Ly6C− monocytes from wild-type control or Nr4a1−/− mice. (e) Relative expression of Cyclin A2 (Ccna2), Cdk1 (Cdc2a), and E2F2 (E2f2) transcripts in Ly6C− monocytes isolated by FACS from bone marrow of Nr4a1−/− or wild-type mice and measured by qRT-PCR. (*P < 0.05, n = 6 mice/group, expressed as a percentage of wild-type transcript).
Nur77-deficient Ly6C− monocytes die in the bone marrow
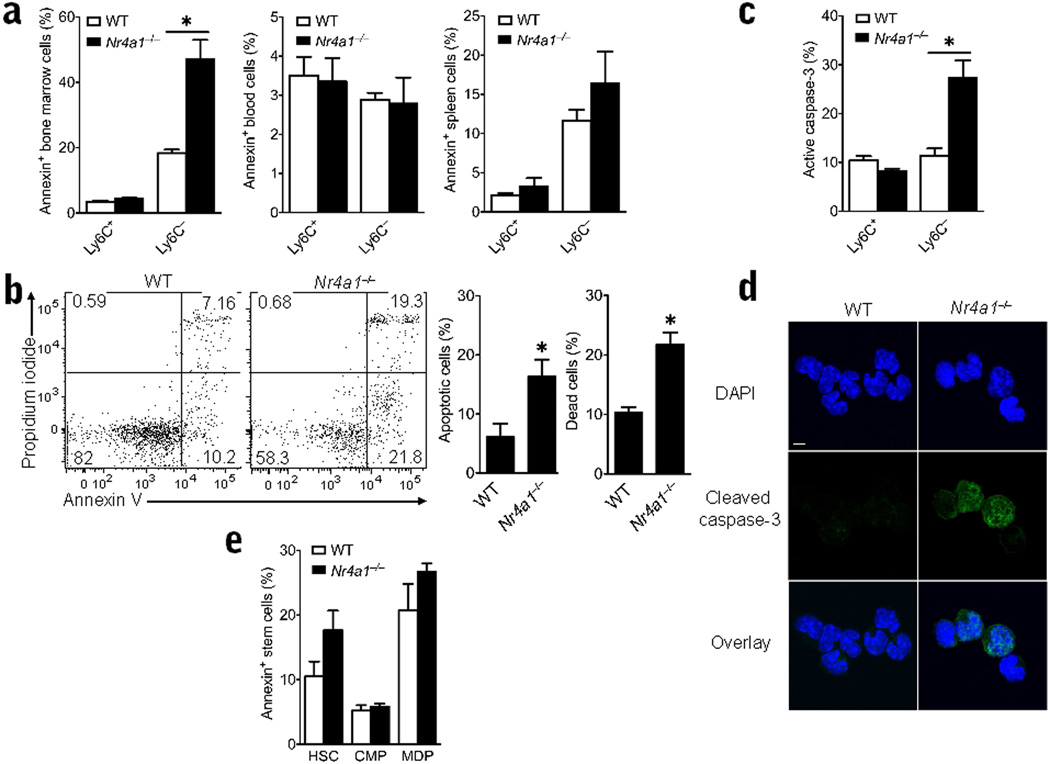
To determine the nature of the defect in monocyte production and cell cycle arrest, we measured apoptosis of Ly6C+ and Ly6C− populations in wild-type and Nr4a1−/− mice. Annexin V and propidium iodide staining of monocyte populations in bone marrow, blood and spleen revealed a significant increase in the number of apoptotic and dead Ly6C− monocytes in Nr4a1−/− bone marrow (Fig. 7a,b). Approximately 2-fold more apoptotic and dead Ly6C− monocytes were present in Nr4a1−/− versus control bone marrow (Fig. 7a,b). Additionally, we observed higher expression of cleaved caspase-3, another hallmark of apoptosis, in Ly6C− monocytes obtained from Nr4a1−/− bone marrow compared to control mouse monocytes (Fig. 7c,d). No changes in Annexin V staining of Ly6C− monocytes in the blood or spleen were observed (Fig. 7a), suggesting that the Ly6C− monocytes, or their precursors, were undergoing apoptosis and death in the bone marrow before they were able to egress to blood. We did not observe changes in apoptosis in myeloid progenitor populations in Nr4a1−/− bone marrow, suggesting that apoptosis may occur during monocyte maturation after the MDP stage (Fig. 7e). Interestingly, we did not observe apoptosis among Ly6C+ monocytes in bone marrow of the Nr4a1−/− mice (Fig. 7a), implying that in the steady state, Nur77 expression is not critical for differentiation or survival of Ly6C+ monocytes.
Figure 7. Increased apoptosis exclusively in Ly6C− bone marrow monocytes from Nr4a1−/− mice.
(a) Apoptosis detection in Ly6C + and – monocytes in bone marrow, spleen and blood in Nr4a1−/− or wild-type (WT) control mice as measured by flow cytometric analysis of Annexin V staining. * P <0.01, n = 6 mice/group). (b) Left panel shows representative scatterplot of Annexin V staining to detect apoptosis and propidium iodide staining to detect cell death in bone marrow Ly6C− monocytes from Nur77−/− or wild-type control mice. Right panel shows quantification of apoptotic and dead cells measured by Annexin V and propidium iodide staining. (*P <0.01, n = 6) (c) Percent cells expressing cleaved (active) caspase-3 in bone marrow monocytes from Nr4a1−/− or wild-type mice using flow cytometry. (*P <0.009 n=8 mice/group) (d) Representative cleaved (active) caspase-3 and DAPI (nuclear) immunofluorescence microscopy of Ly6C− bone marrow monocytes isolated by FACS from Nr4a1−/− or wild-type mice. (scale bar=5µm) (e) Apoptosis detection in myeloid stem cell populations of bone marrow in Nr4a1−/− or wild-type control mice as measured by flow cytometric analysis of Annexin V staining. (n = 6 mice/group).
Phenotype of Ly6C− monocytes in Nr4a1−/− mice
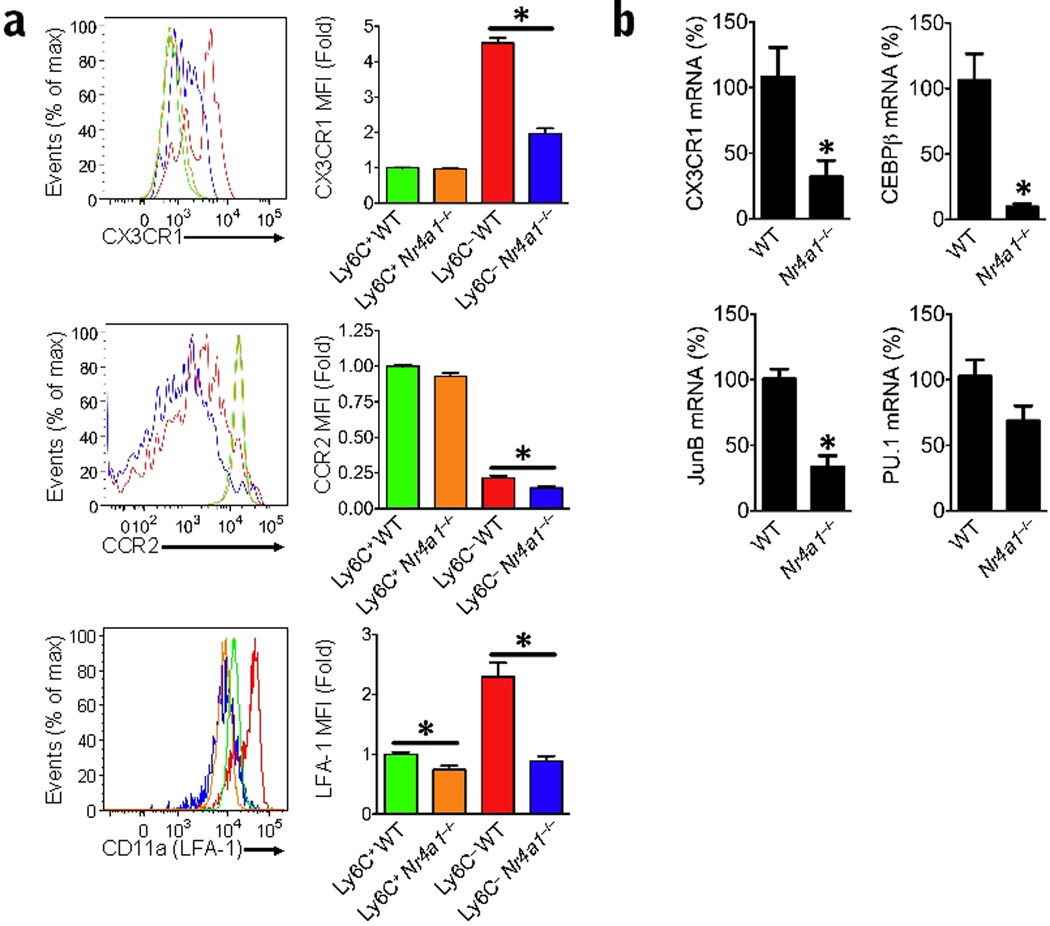
Next, we measured the relative mRNA expression for factors known to be important for monocyte and macrophage differentiation. Significantly lower expression of the chemokine receptor CX3CR1 (Cx3cr1; 70% lower), and the transcription factors CEBP/β (Cebpb; 90% lower) and junB (Junb; 70% lower) were observed in Ly6C− monocytes from Nr4a1−/− mice compared to wild-type controls (Fig. 8a,b), suggesting a reduction in differentiation potential. Expression of the transcription factor PU.1 (Sfpi1) was not significantly different in Ly6C− monocytes from Nr4a1−/− mice compared to controls. Moreover, Ly6C− monocytes from Nr4a1−/− mice had increased NFκB activation compared to Ly6C− monocytes from Nr4a1+/+ mice, suggesting alterations in the inflammatory phenotype of this monocyte subset in the absence of Nur77 (Supplementary Fig. 6). At the protein level, the expression of CX3CR1 and CCR2 in Ly6C− monocytes in bone marrow of Nr4a1−/− mice was also reduced in comparison to wild-type mice (Fig. 8a). Interestingly, Ly6C+ monocytes from Nur77-deficient mice had similar CX3CR1 and CCR2 surface expression compared to wild-type monocytes. CD11a (LFA-1), an integrin that is important for the patrolling function of Ly6C− monocytes, was reduced by about 60% in Ly6C− monocytes and 30% in Ly6C+ monocytes from Nr4a1−/− mice (Fig. 8b). Thus, Ly6C− monocytes in bone marrow of Nr4a1−/− mice did not express phenotypic markers consistent with normal, differentiated Ly6C− monocytes, further supporting the notion of impaired differentiation of this monocyte subset in the absence of Nur77.
Figure 8. Decreased expression of chemokine receptors, adhesion molecules and differentiation factors in Ly6C− monocytes from Nr4a1−/− mice.
(a) Expression of CCR2, CX3CR1, and LFA-1 (CD11a) in monocyte populations from Nr4a1−/− or wild-type (WT) control bone marrow analyzed by flow cytometry. (*P <0.05 n = 5 mice/group; bar graphs show MFI fold change compared to Ly6C+ WT cells) (b) Relative expression of Cx3cr1, Cebpb, Junb and Sfpi1 (PU.1) transcripts in Ly6C− monocytes isolated by FACS from bone marrow of Nr4a1−/− or wild-type mice measured by qRT-PCR. (*P <0.05 n = 6 mice per group, expressed as a percentage of wild-type transcript).
Discussion
In this study, we show that mice deficient in Nur77 have a defect in bone marrow production of Ly6C− monocytes, and as a consequence, these mice lack mature Ly6C− monocytes circulating in the blood and spleen and patrolling the vascular endothelium. The loss of this subset in the absence of Nur77 is likely due to an intrinsic defect in hematopoietic cell differentiation. Resident MDP populations in the bone marrow and dendritic cell populations in the spleen were unaffected by the absence of Nur77, implying a specific role of Nur77 in mediating monocyte differentiation. The few Ly6C− monocytes that remain in the bone marrow in the absence of Nur77 appear unable to proliferate and differentiate properly: they are larger than Ly6C− monocytes from control mice, and have reduced CX3CR1, CCR2, and LFA-1 expression. Nur77-deficient Ly6C− monocytes also accumulate in S phase of the cell cycle, albeit unable to complete cell division, and undergo apoptosis during S and G2 phases of the cell cycle. Thus, our data demonstrate that Nur77 functions in the bone marrow as a critical transcription factor required for the production of monocytes, particularly the Ly6C− subset.
Reduced expression of the transcription factor E2F2, which is considered a critical repressor for cell commitment to S-phase44, may be partially responsible for the premature entry and incompletion of the S-phase observed among Nr4a1−/− Ly6C− monocytes. Recent findings showing that E2F family members are required for myeloid cell survival and their ability to respond properly to CSF-1 signals45 are consistent with our observations. That Nur77 may regulate cell cycle progression in monocytes is, perhaps, not surprising given that investigators have recently shown that the closely related NR4A family member, Nurr1, has a similar regulatory role in maintaining hematopoietic stem cell quiescence via associated upregulation of cell-cycle inhibitors26. Our findings of increased expression of Nurr1 in MDP cells, the closest identified precursor to monocyte populations, imply that Nurr1 may be regulating homeostasis of monocytic and dendritic cell precursor populations. Upregulation of the cell cycle initiators Cyclin A2, Cyclin D2 and Cdk1 in Nr4a1−/− monocytes imply that Nur77 may have a similar role in maintaining Ly6C− and possibly Ly6C+ monocyte production.
An alternative explanation for our findings is that Nur77 may be working as a survival and/or differentiation factor in Ly6C− monocytes by encouraging expression of receptors important for monocyte function, such as CX3CR1. CX3CR1 has been identified as an important factor for Ly6C− monocyte survival46. However, absence of CX3CR1 or its ligand CX3CL1 in mice results in only a modest reduction of Ly6C− blood monocyte numbers under steady-state and inflammatory conditions46, 47. The relationship between Nur77 and the potential monocyte survival factor CX3CR1 is uncertain, as are the identities of additional factors that drive Ly6C− monocyte survival and differentiation. Our data suggest that the defect in monocyte production in Nr4a1−/− mice is more likely upstream from CX3CR1 since Ly6C− monocytes are reduced in the bone marrow of Nr4a1−/− mice, but not in CX3CR1-deficient mice46.
Although it is accepted that both monocyte subsets arise from a common macrophage dendritic precursor (MDP) in the bone marrow47, molecular control of their differentiation, and the putative intermediaries between MDPs and mature monocyte subsets have remained unclear. Specifically, whether Ly6C− monocytes differentiate directly from a bone marrow progenitor or from the conversion of mature Ly6C+ monocytes is debated. Although mature monocytes recirculate in the bone marrow40, our data do not support conversion of mature Ly6C+ into Ly6C− monocytes in the peripheral blood. Rather, our findings suggest that the precursor of Ly6C− monocytes is an immature proliferating cell.
Under conditions of competitive bone marrow transplant, both Ly6C+ and Ly6C− monocyte subsets are reduced in the absence of Nur77. Given that irradiation and reconstitution inevitably enhance inflammation, it is possible that stress or inflammatory factors result in Nur77-mediated regulation of both Ly6C+ and Ly6C− monocyte populations. Expression of Nur77 is present but low in Ly6C+ monocytes and is likely to have some role in Ly6C+ monocyte function. Interestingly, activation of the NF-κB pathway, an important pathway regulating inflammation, was increased in both populations of monocytes in bone marrow of Nr4a1−/− mice. Active Nur77 binding sites have been identified on the promoter of the gene encoding the NF-κB inhibitory regulator, IκBα, and have been suggested as a means of down-regulating NF-κB inflammatory signaling48. These findings suggest that in both Ly6C+ and Ly6C− monocytes, Nur77 plays an important role in regulating NF-κB-mediated inflammatory responses, and that the absence of Nur77 may lead to an exaggerated inflammatory response11.
Inflammation also leads to a significant increase in the number of circulating monocytes30, 49. Nur77 is well known to rapidly respond to early inflammatory events and may partially drive monocyte proliferation and/or extravasation. Ly6C− monocytes have been observed to extravasate and respond rapidly32, 38, 47, in a time frame very similar to that of Nur77 induction in response to early inflammation11. However, inflammation is unlikely to overcome the defect in Ly6C− monocyte subset production in Nr4a1−/− mice. One example of this is the fact that irradiation of the Nur77-deficient mice did not correct the impaired production of the Ly6C− subset in our studies. However, inflammation may indeed modulate the number of Ly6C+ monocytes as well as influence the functions of both monocyte subsets. This may vary depending on the type or model of inflammation studied. Further research is clearly needed to understand exactly how Nur77 mediated signaling pathways are involved in regulating both monocyte subset differentiation and the specific function of these monocyte subsets in disease.
In summary, we have identified Nur77 as a critical regulator of monocyte homeostasis in the bone marrow. Deletion of Nur77 from hematopoietic cells leads to loss of monocytes in vivo, particularly the Ly6C− subset. Thus, Nur77-mediated signaling pathways may be key targets for developing therapeutics to regulate monocyte differentiation and function to control early inflammatory events in disease.
METHODS
Mice
C57BL/6J (stock no. 000664) mice (wild-type) and global Nr4a1−/− mice25 on a congenic C57BL/6J background were purchased from JAX (stock no. 006187) were purchased from The Jackson Laboratory, and B6.SJL-Ptprca/BoyAiTac (stock no. 004007) mice were purchased from Taconic Farms. Mice were fed a standard rodent chow diet and housed in microisolator cages in a pathogen-free facility. The Nur77-GFP reporter mice were generated as described50. All experiments followed the La Jolla Institute for Allergy and Immunology Animal Care and Use Committee guidelines, and approval for use of rodents was obtained from the La Jolla Institute for Allergy and Immunology according to criteria outlined in “Guide for the Care and Use of Laboratory Animals” from the National Institutes of Health. Mice were euthanized using CO2.
Flow Cytometry
Spleens were excised and pushed through a 70-µm strainer, bone marrow cells from both femurs and tibias were collected by centrifugation, and blood was drawn via cardiac puncture with an EDTA coated syringe. All samples were collected in DPBS with 2 mM EDTA and maintained on ice during staining and analysis. Red blood cells were lysed in lysis buffer (BioLegend) according to the manufactures protocol.
2–4 × 106 cells were resuspended in 100 µl staining buffer (1% BSA, 0.1% sodium azide in PBS). Cells were blocked with Fcγ receptor for 15 min and stained for surface antigens with flow cytometry Abs for 30 min at 4°C. A fixable LIVE/DEAD stain (Invitrogen) was used to measure viability, and FSC/SSC parameters were used to excluded doublets from analysis. Specific antibodies used for flow cytometry are listed in Supplementary Fig. 7.
For additional intracellular staining, cells were fixed and permeabilized using the Cytofix/Cytoperm kit from BD Bioscience. Directly conjugated fluorescent antibodies were stained for 30 min at 4°C. Indirect antibodies were stained for 1 h at 4°C, washed two times and then stained for 30 min at 4°C with secondary antibody. CCR2 was stained as previously described51. Nur77 and CX3CR1 were stained intracellularly using the corresponding anti-mouse IgG-FITC (BD Biosciences) or anti-goat IgG-FITC (Santa Cruz Biotechnology) secondary antibodies.
The absolute numbers of cells were calculated by multiplying the percentage of live cells in individual subsets by the total cell count before staining. Calculations of percentages were based on live cells as determined by FSC/SSC and viability analysis. Cell fluorescence was determined using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (version 9.2). The mean fluorescence intensity was quantified, and the expression relative to wild-type control was calculated.
Monocytes were identified as described in Supplementary Fig. 2 and Fig. 3. Hematopoietic stem cells (HSC) were identified as Lin−CD117+Sca-1+; Common Myeloid Precursors (CMPs) as Lin− CD117+Sca-1−CD34+; Macrophage Dendritic Precursors (MDPs) as Lin−CD115+CD117+; dendritic cells (DC) as CD11cHi; Granulocytes as Ly6G+; natural killer T cells (NKTs) as CD49b+; B cells as CD19+; and T cells as CD3ε+.
For staining of monocytes from the Nur77-GFP reporter mice, blood was collected from the tail vein and heparin was added to inhibit clotting. Red blood cells were lysed in hypotonic ACK solution, and 5 × 105 cells were incubated for 30 min at 4°C in 30 µl of FACS buffer with the indicated antibodies in the presence of Fcγ block. Cellular fluorescence was determined using a LSRII, FACSAria II or a FACSCalibur flow cytometer (BD Biosciences), and data analyses were performed with FlowJo software (Tree Star).
Fluorescent Activated Cell Sorting (FACS)
Prior to sorting bone marrow, monocytes and stem cell populations were enriched by negative selection using Miltenyi Biotec isolation kits using the manufacturer’s protocol (CD19, CD4, CD8, Ter119 custom cocktail for monocytes, and lineage cell depletion kit #130090858 for stem cells). Cells were then stained for surface antigens as described above and sorted using a FACSAria cell sorter (BD Biosciences). Approximately 5–10 × 104 events were collected for mRNA samples. Purity of monocyte and stem cell populations were verified by cytospin preps of sorted cells, followed by staining with HEMA 3 (Fisher Scientific) and microscopic analysis.
Quantitative Real-Time PCR (qRT-PCR)
Total cellular RNA was collected from FACS isolated monocyte and stem cell populations using a Qiagen RNeasy Plus Micro Kit following the manufacturer's protocol. RNA purity and quantity was measured using a nanodrop spectrophotometer (Thermo Scientific). Approximately 500 nanograms of RNA was used to synthesize cDNA using an Iscript cDNA synthesis kit (Bio-Rad). Total cDNA was diluted 1:20 in H2O, and 9 µl were used for each real-time condition using a Bio-Rad MyIQ single-color real-time PCR detection system and TaqMan Gene Expression Mastermix. TaqMan primers used for analysis are listed in Supplementary Fig. 8. Data were analyzed and presented on the basis of the relative expression method. The formula for this calculation is as follows: relative expression = 2−(SΔ Ct − CΔC t ) where ΔCt is the difference in the threshold cycle between the gene of interest and the housekeeping gene (GAPDH), S is the Nr4a1−/− mouse, and C is the C57BL/6J control mouse.
Bone Marrow and MDP Transplantation Studies
Recipient mice (wild-type or Nr4a1−/−) were irradiated in two doses of 550 rads each, for a total of 1100 rads, 4 h apart. Bone marrow cells from both femurs and tibias of donor mice (wild-type or Nr4a1−/−) were harvested under sterile conditions. Approximately 50 million nucleated bone marrow cells were obtained from each donor mouse. Bones were centrifugated to collect marrow, and then washed and resuspended for injection in DPBS. Approximately 5 million unfractionated bone marrow cells in 200 µl of media were delivered retro-orbitaly to each recipient mouse. Recipient mice were housed in a barrier facility under pathogen-free conditions before and after bone marrow transplantation. After bone marrow transplantation, the mice were maintained on autoclaved acidified water with antibiotics (Trimethoprim/Sulfa) and fed autoclaved food. Mice were used for experiments after 6 weeks of bone marrow reconstitution.
The 45.1 antigen in the B6.SJL-Ptprca/BoyAiTac mice (wild-type 45.1), and the 45.2 antigen in the Nr4a1−/− and C57BL/6 (wild-type 45.2) mice were used to track cells in mixed chimeric bone marrow transplant mice. For the mixed chimeric transplants 2.5 million cells from Nr4a1−/− mice (45.2) and 2.5 million cells from B6.SJL-Ptprca/BoyAiTac mice (wild-type 45.1) were mixed 1:1 for reconstitution into recipients (wild-type 45.1 or wild-type 45.2) as described above. For MDP chimeras, approximately 1×104 cells were reconstituted from each donor (Nr4a1−/− 45.2 mice and wild-type 45.1 mice) in a 1:1 ratio into recipients (wild-type 45.1 or wild-type 45.2) recipients. MDP transplants were done as described above, except that MDP recipients received only one 600 RAD dose of radiation, and were analyzed after only 7 days of reconstitution.
Annexin V Analysis of Apoptotic Cells
Annexin V and propidium iodide were used to identify apoptotic and dead cell populations by flow cytometry. Bone marrow, blood and spleen cells were stained for annexin V (Invitrogen) and propidium iodide (Invitrogen) according to the manufacturers’ protocols, after surface staining for monocye or stem cell populations.
Cell Cycle Analysis
For cell cycle analysis, monocyte populations in the bone marrow were first stained for identifying surface antigens, and then fixed and permeabilized using the Cytofix/Cytoperm kit from BD Bioscience. Permeabilized cells were then stained with 50 µg/ml propidium iodide (Invitrogen) for 30 min at 25°C followed by one wash and resuspension with flow buffer. For analysis of DNA damage, a fluorescently conjugated H2A.X (pS139) antibody (BioLegend) was also stained during the propidium iodide incubation. Cells were then analyzed on a linear scale for DNA content and then gated and analyzed for cells in G1/0, S and G2 phase of the cell cycle using FlowJo software.
Immunofluorescence
Cytospin preparations of bone marrow monocytes sorted from wild-type and Nr4a1−/− mice by FACS were stained for cleaved (active) caspase 3, p65 (NFκB), and DAPI and visualized using immunofluorescence microscopy. Cytospin preps were dried overnight, fixed with 4% paraformaldehyde for 20 min, washed in PBS, permeablized in 0.1% citrate 0.1% Triton X solution for 5 min, washed 3 times in PBS, blocked in 5% donkey serum with 0.2% Triton X in PBS for 60 min, and then incubated with primary antibody diluted in PBS with 0.2% Triton X and 1% BSA overnight at 4°C. The following day, slides are then washed 3 times in PBS and incubated for 1 hour at room temperature with fluorescent secondary antibodies at a 1:1000 dilution, slides were then washed 3 more times in PBS and mounted with ProlongGold with DAPI (Invitrogen). A 1:500 dilution of rabbit anti-mouse cleaved caspase-3 (Asp175) antibody from Cell Signaling (#9661) was used, followed by staining with a donkey anti-rabbit Alexa488 fluorescently conjugated secondary antibody (Invitrogen). A 1:33 dilution of mouse anti-mouse p65 (NF-κB) antibody from Santa Cruz Biotechnology (sc-8008) was used followed by staining with a donkey anti-mouse Alexa594 fluorescently conjugated secondary antibody (Invitrogen). Images were acquired using an Olympus FV10i confocal microscope and analyzed using Imaris software (Bitplane). The % of p65 fluorescence overlapping with DAPI staining was quantified in wild-type and Nr4a1−/− Ly6C− monocytes from the bone marrow.
Analysis of Patrolling Cells
CD45.1 mice were irradiated (9.5 Gy) and 3 h later injected with 2×107 bone marrow cells from either wild-type mice (CD45.2) or Nr4a1−/− mice (CD45.2) in 150 ml PBS i.v. via tail vein. Before and after the procedure the mice where maintained in a sterile environment and given water containing an antimicrobial (Baytril; Bayer). 6 weeks later blood samples were obtained via tail vein and the mice were assessed for full chimerism and monocyte subset phenotype by flow cytometry. Following blood sampling, mice were anaesthetized with Ketamine/Xylazine/ACP (i.p.) and injected i.v. with 10 µg PE conjugated anti-mouse CD11b (M1/70; Becton Dickinson). Blood vessels in the ear were imaged through a glass cover slip using an inverted confocal microscope (SP5) 561 nm DPSS laser line and ×20 0.7 N. A. water immersion objective. Both the blood vessels (circulating Ab) and CD11b+ cells (bound Ab) were visualized over 1 h in several fields per animal. These data were deconvolved (LSCM blind algorithm; Autoquant X2, Media Cybernetics) and visualized and analyzed using Imaris software (Bitplane). CD11b+ cells in the blood vessels were tracked using the autoregressive motion algorithm and tracks were filtered for a minimum track length of 30 µm from their origin and a minimum track duration of 3 min, then manually assessed and edited for track continuity. For each field the tracks were translated to a common origin in space to allow their comparison (number, direction, displacement). These tracks were then counted and the number (i.e. the number of patrolling cells) plotted per field.
Statistical analysis
Data for all experiments were analyzed using Prism software (GraphPad, San Diego, CA). Unpaired t tests and two-way ANOVA were used to compare experimental groups. All experiments were repeated at least 3 times with 3 or more individuals per group. Data are graphically represented as mean ± SEM. A P value <0.05 was considered significant.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. K.A. Hogquist (University of Minnesota) for providing the Nur77-GFP reporter mice and Dr. M. Mack from Klinikum der Universitat Regensburg for providing us with the CCR2 (MC-21) antibody. We would like to thank Dr. K. Ley and Dr. I. Shaked (LIAI) for helpful discussions, and A. Blatchley, D. Yoakum, and D. Huynh (LIAI) for assistance with mouse colony management.
Footnotes
Author contributions
R.N.H. and L.M.C. designed and performed experiments, analyzed data, and contributed to the writing of the manuscript. H.G.H. performed experiments using the Nur77-GFP mouse. D.N. and A.M.G. performed experiments. J.A.P. conceived of the studies related to the Nur77-GFP mouse, analyzed data, and contributed to the writing of the manuscript. F.G. conceived and directed the research related to the intravital microscopy studies, analyzed data, and contributed to the writing of the manuscript. C.C.H. conceived of the research, directed the study, assisted with experimental design, and contributed to the writing of the manuscript.
References
- 1.Martínez-González J, Badimon L. The NR4A subfamily of nuclear receptors: new early genes regulated by growth factors in vascular cells. Cardiovasc Res. 2005;65:609–618. doi: 10.1016/j.cardiores.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced 'primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. [PubMed] [Google Scholar]
- 3.Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci U S A. 1988;85:8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 5.Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3--an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–540. doi: 10.14670/HH-21.533. [DOI] [PubMed] [Google Scholar]
- 6.Rajpal A, et al. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J. 2003;22:6526–6536. doi: 10.1093/emboj/cdg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JM, Lee KH, Weidner M, Osborne BA, Hayward SD. Epstein-Barr virus EBNA2 blocks Nur77- mediated apoptosis. Proc Natl Acad Sci U S A. 2002;99:11878–11883. doi: 10.1073/pnas.182552499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou T, et al. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med. 1996;183:1879–1892. doi: 10.1084/jem.183.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 10.Cho HJ, et al. Cutting edge: identification of the targets of clonal deletion in an unmanipulated thymus. J Immunol. 2003;170:10–13. doi: 10.4049/jimmunol.170.1.10. [DOI] [PubMed] [Google Scholar]
- 11.Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- 12.Baker KD, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 14.Fahrner TJ, Carroll SL, Milbrandt J. The NGFI-B protein, an inducible member of the thyroid/steroid receptor family, is rapidly modified posttranslationally. Mol Cell Biol. 1990;10:6454–6459. doi: 10.1128/mcb.10.12.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson TE, Fahrner TJ, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 16.Wallen-Mackenzie A, et al. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 2003;17:3036–3047. doi: 10.1101/gad.276003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 18.Lin B, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–1036. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonta P, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol. 2006;26:2288–2294. doi: 10.1161/01.ATV.0000238346.84458.5d. [DOI] [PubMed] [Google Scholar]
- 21.Arkenbout EK, et al. Protective function of transcription factor TR3 orphan receptor in atherogenesis: decreased lesion formation in carotid artery ligation model in TR3 transgenic mice. Circulation. 2002;106:1530–1535. doi: 10.1161/01.cir.0000028811.03056.bf. [DOI] [PubMed] [Google Scholar]
- 22.Bonta PI, et al. Nuclear receptor Nur77 inhibits vascular outward remodelling and reduces macrophage accumulation and matrix metalloproteinase levels. Cardiovasc Res. 2010;87:561–568. doi: 10.1093/cvr/cvq064. [DOI] [PubMed] [Google Scholar]
- 23.Mullican SE, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez-Herrick AM, Mullican SE, Sheehan AM, Conneely OM. Blood. Vol. 117. United States; 2011. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice; pp. 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, et al. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77) Science. 1995;269:532–535. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 26.Sirin O, Lukov GL, Mao R, Conneely OM, Goodell MA. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat Cell Biol. 2010;12:1213–1219. doi: 10.1038/ncb2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randolph G. Emigration of monocyte-derived cells to lymph nodes during resolution of inflammation and its failure in atherosclerosis. Curr Opin Lipidol. 2008;19:462–468. doi: 10.1097/MOL.0b013e32830d5f09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geissmann F, Jung S, Littman D. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 29.Palframan R, et al. Inflammatory chemokine transport and presentation in HEV: a remote control mechanism for monocyte recruitment to lymph nodes in inflamed tissues. J Exp Med. 2001;194:1361–1373. doi: 10.1084/jem.194.9.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narni-Mancinelli E, et al. Memory CD8+ T cells mediate antibacterial immunity via CCL3 activation of TNF/ROI+ phagocytes. J Exp Med. 2007;204:2075–2087. doi: 10.1084/jem.20070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auffray C, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 33.Tacke F, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117:185–194. doi: 10.1172/JCI28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cros J, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nahrendorf M, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landsman L, Varol C, Jung S. Distinct differentiation potential of blood monocyte subsets in the lung. J Immunol. 2007;178:2000–2007. doi: 10.4049/jimmunol.178.4.2000. [DOI] [PubMed] [Google Scholar]
- 37.Shechter R, et al. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 2009;6:e1000113. doi: 10.1371/journal.pmed.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auffray C, Sieweke M, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 39.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varol C, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yrlid U, Jenkins CD, MacPherson GG. Relationships between distinct blood monocyte subsets and migrating intestinal lymph dendritic cells in vivo under steady-state conditions. J Immunol. 2006;176:4155–4162. doi: 10.4049/jimmunol.176.7.4155. [DOI] [PubMed] [Google Scholar]
- 42.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woollard KJ, Geissmann F. Monocytes in atherosclerosis: subsets and functions. Nat Rev Cardiol. 2010;7:77–86. doi: 10.1038/nrcardio.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Infante A, et al. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008;7:3915–3927. doi: 10.4161/cc.7.24.7379. [DOI] [PubMed] [Google Scholar]
- 45.Trikha P, et al. E2f1-3 are critical for myeloid development. J Biol Chem. 2011;286:4783–4795. doi: 10.1074/jbc.M110.182733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landsman L, et al. CX3CR1 is required for monocyte homeostasis and atherogenesis by promoting cell survival. Blood. 2009;113:963–972. doi: 10.1182/blood-2008-07-170787. [DOI] [PubMed] [Google Scholar]
- 47.Auffray C, et al. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You B, Jiang YY, Chen S, Yan G, Sun J. The orphan nuclear receptor Nur77 suppresses endothelial cell activation through induction of IkappaBalpha expression. Circ Res. 2009;104:742–749. doi: 10.1161/CIRCRESAHA.108.192286. [DOI] [PubMed] [Google Scholar]
- 49.Swirski FK, et al. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117:195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011 doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mack M, et al. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol. 2001;166:4697–4704. doi: 10.4049/jimmunol.166.7.4697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.