Abstract
Melanoma inhibitory activity (MIA) is a 12-kDa protein that is secreted from both chondrocytes and malignant melanoma cells. MIA has been reported to have effects on cell growth and adhesion, and it may play a role in melanoma metastasis and cartilage development. We report the 1.4-Å crystal structure of human MIA, which consists of an Src homology 3 (SH3)-like domain with N- and C-terminal extensions of about 20 aa each. The N- and C-terminal extensions add additional structural elements to the SH3 domain, forming a previously undescribed fold. MIA is a representative of a recently identified family of proteins and is the first structure of a secreted protein with an SH3 subdomain. The structure also suggests a likely protein interaction site and suggests that, unlike conventional SH3 domains, MIA does not recognize polyproline helices.
Melanoma inhibitory activity (MIA) or cartilage-derived retinoic acid-sensitive protein (CD-RAP) is a small secreted protein that is normally expressed by cartilage, but is also produced by malignant melanoma (1, 2). Historically, MIA was identified as a factor secreted from melanoma cells that inhibited their growth in vitro (3). MIA's expression has been observed less frequently in other cancers, and it is not typically expressed in normal melanocytes (4). Although the pathological function of MIA in malignant melanoma is not known, it has been implicated in metastasis. Some studies have shown that increased plasma levels of MIA are correlated with a more advanced metastatic disease state (5). Also, increased expression of MIA in melanoma cells enhanced their metastatic potential when injected into hamsters (6). MIA inhibits the adhesion of melanoma cells to the extracellular matrix in vitro, including matrices composed solely of fibronectin or laminin (7). Therefore, MIA might function to promote the formation of metastases by inhibiting the attachment of melanoma cells to the extracellular matrix.
In normal tissues, MIA is expressed primarily by chondrocytes. Its expression begins at chondrogenesis and continues through development and in mature chondrocytes, suggesting that MIA is fundamental to the cartilage cell phenotype (8). Additionally, MIA expression is inhibited in vitro concurrently with repression of cartilage cell phenotype when retinoic acid is added to cartilage cells, hence the bovine homologue is known as cartilage-derived retinoic acid-sensitive protein (CD-RAP) (2). Here, we describe the 1.4-Å crystal structure of MIA and discuss its implications for the biological function of this protein.
Materials and Methods
Protein Expression and Purification.
A synthetic MIA gene was constructed, cloned into the pET3a vector (Novagen), and transformed into the Escherichia coli strain BL21. Unlabeled and selenomethionine-labeled protein were expressed in Mops minimal media (9) where selenomethionine was incorporated as described previously (10). MIA was refolded from inclusion bodies as follows. Washed inclusion bodies from a 1.5-liter culture were dissolved in 15 ml of 8 M guanidinium hydrochloride/2 mM EDTA/50 mM Tris (pH 8.3)/30 mM DTT. The soluble fraction was added dropwise to 300 ml of 50 mM Tris (pH 8.3)/5 mM EDTA/0.6 M guanidinium hydrochloride/2 mM oxidized glutathione/1 mM reduced glutathione. The oxidized protein was further purified by ion exchange with SP-fast flow resin (Amersham Pharmacia) at pH 4.5, followed by size exclusion from a HiLoad 26/60 Superdex 75 column (Amersham Pharmacia). Mass spectral analysis confirmed that MIA retained the initiating methionine, the disulfides were oxidized, and the selenomethionine occupancy was >99%.
Protein Crystallization and Structure Determination.
Hanging drops were prepared by mixing 6 μl of MIA at 6.5 mg/ml in 10 mM acetate (pH 4.5)/0.02% azide, with 2 μl from the reservoir. Native crystals were grown over a reservoir of 100 mM Tris (pH 8.2) and 8% polyethylene glycol (PEG) 8000. The selenomethionine crystal was grown at pH 8.5. The cryoprotectant, 100 mM Tris (pH 8.5)/20% 2-methyl-2,4 pentanediol/8% PEG 8000, allowed flash-cooling in liquid N2. The crystals belong to the space group P212121 (a = 47.534 Å, b = 48.396 Å, c = 87.774 Å), and contain two MIA molecules per asymmetric unit.
Synchrotron x-ray data were collected from one crystal each of native protein and selenomethionine containing protein (Table 1). The structure was determined by using multiwavelength anomalous diffraction (MAD) methods. The automation program elves (J.M.H., unpublished material) was used to direct the following programs for all stages of data analysis. Diffraction data were integrated with mosflm (11) and scaled with scala (12). Seven selenium sites were found by the program solve (13), and phases were refined with mlphare (12). Solvent flattening and histogram matching (14) produced a clearly interpretable electron density map within 11 h of data collection. Automatic model building with arp/warp (15) traced 90% of the amino acid residues. The rest of the structure was built manually into the refined electron density map by using the program o (16). The structure was refined against all native data from 44 Å to 1.4 Å, by using refmac (17). No noncrystallographic symmetry restraints were used in the refinement. The refined model contains 210-aa residues and 212 water molecules. The free-R factor was calculated with 5% of the data (2019 reflections). The atomic coordinates have been deposited in the Protein Data Bank (PDB ID code 1I1J). Figs. 1 and 3 A and B were prepared with the program molmol (18).
Table 1.
Data collection, phasing, and refinement statistics
| SelenoMet
|
Native f | |||
|---|---|---|---|---|
| f′ | f′ | f (low) | ||
| Wavelength, Å | 0.9791 | 0.9794 | 1.0332 | 1.0000 |
| Resolution, Å | 1.29 | 1.29 | 1.29 | 1.39 |
| Rsym* | 0.050 | 0.063 | 0.056 | 0.062 |
| Completeness, % | 92.57 | 92.94 | 85.44 | 99.44 |
| Multiplicity | 8.3 | 8.5 | 8.1 | 6.6 |
| I/SD† | 17.6 (1.9) | 14.2 (1.1) | 18.0 (1.4) | 16.8 (3.0) |
| Phasing power‡ | 0.66/1.26 | 0/1.11 | 1.3/0.17 | |
| Mean figure of merit§ (44–1.29 Å): 0.512 (0.691 after solvent flattening) | ||||
| Rcryst/Rfree¶ (44–1.39 Å) = 0.208/0.231 | ||||
| rms Δbonds, rms Δangles‖: 0.014, 1.5° | ||||
| Average B factor: 23.35 Å2 | ||||
Rsym = Σ|I − 〈I〉|/Σ I; I, intensity.
I, intensity; SD, standard deviation. Parentheses denote I/SD for highest resolution bin.
Phasing power (dis/ano) = [Σn|FH|2/Σn|E|2]1/2; FH, calculated heavy atom scattering factor; E, lack of closure error.
Mean figure of merit = 〈∥∑αP(α)eiα/ΣαP(α)∥〉; αphase; P(α), phase probability distribution.
Rcryst = Σ|Fo − Fcalc|/ΣFo; Fo, observed structure-factor amplitude; Fcalc, calculated structure-factor amplitude.
rms deviations from ideal values.
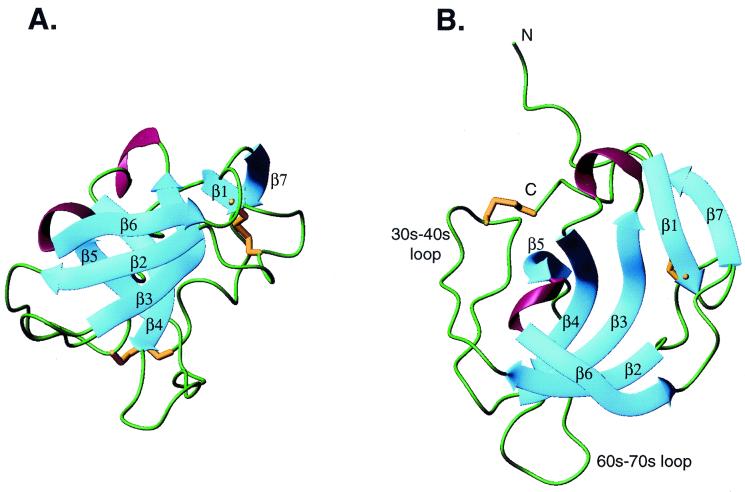
Figure 1.
The structure of melanoma inhibitory activity protein. β-strands are in blue, regions lacking regular secondary structure are in green, 310 helices are in red, and disulfide bonds are in gold. (A) View of the two β sheets that pack at right angles to each other. Sheet I is in front. β1 and β7 add onto sheet II of the SH3 β-sandwich. The top of the barrel is toward the top of the page. (B) View looking into one end (mouth) of the barrel. The 30s-40s loop (analogous to the RT loop of SH3 domains) and the 60s-70s loop, flank one mouth of the barrel. The N-terminal residues preceding β1 and the C-terminal residues following β7 run along the outside face of sheet II. A and B are on the same scale.
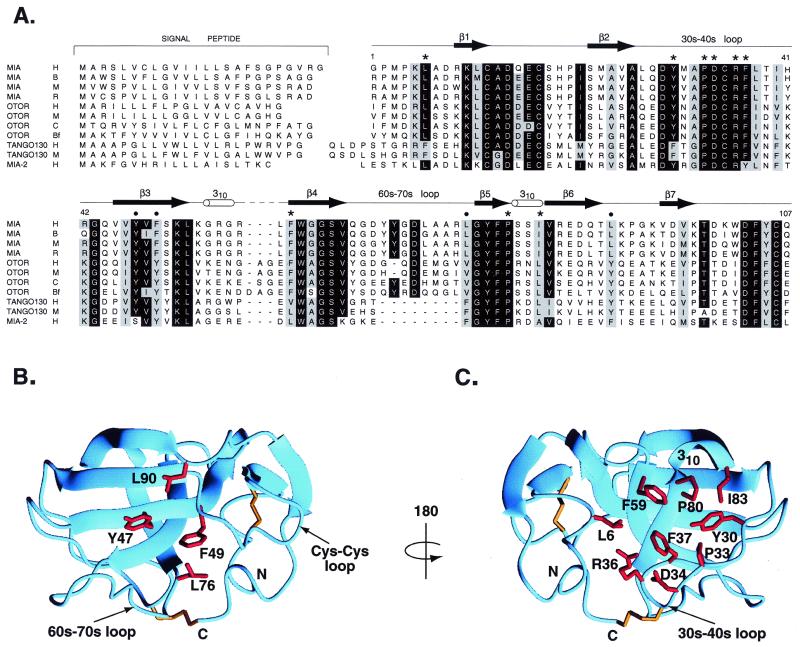
Figure 3.
Sequence alignment of MIA with secreted homologues defines possible ligand binding sites. (A) Sequence alignment of MIA with secreted homologues. Species are abbreviated as follows: H, human; B, bovine; M, mouse; R, rat; C, chicken; and Bf, bullfrog. Numbering corresponds to human MIA, and the MIA secondary structure is shown. Bullets indicate a conserved patch located on one side of the molecule; these residues are displayed on the MIA structure in B. Stars indicate conserved surface residues on the opposite side of the molecule. These residues are shown on the MIA structure in C. In SH3 domains, the structurally analogous region to that shown in C is the polyproline helix binding site. Signal peptides were predicted by using the program signalp (34). MIA-2 is predicted by genscan from GenBank genomic sequence 7708219. tango 130 sequences are from U.S. patent WO 00/12762.
Results and Discussion
The crystal structure of MIA was determined by using multiwavelength anomalous diffraction (MAD) analysis of selenomethionine containing protein (Table 1). The MAD-phased electron density map was of high quality. The model was refined against native data to 1.4 Å. The model contains good bond geometry and has 96% of the residues in the most favored region of the Ramachandran plot. The asymmetric unit contained two independent molecules (A and B). The two molecules were extremely similar, with an overall backbone rms deviation of 0.2 Å. The largest structural differences were found at the termini and in the 60s-70s loop, and the electron density was weakest for these regions. The first two residues in molecule B and the C-terminal residue from both molecules had no electron density and were not included in the model.
Overview of the Structure.
MIA is a small single-domain globular protein consisting of seven β-strands, two short segments of 310 helix, and several loops (Fig. 1). The overall structure is oblong in shape with dimensions of 18 × 18 × 28 Å. Five β-strands (β2-β6) and the connections between them make up a subdomain that is structurally related to Src homology 3 (SH3) domains. Two antiparallel three-stranded β-sheets pack at approximately right angles to each other, and an 18-residue hairpin-like loop, analogous to the Src family of SH3 domains' RT loop, is formed between strands β2 and β3. Sheet I is formed by strands β2, β3, and β6. Strand β3 continues in sheet II, which is formed by strands β3, β4, and β5. These sheets are highly curved, and overall, the SH3 subdomain resembles an open β-barrel. The 30s-40s loop, referred to as the RT loop in SH3 domains, and a 12-residue 60s-70s loop, flank one mouth of this barrel.
At the opposite end of the structure, MIA has N- and C-terminal extensions to the SH3 subdomain of approximately 20 aa each that wrap around the top of the barrel and build outward, extending the barrel's height and widening the mouth. The extensions add two additional β strands to the top of the SH3 barrel that pair with each other and with strand β3. A six-residue loop bounded by a disulfide between Cys-12 and Cys-17 directly follows β1 and protrudes into solvent. Eight residues that lack regular secondary structure follow β7 and run along the outside face of sheet II from the top of the barrel to the bottom. These residues are constrained by a disulfide bond between Cys-106, the penultimate residue, and Cys-35, which is located at the apex of the 30s-40s loop. The N-terminal residues preceding β1 also run along this face of the molecule, whereas residues one and two extend away from the side of the barrel into the solvent.
Comparison to the Src Family of SH3 Domains.
MIA is the first structure of a secreted protein that contains an Src-homologous SH3 subdomain. The MIA SH3 subdomain shares sequence similarity with canonical SH3 domains, suggesting that they are evolutionarily related (Fig. 2). MIA and the closest SH3 homologue, VAV-3, are 35% identical. In addition, many SH3 hydrophobic core residues are conserved (19). Conventional SH3 domains are found abundantly as modular units of intracellular proteins, and they mediate protein–protein interactions in signal transduction cascades and membrane-cytoskeleton structures through recognition of proline-rich ligands (20, 21). In contrast, MIA is a single domain-secreted protein with disulfide bonds that contains an SH3 subdomain, and it probably binds to a different class of ligands (see below).
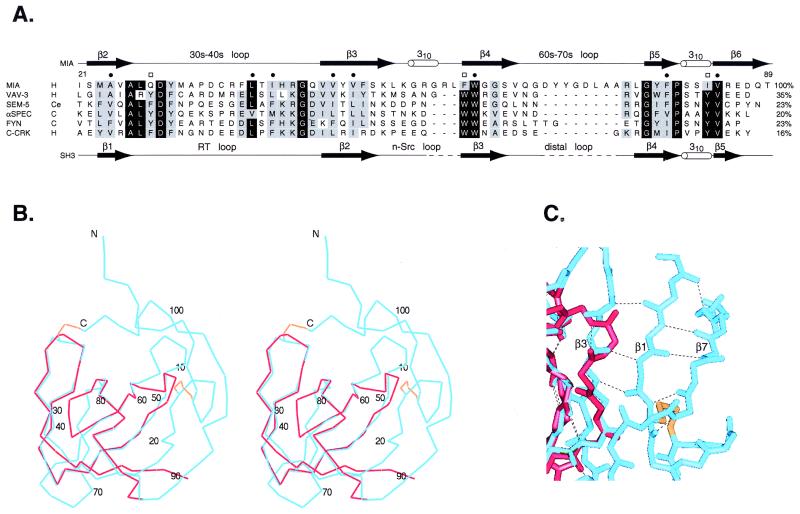
Figure 2.
Comparison of MIA to SH3 domains. (A) Sequence alignment of MIA with several SH3 domains. Species are abbreviated as follows: H, human; Ce, Caenorhabditas elegans; and C, chicken. Residues labeled with a bullet make up the conserved hydrophobic core in SH3 domains. Conserved SH3 residues labeled with a box make up a triad of aromatic residues arranged approximately linearly at the polyproline helix binding site. The secondary structure of MIA is shown above the alignment, and the secondary structure of the C-Crk SH3 domain is shown below the alignment. The percent identity relative to MIA is listed at the right of each sequence. (B and C) Comparison of the MIA structure with an SH3 domain. MIA is in blue, the Sem-5 SH3 domain is in red, and the MIA disulfide bonds are in gold. (B) Stereo view of the superposition of MIA with the Sem-5 SH3 domain. The rms deviation of the carbon-α positions is 0.61 Å, where 37 α-carbon pairs were used in the superposition. The numbering corresponds to human MIA. (C) View of the β3 strand extension in MIA relative to SH3 domains. Hydrogen bonding between β3 and β1 extends the length of strand β3.
The SH3 subdomain of MIA is strikingly similar to the canonical structure of SH3 domains. The superposition of MIA with the Sem-5 SH3 domain is shown in Fig. 2B. Sem-5 was chosen as a representative SH3 domain because of the primary and tertiary structural similarities between Sem-5 and MIA and because the ligand-bound structure of Sem-5 is known. These superimposed regions include the shared secondary structure elements and contain 37 of the 56 possible α-carbon pairs. The rms deviation of the superimposed α-carbons is 0.61 Å.
The MIA 30s-40s loop is similar in structure to the RT loop of SH3 domains. The RT loop, a characteristic feature of members of the Src-family of SH3 domains, contains residues that are involved in achieving ligand specificity (21). The length and composition of RT loops vary, but they typically adopt an irregular antiparallel β-hairpin structure. Conserved backbone–backbone and sidechain–backbone cross-strand hydrogen bonds stabilize the loop structure (19, 22). The 30s-40s loop of MIA is similar in structure to the RT loop and contains two analogous cross-strand backbone–backbone hydrogen bonds between residues 30 and 40, located at its base. However, the 30s-40s loop does not contain the conserved sidechain–backbone cross-strand hydrogen bonds found in many SH3 domains. Instead, MIA has several unique features that stabilize the loop structure. These include the disulfide Cys-35—Cys-106 that tethers the C terminus to the apex of the 30s-40s loop and a network of hydrogen bonds at the apex of the loop. The hydrogen bonds are between residues 34 and 36, between the backbone carbonyl oxygen of Cys-35 and the sidechain of Arg-75, and between the backbone amide of residue 36 and the following possible acceptors: the sulfur of Cys-35, the sulfur of Cys-106, the backbone carbonyl oxygen of Asp-34, and the sidechain of Asp-34 (not shown).
Several other subtle differences between the MIA SH3 subdomain and the Src-family of SH3 domains are apparent. MIA's β3 strand is lengthened relative to the corresponding strand in canonical SH3 domains because of additional hydrogen bonds that form between β3 and β1 (Fig. 2C). Between β3 and β4 is the region that corresponds to the n-Src loop in SH3 domains. This loop flanks the ligand binding site of SH3 domains and varies in length and composition (21, 23), but in most SH3 domains solved to date, the n-Src loop lacks defined secondary structure. In contrast, this region of MIA adopts a 310 helix. Between β4 and β5 is the 60s-70s loop, which corresponds to the distal loop (distant from the binding site) in SH3 domains (21). The distal loop varies in length and composition, and, in many SH3 domains, it adopts a β-turn conformation (19). The MIA 60s-70s loop is longer than the corresponding loop in canonical SH3 domains, and, in the solution structure, this loop has multiple conformations from amino acid residues 66 to 76 (J.C.L., P.J.D., and J.M.H., unpublished results).
Functional Implications.
MIA is a member of a recently identified protein family whose biological functions are only beginning to emerge. We are unaware of any fold that has similarity to the entire molecule, and MIA is the first structure of a secreted protein with an Src-homologous SH3 subdomain. Recently two new paralogs were discovered, otoraplin (OTOR; ref. 24) and TANGO 130 (25), and a fourth putative paralog, which we will refer to as MIA-2, is predicted from human genomic sequences (Fig. 3). As with SH3 domains, the MIA fold can occur as a domain within a larger protein; however, unlike SH3 domains, the known MIA domains appear only at N-terminal locations in proteins. In the murine variant of TANGO 130, the MIA-homologous portion is at the N terminus of an ≈700-aa protein. Alternatively spliced forms may also exist. For MIA-2, genscan (26) predicts both a long form, with the MIA domain at the N terminus, and a short form consisting of only the MIA domain. The presence of MIA-like modules as isolated domains or as domains within larger proteins suggests the possibility of wide functional diversity.
Of the four paralogs, MIA is the most characterized. Effects on cell growth and adhesion and a localized expression pattern in cartilage and malignant melanoma have been described. MIA has been postulated to competitively mask integrin binding sites by specifically associating with the extracellular matrix proteins fibronectin and laminin, leading to decreased adhesiveness of the connective tissue to melanoma cells (7); however, the experimental data to support this hypothesis have not yet been published. Additionally, the reported in vitro effects on cell growth and adhesion occur at nanomolar concentrations (3, 7) and are consistent with a high affinity receptor-mediated response. However, neither a receptor nor a cell-signaling activity of MIA has been reported. Thus, the biological functions of MIA and the biochemical mechanisms leading to the observed effects remain elusive. As a first step in characterizing the biochemical mechanism of action, we analyzed the molecular surface of MIA to explore its possible ligand interaction site(s). In particular, we considered whether or not MIA might recognize similar ligands as conventional SH3 domains.
The surface of MIA has two large hydrophobic areas that have solvent-accessible surface areas of ≈350 Å2 and 340 Å2 and several basic patches. The basic clusters are not conserved in the homologues. In contrast, both of the hydrophobic patches contain multiple conserved residues, raising the possibility that these areas may be ligand interaction sites. Indeed, analyses of protein complexes indicate that often one of the most hydrophobic clusters on the surface of a protein is found at the interface (27).
The larger of the two hydrophobic areas is comprised of residues Pro-20, Met-23, Val-25, Val-45, Tyr-47, Phe-49, Leu-72, Leu-76, and Leu-90, and it is located in a groove between the 60s-70s loop and the conserved Cys-12–Cys-17 loop (Fig. 3B). Of these residues, only Leu-76, Leu-90, and Phe-49 are conserved as hydrophobic amino acids among MIA homologues, whereas Tyr-47 is conserved in all homologues except for MIA-2. These residues expose about 170 Å2 of solvent-accessible hydrophobic surface and form a curved strip on the surface of the molecule. Because the conserved patch is comprised of only four residues, it is difficult to predict whether or not it is a ligand recognition site.
The other hydrophobic patch is part of a relatively flat surface between the 30s-40s loop and the first turn of 310 helix (Fig. 3C), and is a likely candidate for a ligand interaction site. This surface corresponds to the polyproline helix binding region in SH3 domains (21) and is the most conserved area of the molecular surface among MIA homologues. The hydrophobic residues include Leu-6, Tyr-30, Pro-33, Phe-37, Leu-58, Phe-59, Pro-80, and Ile-83. All of these are conserved except for Leu-58, which lies on the edge of the patch. The conserved residues expose about 210 Å2 of solvent-accessible hydrophobic surface. Additionally, four residues at the apex of the 30s-40s loop, 33PDCR36, are strictly conserved. Here Pro-33 contributes to the hydrophobic patch whereas Asp-34 and Arg-36 flank it. This putative MIA ligand binding site lies within a larger platform-like surface which is extended by ≈50% relative to SH3 domains. The extension is in the direction that a putative polyproline helix axis would run if MIA bound polyproline helices analogously to SH3 domains.
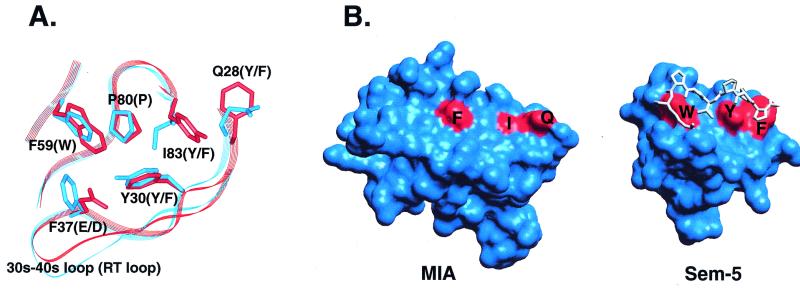
Although MIA has conserved residues located at the putative polyproline helix binding site, we predict that it has evolved its own distinct mechanism of ligand recognition. Previous structural work has revealed the basis for recognition of left-handed polyproline type II (PPII) helices by SH3 domains (28, 29). PPII ligands contact conserved SH3 domain residues that form several grooves and ridges (Fig. 4B). Each of two hydrophobic grooves recognizes one X-P unit of the PPII helix. The hydrophobic grooves are formed by a conserved set of three aromatic residues, Tyr/Phe, Tyr/Phe, and Trp, which are arranged approximately linearly on the molecular surface. Similarly, WW domains, which also bind to PPII helices, have two conserved aromatics that form one X-P binding groove, emphasizing the importance of these grooves in PPII helix recognition (30, 31). In MIA, four of six of the conserved SH3 binding site residues differ from canonical SH3 domains (Fig. 4A). The four conserved SH3 binding site residues that differ in MIA are a Glu/Asp and the three aromatics that form the X-P grooves. The substitution of Glu/Asp by Phe-37 may not affect selection for the PPII helix because this residue is not involved in forming the X-P grooves. Instead, it specifically recognizes a potentially variable side chain of the bound PPII helix (23). However, the replacements of the Tyr/Phe, Tyr/Phe, Trp in SH3 domains by a Gln, Ile, Phe triad in MIA results in a relatively flat molecular surface that lacks complementary packing with polyproline helices (Fig. 4B). Also, the conservative replacement of Trp by Phe in the Sem-5 SH3 domain has been shown to abolish binding to a polyproline ligand (32). We also tested the ability of MIA to bind proline-rich peptides by using phage display, as has been done for many SH3 domains (33), but we were unable to identify significant peptide binders out of a biased polyproline helix phage display library (J.C.L., J. D. Kasanov, T.M.H., and B. K. Kay, unpublished results). Together, these data suggest that the constellation of residues in MIA required for receptor or ligand recognition differs from SH3 domains.
Figure 4.
Comparison of the SH3 polyproline helix binding site to the analogous region in MIA. (A) Superposition of the conserved residues in the polyproline helix binding site of Sem-5 (red) and the corresponding residues in MIA (blue). Four of six residues conserved in SH3 domains differ in MIA. Residue numbering corresponds to MIA. Residues listed in parentheses are those found in canonical SH3 domains. (B) View of the molecular surfaces of MIA (Left) and the Sem-5 SH3 domain (Right). The structure of Sem-5 is shown with its polyproline ligand bound. Sem-5 residues colored in red are those that make up a triad of conserved aromatic residues arranged approximately linearly on the molecular surface. The dissimilarity of the corresponding residues in MIA, F59, I83, and Q28 result in a much smoother molecular surface that most likely does not recognize polyproline helices.
MIA is a member of a recently identified family of proteins and is the first structure of a protein with a secreted SH3 subdomain. Despite the striking similarity between the SH3 subdomain of MIA and canonical SH3 domains, MIA most likely does not recognize polyproline helices. MIA has N- and C-terminal extensions that add structural elements to the SH3 domain, forming a previously unobserved fold. The resulting overall fold, its extracellular location, and our structural results suggest that nature has elaborated a common scaffold to create additional functional diversity.
Acknowledgments
We thank R. Rose, S. McWhirter, and members of the Alber lab for useful suggestions and help with x-ray data collection. Data were collected at the Advanced Light Source (ALS), Lawrence Berkeley Laboratory, with the help of T. Earnest, and at the Stanford Synchrotron Radiation Laboratory (SSRL) with the help of P. Ellis. Both facilities are operated by the Department of Energy, and SSRL is supported by the National Institutes of Health. We also thank J. Kasanov and B. Kay for their help with phage display experiments, D. King for mass spectrometry, and N. Pokala and members of the Handel group for helpful discussions. This work was supported by grants awarded to T.M.H. from the National Institutes of Health, the Pew Scholars Program in the Biomedical Sciences, and the University of California Campus-Laboratory Collaborations program and by a National Institutes of Health training grant to J.C.L.
Abbreviations
- MIA
melanoma inhibitory activity
- PPII
polyproline type II
- SH3
Src homology 3
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank (PDB ID code 1I1J).
References
- 1.Bosserhoff A K, Hein R, Bogdahn U, Buettner R. J Biol Chem. 1996;271:490–495. doi: 10.1074/jbc.271.1.490. [DOI] [PubMed] [Google Scholar]
- 2.Dietz U H, Sandell L J. J Biol Chem. 1996;271:3311–3316. doi: 10.1074/jbc.271.6.3311. [DOI] [PubMed] [Google Scholar]
- 3.Blesch A, Bosserhoff A K, Apfel R, Behl C, Hessdoerfer B, Schmitt A, Jachimczak P, Lottspeich F, Buettner R, Bogdahn U. Cancer Res. 1994;54:5695–5701. [PubMed] [Google Scholar]
- 4.Golob M, Buettner R, Bosserhoff A K. J Invest Dermatol. 2000;115:42–47. doi: 10.1046/j.1523-1747.2000.00028.x. [DOI] [PubMed] [Google Scholar]
- 5.Bosserhoff A K, Kaufmann M, Kaluza B, Bartke I, Zirngibl H, Hein R, Stolz W, Buettner R. Cancer Res. 1997;57:3149–3153. [PubMed] [Google Scholar]
- 6.Guba M, Bosserhoff A K, Steinbauer M, Abels C, Anthuber M, Buettner R, Jauch K W. Br J Cancer. 2000;83:1216–1222. doi: 10.1054/bjoc.2000.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosserhoff A K, Golob M, Buettner R, Landthaler M, Hein R. Hautarzt. 1998;49:762–769. doi: 10.1007/s001050050822. [DOI] [PubMed] [Google Scholar]
- 8.Bosserhoff A K, Kondo S, Moser M, Dietz U H, Copeland N G, Gilbert D J, Jenkins N A, Buettner R, Sandell L J. Dev Dyn. 1997;208:516–525. doi: 10.1002/(SICI)1097-0177(199704)208:4<516::AID-AJA7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 9.Neidhardt F C, Bloch P L, Smith D F. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 11.Leslie A G W, Brick P, Wonacott A. Daresbury Lab Info Q Protein Crystallogr. 1986;18:33–39. [Google Scholar]
- 12.Collaborative Computational Project Number 4. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- 13.Terwilliger T C, Berendzen J. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowtan K. Joint CCP 4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 15.Perrakis A, Morris R, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 16.Jones T A, Zou J Y, Cowan S W, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Murdushov G N, Dodson E J, Vagin A A. Proceedings of Daresbury Study Weekend. Warrington, U.K.: CLRC Daresburg Lab.; 1996. pp. 93–104. [Google Scholar]
- 18.Koradi R, Billeter M, Wuthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. , 29–32. [DOI] [PubMed] [Google Scholar]
- 19.Larson S M, Davidson A R. Protein Sci. 2000;9:2170–2180. doi: 10.1110/ps.9.11.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 21.Musacchio A, Wilmanns M, Saraste M. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 22.Cordier F, Wang C, Grzesiek S, Nicholson L K. J Mol Biol. 2000;304:497–505. doi: 10.1006/jmbi.2000.4274. [DOI] [PubMed] [Google Scholar]
- 23.Chen J K, Schreiber S L. Angew Chem Int Ed Engl. 1995;34:953–969. [Google Scholar]
- 24.Robertson N G, Heller S, Lin J S, Resendes B L, Weremowicz S, Denis C S, Bell A M, Hudspeth A J, Morton C C. Genomics. 2000;66:242–248. doi: 10.1006/geno.2000.6224. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y. A Novel Protein Related to Melanoma-Inhibiting Protein and Uses Thereof. U.S. Patent Office; 2000. , International Publication Number WO 00/12762. [Google Scholar]
- 26.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 27.Young L, Jernigan R L, Covell D G. Protein Sci. 1994;3:717–729. doi: 10.1002/pro.5560030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim W A, Richards F M, Fox R O. Nature (London) 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck M J. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 31.Verdecia M A, Bowman M E, Lu K P, Hunter T, Noel J P. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 32.Lim W A, Richards F M. Nat Struct Biol. 1994;1:221–225. doi: 10.1038/nsb0494-221. [DOI] [PubMed] [Google Scholar]
- 33.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quillam L A, Kay B K. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]