Abstract
Background. Little is known about the clonality of Staphylococcus epidermidis in the United States, although it is the predominant pathogen in infections involving prosthetic materials, including ventricular assist devices (VADs).
Methods. Seventy-five VAD recipients at 4 geographically diverse US cardiac centers were prospectively followed up to 1 year of VAD support. The anterior nares, sternum, and (future) driveline exit site were cultured for S. epidermidis before VAD insertion and at 7 times after surgery. Infection isolates were also collected. Isolates were typed by pulsed-field gel electrophoresis. A subset underwent susceptibility testing and staphylococcal chromosomal cassette mec and multilocus sequence typing.
Results. A total of 1559 cultures yielded 565 S. epidermidis isolates; 254 of 548 typed isolates (46%) belonged to 1 of 7 clonal types as defined by pulsed-field gel electrophoresis. These clones were identified in up to 27 people distributed across all 4 cardiac centers. They caused 3 of 6 VAD-related infections. Disseminated clones were more antibiotic resistant than were less prevalent isolates (eg, 79% vs 54% methicillin resistant; P = .0021).
Conclusions. This study revealed that healthcare–associated S. epidermidis infection is remarkably clonal. We describe S. epidermidis clones that are highly resistant to antibiotics distributed across US cardiac centers. These clones may have determinants that enhance transmissibility, persistence, or invasiveness.
Clinical Trials Registration. NCT01471795.
Staphylococcus epidermidis is a common colonizer of the skin, oral, and nasal mucosa. Although it is usually part of the normal commensal flora, it is one of the most frequent causes of catheter- and device-related infections, including ventricular assist devices (VADs) [1–3]. More than 70% of hospital isolates may be methicillin-resistant S. epidermidis (MRSE), and the majority of them are multidrug resistant, which make these infections more difficult and expensive to treat [4, 5].
The clonality of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) has been extensively studied, and it is well-recognized that MRSA has evolved from a discrete number of major clonal lineages [6]. In contrast, the clonality of S. epidermidis and MRSE is less clear. Clinical isolates frequently belong to clonal complex 2 (CC2), which includes the most commonly isolated sequence type 2 (ST2) [7]. However, although there has been the recognition of clonal outbreaks or dominant clones in particular clinical settings [8–15], data have suggested that S. epidermidis strains, including MRSE, are generally highly diverse, especially in the community [16].
A limited number of studies have indicated that there may be geographic dissemination of particular MRSE strains, either at the country level [17] or on a global scale [18, 19]. To our knowledge, the clonality of S. epidermidis colonization has not been studied on a broad scale in hospitals in the United States. Identifying highly successful S. epidermidis clones in specific patients, such as those with VADs, beyond a specific healthcare center would both have clinical relevance and may provide insight into S. epidermidis epidemicity.
As a part of a multicenter, prospective, observational study of infection and colonization of VAD recipients, we studied the clonality of S. epidermidis carriage and infection at and across 4 geographically diverse cardiac centers in the United States.
METHODS
Study Design and Participants
The VAD Infection Study was a multicenter, prospective, observational study conducted from 2006 through 2008 and was coordinated by the International Center for Health Outcomes and Innovation Research at Columbia University. One hundred fifty participants with advanced heart failure scheduled to receive a VAD were enrolled in the study at 11 cardiac centers and were followed up for 1 year (or until death, device explant, and/or transplantation). All participants gave informed consent, and the institutional review board at each clinical center approved the study.
To study S. epidermidis colonization, samples were obtained for surveillance culture from the following body sites with premoistened culturettes (Becton Dickinson BBL CultureSwabs) before surgery: anterior nares, sternum, and future driveline exit site (usually right, lateral, abdomen). After VAD placement, the nares, sternal wound/site, driveline exit site, and 10 inches of the VAD driveline were swabbed at the following times: 24–48 hours after the discontinuation of perioperative antibiotics (or at 1 week if antibiotics were still being administered), 2 weeks, 4 weeks (or at hospital discharge if this came sooner), and 3, 6, 9, and 12 months.
To study S. epidermidis infection, S. epidermidis and other coagulase-negative staphylococci not identified at the species level that were isolated from clinical cultures (ie, blood cultures) were sent to our laboratory. VAD infections were reported by cardiac center investigators and confirmed by a review of the medical record by an infectious diseases specialist (R. G.). All infected individuals had clinical signs of infection, and coagulase-negative staphylococci were either cultured from a sterile site (eg, the pump pocket in the operating room) or from multiple sites (eg, ≥2 blood cultures, driveline drainage, and drainage from the pump pocket), as reported by the centers’ clinical microbiology laboratories.
We analyzed 1513 surveillance cultures and 46 clinical specimens obtained from all 75 left ventricular assist device (LVAD) recipients enrolled at 4 of the 11 clinical centers. These centers were chosen for their geographic diversity: New York–Columbia Presbyterian Medical Center, New York (center 1); Aurora St Luke’s Medical Center, Wisconsin (center 3); Advocate Christ Medical Center, Illinois (center 12); and Sacred Heart Medical Center, Washington State (center 13).
Specimen Processing
Samples were processed for S. epidermidis. Swab tips were sterilely placed into 1.5-mL Eppendorf tubes, vortexed with 1 mL of Todd-Hewitt broth, and incubated at 37°C (>200 rpm) for 3 hours; 100 μL was then plated onto Columbia CNA agar plus 5% sheep blood. CNA plates were incubated at 37°C for 72 hours and then kept at room temperature for 48 hours. Each colony with a unique morphology consistent with coagulase-negative staphylococci was randomly selected and isolated on blood agar plates for further species identification [20]. Overnight cultures of all isolates were frozen with 20% glycerol and stored at −80°C.
Species Identification
Single colonies were confirmed to be S. epidermidis by amplification by polymerase chain reaction of the tuf gene using specific primers for the species, according to Martineau et al [21].
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) was the primary method used to define clonality in this study. All isolates were typed using PFGE with the exception of 17 isolates that were not retrievable. DNA from S. epidermidis isolates was prepared in agarose disks as previously described [22]. After SmaI digestion, DNA fragments were separated in a contour-clamped homogeneous electric field apparatus (CHEF-DRII; Bio-Rad). PFGE images were captured, archived, and analyzed using Bionumerics software coupled with the GelDoc 100 System (Bio-Rad). We constructed dendrograms showing percentage of relatedness with use of the Dice coefficient method with 0.8 optimization and 1.3% position tolerance. Clusters were designated by Bionumerics using a >79% cutoff and confirmed by manual visualization [23] and were identified by arabic numbers. When a single culture yielded S. epidermidis isolates with different morphologies but with an identical PFGE pattern, only 1 of the isolates was included in further analyses. Sixteen isolates, corresponding to 2–3 representatives of each of the 7 most predominant clusters, were also compared with a large non-US geographically diverse sample of S. epidermidis isolates [18] by PFGE, multilocus sequence typing (MLST), and SCCmec typing.
Multilocus Sequence Typing
Sixteen isolates representative of the 7 most predominant clonal types, as defined by PFGE, were selected for characterization by MLST using the method described by Thomas et al [24] goeBURST (http://goeburst.phyloviz.net) was used to assess the relationships among the predominant sequence types identified in the study with all available data in the database (www.mlst.net) as of 25 October 2011.
Antibiotic Susceptibility Testing
One hundred isolates representative of the 7 most prevalent clones and 50 randomly selected, less prevalent isolates underwent testing using the Kirby-Bauer disk diffusion method according to guidelines of the Clinical Laboratory Standards Institute [25]. The following antibiotics were tested: penicillin, cefoxitin, oxacillin, tetracycline, gentamicin, rifampin, levofloxacin, linezolid, and trimethoprim-sulfamethoxazole.
SCCmec Typing
MRSE isolates that underwent MLST (representatives of the 7 most predominant clonal types) were also selected for SCCmec typing. SCCmec types were defined using a combination of the class of the mec complex and the ccr type [7, 26]. Isolates were considered to be nontypeable when the results did not correspond to any of the SCCmec types I–XI.
Statistical Analysis
A robust repeated-measures analysis was performed to determine whether cultures were more likely to be positive for S. epidermidis at certain clinical centers and/or culture sites. The GENMOD procedure, which uses generalized estimating equations to account for correlated data collected over time, was used to investigate associations and generate odds ratios (ORs) and P values [27]. The same procedure was used to test the hypothesis that particular clones of S. epidermidis would be more prevalent at particular clinical centers, culture sites, or times. The GENMOD procedure was also used to test the hypothesis that the 7 most prevalent isolates would be more resistant to antibiotics, compared with less prevalent isolates.
Samples were collected from (1) different culture sites from the same person, (2) different times from the same person, and (3) different people at the same clinical center. Because these repeated measurements were likely correlated, the GENMOD procedure was used to model a correlation structure, commonly referred to as a covariance pattern. Of the various covariance structures to choose from, the exchangeable structure provided the best fit. This model allows for improved estimates of the standard errors of measurement and also allows for estimates at all times if data can be assumed to be missing at random [27].
P values < .05 were considered statistically significant but were not adjusted for multiple comparisons and type I error. Data were analyzed using SAS system software, version 9.1.3 (SAS Institute).
RESULTS
S. epidermidis Frequency Among Cardiac Centers
Although 75 participants (Table 1) were enrolled in the study, only 74 had cultures performed for S. epidermidis. Overall, 1513 surveillance samples were collected, of which 425 (28.1%) were positive for at least 1 S. epidermidis isolate (Table 2). Surveillance samples from center 3 were more likely to be positive for S. epidermidis than were those from center 1 (OR, 1.90; P = .006), and sample positivity across all 4 clinical centers differed with near statistical significance (χ2 = 7.65; P = .05). In total, 565 S. epidermidis isolates were obtained from the 4 selected cardiac centers, including 519 isolates from surveillance and 46 clinical isolates. Eighty-nine samples yielded >1 S. epidermidis strain according to PFGE cluster types.
Table 1.
Patient Characteristics (n = 75)
| Characteristic | No. (%) |
| Age, years, mean ± SD (range) | 58 ± 13 (21–82) |
| Male sex | 65 (87) |
| Race | |
| White | 52 (69) |
| Black | 21 (28) |
| Other | 2 (3) |
| Hispanic or Latino ethnicity | 3 (4) |
| Ischemic etiology of heart failure | 38 (51) |
| History of: | |
| Myocardial infarction | 33 (44) |
| Coronary artery bypass graft surgery | 21 (28) |
| Chronic obstructive pulmonary disease | 13 (17) |
| Chronic renal insufficiency | 23 (31) |
| Diabetes | 23 (31) |
| Valve replacement surgery | 20 (13) |
| Currently has or was on: | |
| Central line | 25 (33) |
| Permanent pacemaker | 47 (63) |
| Automatic intracardiac defibrillator | 59 (79) |
| Intraaortic balloon pump | 16 (21) |
| Mechanical ventilation | 7 (9) |
Table 2.
Staphylococcus epidermidis Culture Data, by Clinical Center
| Clinical Center ID No. | Participants (n = 74) | Surveillance Culture Swabs (n = 1513) | Surveillance Cultures Positive for ≥1 SE Isolate (425; 28.1%), No. (%)a | Surveillance SE Isolates (n = 519)b | Other SE Isolates (n = 46)c |
| 1 | 23 | 388 | 96 (24.7) | 113 | 6 |
| 3 | 10 | 246 | 93 (37.8)d | 116 | 26 |
| 12 | 28 | 624 | 170 (27.2) | 199 | 10 |
| 13 | 13 | 255 | 66 (25.9) | 91 | 4 |
Abbreviations: SE, Staphylococcus epidermidis; VAD, ventricular assist device.
Surveillance culture sites: the anterior nares, sternum (preimplantation) or sternal wound, future (preimplantation) or current driveline exit site, and the first 10 inches of the VAD driveline.
Cultures yielded from 1–3 different SE strains.
Other SE isolates included 39 clinical isolates and 7 cultures performed at VAD explantation.
Surveillance cultures from center 3 were more likely to be positive, compared with cultures obtained at center 1 (odds ratio, 1.90; P = .006).
S. epidermidis Distribution Among Different Collection Sites
A total of 398 anterior nares, 396 sternum/sternal wound, 397 driveline/driveline exit site, and 322 driveline (the first 10 inches) surveillance samples were obtained. The percentage of positivity differed among the different culture sites (χ2 = 46.19; P < .0001). Samples from the nares were most likely to be positive (48%), followed by the sternum/sternal wound (34%), the LVAD driveline (18%), and the driveline exit site (11%).
Molecular Characterization of S. epidermidis
Five hundred forty-eight of the 565 S. epidermidis isolates were successfully typed using PFGE. A total of 491 (89.6% of isolates) were grouped into 71 PFGE clusters (cluster size ≥2), whereas 57 (10.4%) were found to be sporadic isolates. Four clusters were particularly large (eg, PFGE cluster 22 [77 isolates], 11 [61 isolates], 12 [51 isolates], and 33 [31 isolates]), whereas the other 67 clusters included 2–17 isolates.
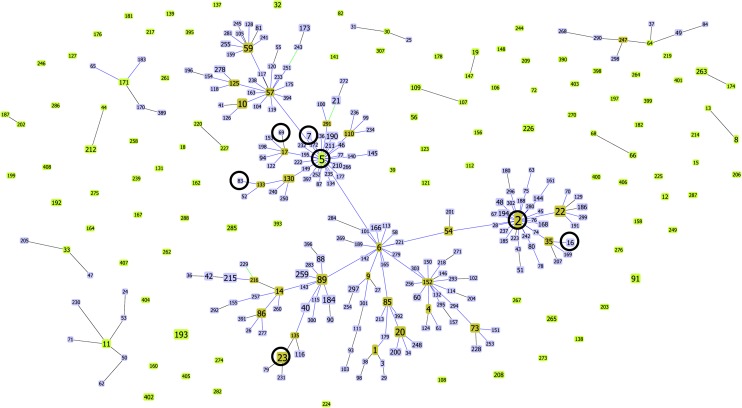
Molecular characterization of representative isolates belonging to the 7 most widely distributed PFGE clusters (see below) by MLST and eBURST showed that a total of 7 different STs were found (Figure 1). Four STs belonged to former CC2, which is now CC5 (because of a change in the defined ancestor of this big clonal complex from ST2 to ST5). Moreover, we verified that isolates belonging to these more prevalent clusters either carried SCCmec type IV (5 isolates) or a nontypeable SCCmec structure (4 isolates) (Figure 2).
Figure 1.
Analysis of multilocus sequence typing data with goeBURST (http://goeburst.phyloviz.net). This figure includes all available data in the database (www.mlst.net) as of 25 October 2011. Predicted founders of each clonal complex are shown in light green. Subgroup founders are depicted in dark green. Single-locus variants are shown in blue. Dark circles indicate sequence types that were identified in this study.
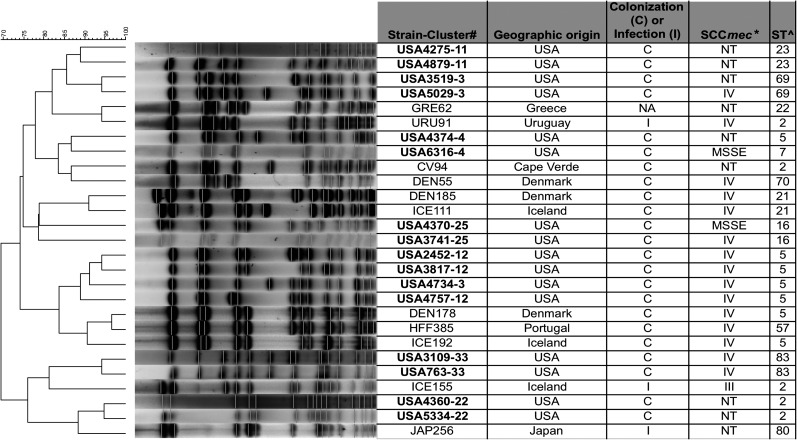
Figure 2.
Similarity of pulsed-field gel electrophoresis (PFGE) macrorestriction profiles of representative US Staphylococcus epidermidis isolates and isolates collected internationally. Strain names, geographical origin, relevance (infection [I] or colonization [C]), SCCmec type, and sequence types (STs) are shown. A dendrogram was performed to compare the PFGE profiles using Bionumerics software. e-BURST was used to assess clonal complexes (www.mlst.net). *mec complex/ccr type; ^All isolates belonged to clonal complex 5 (CC5). Abbreviation: NT: nontypable.
Dissemination of S. epidermidis Clonal Types
We investigated which clusters were identified in the greatest number of individuals, because this cluster analysis did not control for multiple identical specimens from the same person (Table 3). PFGE cluster types 3, 4, 11, 12, 22, 25, and 33 were the 7 most commonly shared clusters and were detected in 8–27 individuals at least once. These 7 cluster types accounted for 254 (48%) of the S. epidermidis isolates collected in the study. Of interest, these S. epidermidis clusters were not restricted to particular clinical centers. Five of these commonly shared cluster types were identified in all 4 clinical centers, whereas cluster types 3 and 4 were detected in 3 centers. Nine less disseminated clusters (identified in <8 individuals) were also identified in persons at multiple clinical centers: cluster 19 at 4 geographic locations and clusters 1, 5, 7, 20, 23, 47, 50, and 51 from 3 geographic locations. Isolates from 36 other clusters were identified in at least 2 of the clinical centers.
Table 3.
Dissemination of the 7 Most Common Pulsed-Field Gel Electrophoresis Cluster Types
| PFGE Cluster Type | Centers Where the Cluster Type Was Isolateda | No. of Persons in Whom the Cluster Type Was Isolated |
| 11 | 1, 3, 12, 13 | 27 |
| 12 | 1, 3, 12, 13 | 27 |
| 22 | 1, 3, 12, 13 | 24 |
| 33 | 1, 3, 12, 13 | 15 |
| 3 | 1, 3, 12 | 10 |
| 4 | 1, 12, 13 | 8 |
| 25 | 1, 3, 12, 13 | 8 |
Abbreviation: PFGE, pulsed-field gel electrophoresis.
Center 1, New York–Columbia Presbyterian Medical Center, New York; center 3, Aurora St Luke’s Medical Center, Wisconsin; center 12, Advocate Christ Medical Center, Illinois; and center 13, Sacred Heart Medical Center, Washington.
Colonization with S. epidermidis within individuals was complex. Participants could be colonized with different cluster types at the same time, and individual carriage with different cluster types was variable over time.
Each cluster type was examined in a repeated-measures analysis to determine whether particular clusters were associated with certain clinical centers or collection sites. Cluster type 22 was more likely to be isolated from the sternum than from the nares (OR, 1.65; P = .0093); however, there were no other statistically significant associations between cluster type and culture site. There were, however, certain clusters that were more predominant at certain clinical centers. PFGE cluster types 3 (OR, 9.04; P = .04), 4 (OR, 9.16; P = .05), and 11 (OR, 10.12; P = .003) were more likely to be isolated from center 12 than from center 1. Cluster type 11 was also more likely to be associated with center 13 than with center 1 (OR, 6.25; P = .04), whereas cluster 33 was associated with center 1, compared with center 3 (OR, 12.50; P = .02). Cluster types 12, 22, and 25 were not more likely to be isolated from one clinical center than from another.
The PFGE macrorestriction patterns of representative isolates of the most prevalent cluster types identified in our study were compared with the macrorestriction patterns of S. epidermidis strains from a large international database to determine whether they existed on a broader scale outside the United States. All of these isolates were closely related (similarity index, >79%) to isolates obtained from other countries in Europe (Denmark, Iceland, Portugal, and Greece), South America (Uruguay), Asia (Japan), and Africa (Cape Verde), although they could be different STs and carry different SCCmec types (see Figure 2).
Antimicrobial Susceptibility of S. epidermidis Isolates: Comparing Highly Disseminated With Less Disseminated Clonal Types
One hundred fifty isolates, including isolates of the 7 most disseminated cluster types (100 isolates) and less disseminated cluster types (50 isolates), were compared for their susceptibility to antimicrobials (Table 4). There were high rates of resistance to all of the antibiotics listed in Table 4, with the exception of tetracycline (14% resistant). Overall, 71% of the isolates were considered to be methicillin resistant because they were resistant to cefoxitin as assessed by disk diffusion. Isolates belonging to the 7 most prevalent cluster types were more likely to be methicillin resistant, compared with the less prevalent isolates (79% vs 54% MRSE; P = .0021). The most prevalent cluster types were also more likely to be resistant to clindamycin, erythromycin, and levofloxacin. There was increased resistance to trimethoprim-sulfamethoxazole with near significance. There were no statistically significant differences in resistance to penicillin, gentamicin, rifampin, and tetracycline. Two isolates were resistant to linezolid.
Table 4.
Antibiotic Susceptibility: Comparing Staphylococcus epidermidis Isolates From the 7 Most Commonly Shared Cluster Types to Less Disseminated Isolates
| Antibiotic | Most Disseminated Isolatesa (n = 100), % Resistantb | Less Disseminated Isolates (n = 50), % Resistant | P Value |
| Cefoxitinc | 79 | 54 | .0021 |
| Clindamycind | 74 | 44 | .0004 |
| Erythromycin | 86 | 58 | .0019 |
| Gentamicin | 46 | 36 | .3588 |
| Levofloxacin | 77 | 48 | .0010 |
| Penicillin | 98 | 92 | .1070 |
| Rifampin | 35 | 32 | .1326 |
| Tetracycline | 18 | 6 | .1043 |
| Trimethoprim-sulfamethoxazole | 73 | 56 | .0680 |
Most disseminated cluster types: 3, 4, 11, 12, 22, 25, and 33.
Isolates with intermediate resistance were considered resistant.
Cefoxitin resistance is the marker for methicillin resistance.
Isolates with inducible resistance were considered resistant.
S. epidermidis Isolates Collected From Infections
Six participants had confirmed coagulase-negative staphylococcal VAD infections. One infection solely involved the pump pocket, and the remaining 5 infections involved multiple sites, including the bloodstream. Sixteen clinical isolates from 5 of these participants were sent to our laboratory and confirmed to be S. epidermidis. Three (50%) of the infected participants were infected with 1 of the most prevalent clonal types in the study (1 with cluster 11, 1 with cluster 12, and another with a cluster 22 strain). The participants infected with cluster 11 and 12 strains were previously colonized with the same strain. We were unable to show that the participant infected with a cluster 22 strain was previously colonized with the same strain. Two other subjects were infected with less prevalent strains, and neither was previously colonized with the same strain.
DISCUSSION
Staphylococcus epidermidis has been regarded as a highly diverse species, although particular strains have been recognized as colonizing patients and causing infections in particular clinical settings. Several studies have identified similar strains in different wards in a hospital [9–15, 28], and few studies have identified clones in different hospitals in a country [17] or clones distributed across different countries [18, 19]. To our knowledge, this is the first observational study to examine S. epidermidis clonality in a cohort of patients at geographically diverse sites that included epidemiologic, microbiologic, and clinical data. This prospective, multicentered study of the epidemiology of VAD infection offered the opportunity to examine the diversity of S. epidermidis among hospitals in the United States during a defined period.
We observed that, similar to S. aureus, clones of multidrug-resistant S. epidermidis have disseminated across the United States. The 7 most prevalent clonal types accounted for almost half of the isolates that were obtained from 74 participants, 3 of which were identified in approximately one-third of the study participants. Although some of the strains were more likely to be found at certain clinical centers, the strains were distributed among 3–4 of the centers.
All of the most prevalent strains have also been identified in other countries, indicating the international dissemination of these S. epidermidis clones. These strains belong to the most successful S. epidermidis genetic lineage, CC5 (formerly CC2). The extent of dissemination observed may be explained, in part, by the presence of virulence factors, such as the arginine catabolic mobile element and biofilm, already described to be associated with this lineage [19, 29].
The geographic dissemination of particular MRSE strains was previously observed. A study in Sweden demonstrated that 3 MRSE clones spread throughout the wards of a county hospital. One of the strains had been previously identified at a referral hospital, indicating either possible transmission or wider dissemination of this strain [17]. In another comprehensive study, in which healthcare–associated S. epidermidis isolates from Denmark and Iceland were compared, 12 PFGE macrorestriction patterns were found to be common to both countries, some of which were also identified in isolates collected from other countries, such as Greece, Cape Verde, Mexico, and Uruguay, suggesting the international dissemination of different MRSE clones [18]. Other investigators used MLST to compare isolates from Germany with those from Ireland, Norway, and 5 areas in North America. With the caveat that MLST is less discriminatory than PFGE, 5 sequence types were identified as being present in >1 country [19].
Although MLST with SCCmec typing would be the most appropriate method to study global epidemiology [24], in this study, we aimed to study clonality in individuals at a variety of cardiac centers. Therefore, we chose PFGE as the primary typing method in our study because of its high discriminatory power. Figure 2 demonstrates that MLST and SCCmec typing generally supported the PFGE cluster assignments. There is some variability of PFGE patterns in a sequence type, which is common in S. epidermidis [7, 23]. This has been previously shown to result from the insertion and excision of mobile genetic elements at specific loci, such as SCCmec elements [30]. Furthermore, several isolates had a nontypeable SCCmec type, which is consistent with past studies that showed considerable variability in the SCCmec cassette in S. epidermidis [31–33].
The most prevalent strains in our study were also more antibiotic resistant than the other isolates. The VAD recipients in our study all had a history of hospitalization and were exposed to antibiotics (perioperatively, for VAD and other healthcare-associated infections, and in some cases, for prophylaxis after VAD implantation). It has been previously demonstrated that perioperative antibiotics administered to cardiac surgery patients is associated with increased resistance in colonizing coagulase-negative staphylococci after surgery [34, 35]. Therefore, antibiotic pressure may contribute to the emergence and persistence of these antibiotic-resistant clonal types.
This study is limited because it is unknown how and when these S. epidermidis clonal types have spread across the United States. Our data are restricted to cardiac patients; however, because of the location of the clinical sites (East Coast, Midwest, and West Coast) and the fact that all of the clonal types have been identified internationally, it seems to be unlikely that the type or transfer of patients or clinical personnel are entirely responsible for this observation. Nevertheless, healthcare workers and the physical environment in the cardiac centers may harbor these strains and have not been cultured. It would also be useful to know whether these strains are present in other units in these hospitals and whether a reservoir of these strains exists in the community.
This study demonstrates that clones of S. epidermidis cause infection and colonization both across the United States and internationally. The most prevalent clones are highly antibiotic resistant and accounted for almost half of the isolates obtained from 74 VAD recipients at 4 different cardiac centers. They caused at least 3 of the 6 S. epidermidis VAD infections. Antibiotic pressure may have helped select for these strains. However, whether these strains were around accidental pathogens [4] or have enhanced invasiveness, compared with the less prevalent strains, is unknown. We suggest that these strains have characteristics that better enable them to spread, persist, and/or cause infection.
Notes
Acknowledgments.
We thank Peter Vavagiakis for computer and technical support.
Financial support.
This work was supported by the National Institutes of Health (K08A1072043 to R. G. and HL077096 to A. D. W., F. D. L., J. C. G., M. S. S., P. P., Y. N., and A. J. T.), the European Commission (007/BI/2009 to J. R. and 222718 to H. de L.), and Fundação Calouste Gulbenkian (P-99911 to H. de L.).
Potential conflicts of interest.
M. S. S. receives research funding from Thoratec Corp. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–9. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 2.von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2:677–85. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 3.Gordon RJ, Quagliarello B, Lowy FD. Ventricular assist device-related infections. Lancet Infect Dis. 2006;6:426–37. doi: 10.1016/S1473-3099(06)70522-9. [DOI] [PubMed] [Google Scholar]
- 4.Otto M. Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diekema DJ, Pfaller MA, Schmitz FJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–32. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 6.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–52. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nouwen JL, van Belkum A, de Marie S, et al. Clonal expansion of Staphylococcus epidermidis strains causing Hickman catheter-related infections in a hemato-oncologic department. J Clin Microbiol. 1998;36:2696–702. doi: 10.1128/jcm.36.9.2696-2702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly S, Collins J, Maguire M, et al. An outbreak of colonization with linezolid-resistant Staphylococcus epidermidis in an intensive therapy unit. J Antimicrob Chemother. 2008;61:901–7. doi: 10.1093/jac/dkn043. [DOI] [PubMed] [Google Scholar]
- 10.Klingenberg C, Ronnestad A, Anderson AS, et al. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin Microbiol Infect. 2007;13:1100–11. doi: 10.1111/j.1469-0691.2007.01818.x. [DOI] [PubMed] [Google Scholar]
- 11.Villari P, Sarnataro C, Iacuzio L. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a three-year period. J Clin Microbiol. 2000;38:1740–6. doi: 10.1128/jcm.38.5.1740-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Silva GD, Justice A, Wilkinson AR, et al. Genetic population structure of coagulase-negative staphylococci associated with carriage and disease in preterm infants. Clin Infect Dis. 2001;33:1520–8. doi: 10.1086/323338. [DOI] [PubMed] [Google Scholar]
- 13.Burnie JP, Naderi-Nasab M, Loudon KW, Matthews RC. An epidemiological study of blood culture isolates of coagulase-negative staphylococci demonstrating hospital-acquired infection. J Clin Microbiol. 1997;35:1746–50. doi: 10.1128/jcm.35.7.1746-1750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muldrew KL, Tang YW, Li H, Stratton CW. Clonal dissemination of Staphylococcus epidermidis in an oncology ward. J Clin Microbiol. 2008;46:3391–6. doi: 10.1128/JCM.00115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liakopoulos V, Petinaki E, Efthimiadi G, et al. Clonal relatedness of methicillin-resistant coagulase-negative staphylococci in the haemodialysis unit of a single university centre in Greece. Nephrol Dial Transplant. 2008;23:2599–603. doi: 10.1093/ndt/gfn101. [DOI] [PubMed] [Google Scholar]
- 16.Jamaluddin TZ, Kuwahara-Arai K, Hisata K, et al. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol. 2008;46:3778–83. doi: 10.1128/JCM.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widerstrom M, Monsen T, Karlsson C, Wistrom J. Molecular epidemiology of meticillin-resistant coagulase-negative staphylococci in a Swedish county hospital: evidence of intra- and interhospital clonal spread. J Hosp Infect. 2006;64:177–83. doi: 10.1016/j.jhin.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Miragaia M, Couto I, Pereira SF, et al. Molecular characterization of methicillin-resistant Staphylococcus epidermidis clones: evidence of geographic dissemination. J Clin Microbiol. 2002;40:430–8. doi: 10.1128/JCM.40.2.430-438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–7. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke BJ, Lowy FD. Use of colony morphology to characterize carriage profiles of coagulase negative staphylococci. Eur J Clin Microbiol Infect Dis. 2007;26:895–9. doi: 10.1007/s10096-007-0387-0. [DOI] [PubMed] [Google Scholar]
- 21.Martineau F, Picard FJ, Ke D, et al. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. 2001;39:2541–7. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung M, de Lencastre H, Matthews P, et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–98. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 23.Miragaia M, Carrico JA, Thomas JC, Couto I, Enright MC, de Lencastre H. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J Clin Microbiol. 2008;46:118–29. doi: 10.1128/JCM.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol. 2007;45:616–9. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindler JF, Barton M, Callihan DR, et al. Performance standards for antimicrobial susceptibility testing. 19th Informational Supplement. Vol. 29. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. pp. M100–S19. [Google Scholar]
- 26.Okuma K, Iwakawa K, Turnidge JD, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–94. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SAS/STAT® 9.2 User’s Guide. SAS/STAT 9.2 User’s Guide to The GENMOD Procedure. http://support.sas.com/documentation/cdl/en/statuggenmod/61787/PDF/default/statuggenmod.pdf. Cary, NC: SAS Institute Inc, 2008. [Google Scholar]
- 28.Nouwen JL, Wielenga JJ, van Overhagen H, et al. Hickman catheter-related infections in neutropenic patients: insertion in the operating theater versus insertion in the radiology suite. J Clin Oncol. 1999;17:1304. doi: 10.1200/JCO.1999.17.4.1304. [DOI] [PubMed] [Google Scholar]
- 29.Miragaia M, de Lencastre H, Perdreau-Remington F, et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One. 2009;4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miragaia M, Couto I, de Lencastre H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE) Microb Drug Resist. 2005;11:83–93. doi: 10.1089/mdr.2005.11.83. [DOI] [PubMed] [Google Scholar]
- 31.Ruppe E, Barbier F, Mesli Y, et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother. 2009;53:442–9. doi: 10.1128/AAC.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanssen AM, Sollid JU. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob Agents Chemother. 2007;51:1671–7. doi: 10.1128/AAC.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbier F, Ruppe E, Hernandez D, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:270–81. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- 34.Archer GL, Armstrong BC. Alteration of staphylococcal flora in cardiac surgery patients receiving antibiotic prophylaxis. J Infect Dis. 1983;147:642–9. doi: 10.1093/infdis/147.4.642. [DOI] [PubMed] [Google Scholar]
- 35.Archer GL. Alteration of cutaneous staphylococcal flora as a consequence of antimicrobial prophylaxis. Rev Infect Dis. 1991;13(Suppl 10):S805–9. doi: 10.1093/clinids/13.supplement_10.s805. [DOI] [PubMed] [Google Scholar]