Abstract
Background
Female sexual dysfunction (FSD) is an important but controversial problem with serious negative impact on women’s quality of life. Data from twin studies have shown a genetic contribution to the development and maintenance of FSD.
Methodology/Principal Findings
We performed a genome-wide association study (GWAS) on 2.5 million single-nucleotide polymorphisms (SNPs) in 1,104 female twins (25–81 years of age) in a population-based register and phenotypic data on lifelong sexual functioning. Although none reached conventional genome-wide level of significance (10×-8), we found strongly suggestive associations with the phenotypic dimension of arousal (rs13202860, P = 1.2×10−7; rs1876525, P = 1.2×10−7; and rs13209281 P = 8.3×10−7) on chromosome 6, around 500kb upstream of the locus HTR1E (5-hydroxytryptamine receptor 1E) locus, related to the serotonin brain pathways. We could not replicate previously reported candidate SNPs associated with FSD in the DRD4, 5HT2A and IL-1B loci.
Conclusions/Significance
We report the first GWAS of FSD symptoms in humans. This has pointed to several “risk alleles” and the implication of the serotonin and GABA pathways. Ultimately, understanding key mechanisms via this research may lead to new FSD treatments and inform clinical practice and developments in psychiatric nosology.
Introduction
People vary in their enjoyment of sexual activity and relationships – a source of significant mental wellbeing or problems. Female sexual dysfunction (FSD) describes a cluster of sexual symptoms including desire, arousal, orgasm and pain. It appears relatively common in the general community and population-level samples and is associated with a severe decrease in quality of life in women [1]–[3]. The etiology of FSD is largely unknown although several biological and psychological correlates have been reported [3]–[5]. Nevertheless, no clear disease mechanisms have emerged and this lack of knowledge has hampered progress in both, psychiatric nosology and treatment strategies for this growing burden on women’s psychiatric health. Both, the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition and International Classification of Diseases, Tenth Revision [6] have arranged FSD into categories based largely on clinical similarities, while in 1998 a consensus based definition and classification system was designed by a panel of experts during the International Consensus Development [7].
Biological research into FSD is woefully inadequate. Recent twin studies suggest FSD is familial, with genetic factors accounting for up to 51% of the phenotypic variance [8]–[10]. Twin studies also show evidence of genetic and environmental contributions to psychological factors previously linked to FSD (such as depression, anxiety, personality traits) [11]–[13]. Thus, it is possible that some of the covariation between FSD and these psychological correlates is explained by shared genetic and non-genetic factors. However, there have been no large-scale studies to identify single genes or gene variants robustly associated with FSD-phenotypes (and no genome-wide association study - GWAS - has ever been performed). To date, only a handful of candidate gene studies of sexual desire and function exist. One candidate gene study has linked serotonin polymorphisms (5HT2A) to reduced sexual desire as a side-effect of SSRI-medication in 89 adult men and women [14]. A further study reported an association between the dopamine D4 receptor gene (DRD4) with self-reports of sexual desire and arousal in 52 men and 92 women [15]. Interleukin-1beta gene (IL-1B) has been correlated with variation in vulvar vestibulitis syndrome scores, a broader phenotype for sexual pain symptoms [16]. All these studies have methodological shortcomings that limit their interpretation, primarily the candidate gene design, small samples in mostly clinical populations (thus lacking power to detect phenotype – DNA variant associations), and the use of non-standardized instruments that lack coverage of the phenotypic heterogeneity in sexual function. Moreover, none of these studies examine women and FSD directly, making them unsuitable for clarifying the etiological mechanisms under FSD at the population-level.
We present here the results of the first GWAS of FSD in a female population-sample published to date. By scanning a dense set of genetic variants throughout the whole genome, we can test replication of previously located genes from candidate gene investigations and also identify novel genes that may lead to the discovery of unknown biological pathways involved in the development of FSD.
Materials and Methods
Participants
The TwinsUK adult twin registry based at St. Thomas’ Hospital in London is a volunteer cohort of over 10,000 twins from the general population [17]. This twin population has been involved in a wide range of studies on common traits and diseases and has been shown to be representative of the general population for a wide variety of medical, behavioral, and sexual traits [3], [18], [19]. The twins were not selected on the basis of the phenotypes being studied and were unaware of any hypothesis being tested. All twins provided written informed consent and the study was approved by St. Thomas’ Hospital Research Ethics Committee.
All participants were dizygotic (DZ) pairs and monozygotic (MZ) singleton twins of white European ancestry. A total of 1,489 subjects were included, all sexually active, heterosexual with no history of any major psychological or medical condition (depression, bipolar disorder, anxiety disorder, diabetes, multiple sclerosis, endometriosis) and with all items on the Female Sexual Function Index-Lifelong (FSFI-LL) available.
Sexual Dysfunction Phenotype
We used the recently developed 19-item Female FSFI-LL questionnaire to measure long-term variation in female sexuality, including periods of dysfunction and healthy function [20], [21]. For genetic analysis, the FSFI-LL is preferable to the “snapshot” measures used in some previous research and it better captures the variation in enduring female sexual functioning required for resolving the underlying genetic and non-genetic mechanisms of FSD symptoms. The FSFI-LL assesses 6 dimensions of women’s average sexual functioning “since they have been sexually active” including desire (2 items), arousal (4 items), lubrication (4 items), orgasm (3 items), satisfaction (3 items), and pain (3 items). Desire items are rated on a Likert-type scale ranging from 1 to 5. The remaining items are rated from 0 to 5 with the supplementary option “no sexual activity”. Dimension scores are derived by summing the item scores within each dimension and multiplying the sum by the dimension factor weight [20]. The dimension factor weighting converts the dimension scores to a consistent range from 0 to 6, except for the desire, which has a dimension score range from 1.2 to 6. Total scores are calculated via a simple computer algorithm and low scores on the FSFI-LL indicate more sexual problems and high scores indicate fewer problems. The FSFI-LL has excellent psychometric properties for both, total- and dimensions-specific scores, including test-retest reliability, internal consistency, external/discriminant validity [20], [21]. Exploratory and confirmatory factor analyses have successfully reproduced the original factor. According to response operator curve (ROC)-derived cut-off scores, all dimensions and the total FSD score displayed a good sensitivity to 1-specificity profile (as measured by the area under the curve = AUC), with arousal (AUC = 0.92) displaying the best trade-off and desire the lowest (AUC = 67.55%). Overall, the FSFI-LL demonstrates excellent comparability to the standard FSFI in terms of factor structure and psychometric properties [21].
Detailed information on prevalences, potential environmental risk factors and heritability estimates for FSD-symptoms in this panel are reported elsewhere (3).
Genotyping, Quality Control, and Imputation
Genotyping was carried out using two genotyping platforms from Illumina: the HumanHap 300k Duo for a part of the TwinsUK Cohort (n = 505) and the HumanHap610-Quad array for the remainder of the TwinsUK Cohort (n = 599). Genotyping with the HumanHap 300k Duo was conducted at the Centre National de Génotypage, Duke University, NC, USA; Helsinki University, Finland; and the Wellcome Trust Sanger Institute, Cambridge, UK. Genotyping with the Infinium 610k assay (Illumina, Inc., San Diego, USA) for the remaining individuals was conducted at the Centre for Inherited Diseases Research (USA) and the Wellcome Trust Sanger Institute.
We applied stringent quality control (QC) criteria to the genotype data. Genotypes were cleaned before analysis by removing single-nucleotide polymorphisms (SNPs) or individuals not fulfilling the QC criteria. The following QC filters were applied for samples: call rate at least 95%; autosomal heterozygosity between 33 and 37%. At the SNP level, Hardy Weinberg Equilibrium (HWE) with P-value >10−4, Minor Allele Frequency (MAF) at least 1%, and call rate at least 95% for SNPs with MAF 0.05 and above or at least 99% for SNPs with MAF less than 0.05. We further visually inspected all intensity cluster plots of SNPs that showed either an association for over-dispersion of the clusters, biased no calling, or erroneous genotype assignment and discarded all SNPs with any of these characteristics.
Genotypes from the TwinsUK were imputed using the genotypes from the 3,855,687 autosomal markers available from the HapMap Phase 2 CEPH population [22]. After imputation using IMPUTE2 a total of 2,558,978 non-monomorphic autosomal markers became available. After removing very rare markers (MAF<0.1) and markers in Hardy-Weinberg Disequilibrium (p<e−6) and individual with poor imputation scores (<0.5), we obtained results from 2,287,762 loci across the chromosome.
Statistical Analysis
Of the 1,489 women with recorded phenotype, genotype data was available for 1,104 subjects after QC check. Data handling and preliminary analyses were conducted using STATA software (StataCorp., College Station, TX) and Merlin (PMID 11731797) [23]. Association analyses were performed using MERLIN. Ancestry was determined through principal component analysis of individual genotypes (compared with subjects participating in the HapMap Phase II standard populations).
All traits were included in multiple regression models, with age and menopausal state included as covariates. Traits were inverse-normalized to avert undue effects of non-normality of their distributions. Regression slopes (β) are given as numbers of standard deviation units per each additional copy of the effect allele from this point onwards in the text. Given the experimental size with hundreds of thousands of SNPs being analysed individually, the commonly used “genome wide significance” (GWS) threshold was used which is the standard approximation routinely set at 5×10−8 [24]. However, given the cost of a strict Bonferroni adjustment in results from relatively small datasets, we considered all the associations with P≤5×10−5, as others have done in other studies of similar sizes (UK IBD Genetics Consortium, 2009; Amundadottir et al., 2009) [25], [26].
Results
Racial and ethnicity stratification was checked through eigenvector analysis and the above mentioned samples only contained individuals of certified and pure European ancestry. The genotyped samples were tested for population stratification, by comparison to the three HapMap phase 2 reference populations (CEU, YRI, CHB+JPT; www.hapmap.org) using principal component analysis.
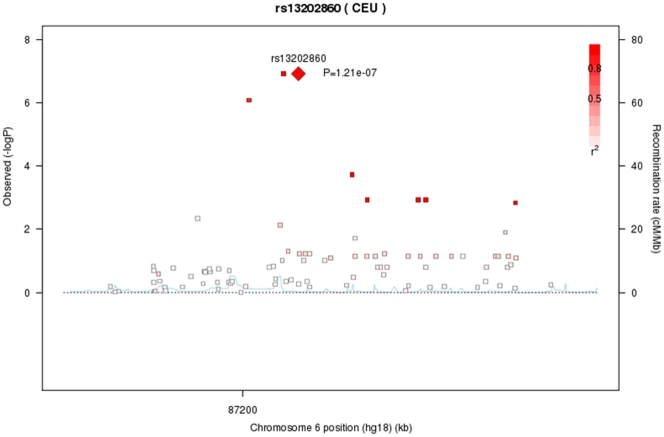
The mean age of participants in the study was 57 years (range 25–81 years). The GWAS analysis was performed using both observed and imputed genotypes. The genomic inflation factor (λ) ranged from 0.98 to 0.99 for the different phenotypes, showing no evidence for population stratification or inflated results due to imputation. We identified 34SNPs with P-values <10−5 of association with FSD-related measures. These results are summarized in Table 1. The most significant association was found between rs13202860 on chromosome 6 and sexual arousal, with P = 1.2×10−7. Two additional nearby SNPs showed P-values less than 4×10−7, (rs1876525, rs13209281; P = 1.2×10−7 and 8.3×10−7, respectively; Table 1 and Figure 1). All three SNPs were associated with arousal levels and spanned a region of 11 kb, around 500 kbp upstream from the HTR1E (5-hydroxytryptamine receptor 1E) locus (Figure 2 and Figure 3).
Table 1. Top SNPs associated with sexual function related measures in a cohort of females of European ancestry.
| Phenotype | SNP | CHR | Position | Locus | Allele | Effect * | s.e.m | P |
| Arousal | rs13202860 | 6 | 87211973 | A | −0.421 | 0.08 | 1.213E−07 | |
| Arousal | rs1876525 | 6 | 87208811 | C | −0.421 | 0.08 | 1.213E−07 | |
| Arousal | rs13209281 | 6 | 87201368 | A | −0.443 | 0.09 | 8.329E−07 | |
| Overall FSD | rs4820255 | 22 | 35533796 | PVALB | C | −0.366 | 0.076 | 1.687E−06 |
| Overall FSD | rs4821535 | 22 | 35533452 | PVALB | G | −0.366 | 0.076 | 1.687E−06 |
| Overall FSD | rs2284024 | 22 | 35528729 | PVALB | T | −0.366 | 0.077 | 1.838E−06 |
| Overall FSD | rs5750311 | 22 | 35533286 | PVALB | G | −0.367 | 0.077 | 2.104E−06 |
| Overall FSD | rs739031 | 22 | 35532649 | PVALB | T | −0.364 | 0.077 | 2.144E−06 |
| Overall FSD | rs4821536 | 22 | 35533947 | PVALB | T | −0.355 | 0.076 | 3.196E−06 |
| Lubrication | rs2370759 | 22 | 32674978 | EPC1 | G | 0.237 | 0.05 | 1.7E−06 |
| Lubrication | rs11594963 | 10 | 32665742 | EPC1 | G | 0.236 | 0.05 | 1.95E−06 |
| Lubrication | rs11599044 | 10 | 32655451 | EPC1 | A | 0.236 | 0.05 | 1.95E−06 |
| Lubrication | rs10508773 | 10 | 32615450 | EPC1 | C | 0.234 | 0.05 | 1.952E−06 |
| Lubrication | rs16933243 | 10 | 32655141 | EPC1 | T | 0.233 | 0.05 | 2.565E−06 |
| Lubrication | rs11008865 | 10 | 32614848 | EPC1 | C | 0.232 | 0.05 | 2.704E−06 |
Effect, β coefficient of linear regression. The effect sizes denote changes in phenotype unit per each additional copy of the reference allele.
Figure 1. Manhattan plots describing the association of 2.5 M SNPs with sexual arousal, lubrication and overall sexual functioning.
SNPs with P≤10–6 are highlighted with a red circle (n = 1104 females).
Figure 2. Association scatter plot for SNPs in the gene desert approximately 1Mbp upstream of the HTR1E gene.
TwinsUK discovery cohort. Negative logarithms of the P values for the association of each SNP with spherical equivalence are plotted. The lead SNP is plotted in diamond shape, with the GWAS-analysis P value for that SNP indicated. Genotyped SNPs are plotted as squares, with the colour indicating the degree of pairwise LD between the lead and neighbouring SNPs. Red indicates strong pairwise LD, with r2≥0.8; orange indicates moderate LD, with 0.5<r2<0.8 yellow indicates weak LD, with 0.2<r2<0.5; and white indicates no LD, with r2<0.2. The recombination rates are shown as light blue line.
Figure 3. Haploview LD plot. The plot uses the hapmapPhase3 data on the CEU and the TSI Caucasian populations and encompasses an 800 kbp segment containing the associated SNP on chromosome 6 and HTR1E.
The LD blocks were defined by confidence intervals according to Gabriel and colleagues [45]. The x-axis corresponds to the genomic position in kb and the red triangles defined by black lines represent LD blocks. The yellow arrow and the red box indicate the location of three GWAS associated SNP and the position of HTR1E, respectively.
We also identified a locus on chromosome 22 with multiple adjacent SNPs showing similar, albeit modest levels of associations with overall sexual function (Table 1; Figure 1). Association was maximal for rs4820255 and rs4821535 (both P = 1.2×10−6), two SNP located 344 bp apart within intron 3 of the parvalbumin (PVALB) gene. Similarly, six SNPs associated with lubrication levels could be detected (P<3×10−6 for all) on chromosome 10 near the EPC1 gene (Table 1; Figure 1).
Previous association studies have suggested several potential candidates to be associated with FSD. More specifically, earlier studies identified several variants on the dopamine D4 receptor (DRD4) and the serotonin 2A receptor gene (5HT2A) to be linked with levels of sexual desire and arousal (14,15). In this GWAS, observed and/or imputed genotype information was available for 2 SNPs in the DRD4, 5 SNPS in the 5HT2A and 3 SNPS in the IL-1B gene, and were hence evaluated for the replication of previously reported associations. However, none of the markers showed significant associations with the previously suggested phenotypes (or with any of the outcome variables), as displayed in Table 2.
Table 2. P-values of available markers in our GWAS, reported to be associated with specific sexual problems in previous candidate gene studies.
| Genes | SNP | Associated phenotypes | |
| P * | P * | ||
| DRD4 | Desire | Arousal | |
| rs3758653 | 0.7495 | 0.4242 | |
| rs11246226 | 0.2303 | 0.7294 | |
| HTR2A | Desire | Arousal | |
| rs2760345 | 0.6594 | 0.1266 | |
| rs7326071 | 0.795 | 0.3783 | |
| rs2770293 | 0.03697 | 0.9446 | |
| rs2760347 | 0.9225 | 0.1493 | |
| rs4941570 | 0.04381 | 0.4987 | |
| Il-B | Pain | ||
| rs1143643 | 0.6775 | ||
| rs1143634 | 0.7558 | ||
| rs1143633 | 0.6775 | ||
Bonferroni corrected p-value.
Discussion
Here we reported the results of the first ever GWAS of female sexual function levels in an unselected population-based cohort of British women. There were no associations at conventional genome-wide level of significance (P<5×10−8), but we found strongly suggestive associations. Several studies of similar size have considered any association with P-values ≤1×10−5 as being suggestive [25], [26]. Moreover, numerous suggestive associations below the genome-wide cut-off have been replicated in independent studies, strongly suggesting that these are indeed real associations rather than spurious results. For example, the Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 in a GWAS study for psoriasis, replicated many of the suggestive associations found by Nair and colleagues including SNP nearby IL-23A and TNFIP3 with P≤2×10−5 and P≤1×10−5, respectively [27], [28]. Here, we identified several strong suggestive associations with much stronger levels of significance. Our strongest association (P 1.2x10−7), was on the phenotypic dimension of arousal with a serotonin receptor gene (HTR1E) which represents a strong biological candidate previously shown to be involved in female sexuality. This potential susceptibility locus resides within a ∼1 Mbp segment of the genome devoid of annotated genes, located about 500 kbp downstream of the HTR1E gene. To assess the relationship between rs13202860 and HTR1E we plotted the LD pattern of the region. HapMap 3 data from two Caucasian populations (CEU and TSI) shows that rs13202860, rs1876525 and rs13209281, which show association with arousal in our study, are located in different LD blocks than HTR1E (Figure 3). Although HTR1E is an interesting candidate gene because of its known physiology, the large distance between both loci, together with the evidence that the significant SNP lie in other LD blocks than HTR1E clearly suggest that these three SNPs are tagging independent associations and that the causal polymorphism is more likely to regulate gene expression rather than the protein structure of HTR1E. Enhancers are elements of the genome that regulate gene expression of nearby or distant genes and which can be located within gene deserts [29]. Recent research suggests that polymorphisms in gene deserts could impact on disease by altering an enhancer element [29]–[31]. Thus, it is possible that our causal variant is altering an enhancer element located in the gene desert influencing HTR1E.
HTR1E encodes one of the families of highly conserved serotonin receptor genes and is strongly expressed in neurons, primarily in limbic brain regions (including caudate putamen, claustrum, hippocampus, and amygdala) [32]–[33]. This high degree of evolutionary conservation of genetic sequence suggests an important physiological role of the HTR1E receptor in humans. However, the actual function of the HTR1E receptor remains unknown. Nevertheless, HTR1E is a gene with considerable biological relevance to our phenotype of sexual functioning as it shares amino acid sequence homologies and some pharmacological characteristics with other 5-HT receptors (serotonin) and is therefore closely related. Comparative research has documented the critical role of serotonin receptors in modulating human and non-human mammalian sexual behavior and functioning acting on both central and peripheral (genital) sites [34]–[35]. SSRI (selective serotonin reuptake inhibitors)-associated sexual side effects, which include peripheral dysfunctions (e.g., erectile) as well as central problems in desire and arousal are well documented at high prevalence among users (up to 60%) [36]. These post-SSRI sexual dysfunctions (PSSD) point strongly to the involvement of serotonin receptors in human sexual behavior. Central serotonergic activity affects female sexual functioning via limbic projections of serotonin neurotransmitter are co-localized with norepinephrine receptors – and both transmitters seem to work in conjunction in the regulation of arousal and lubrication [35], [37]. However, the involvement of serotonin receptors in several reward-related behavioral functions (e.g. satiety, sexual behavior, nociception, escape, and stress) suggests that these receptors may function in the “higher-order” integration of rewarding behavior.
Our finding of a putative association between PVALB and overall sexual functioning scores on the FSFI-LL is entirely new. PVALB is a calcium-binding albumin protein present in GABAergic interneurons, expressed predominantly in the prefrontal cortex. Similar to serotonin, GABA is a major inhibitory neurotransmitter. Several lines of research demonstrate that GABA levels are associated with sexual function. Animal studies show that GABA(A) and GABA(B) receptors are involved in the inhibition of lordosis (a response shown by female animals indicating sexual receptivity) as well as mediating the effects of sex steroids such as estrogen in appetitive sexual behavior (e.g., sexual exploration) [37], [38]. Elevated levels of stress have also been shown to dampen sexual response in animal models as well as being a significant psychological correlate of FSD in women [39], [40]. The number of hippocampal PV-containing GABAergic interneurons is highly responsive to chronic external stressors, offering the potential of stress-induced neuro-structural alterations.
The EPC1 gene encodes the enhancer of polycomb homolog 1 and is a component of the NuA4 histone acetyltransferase complex. Previous research has suggested that this complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, and apoptosis. It has also shown to be involved in skeletal muscle differentiation [41], [42]. At this stage it is unclear through which mechanisms EPC1 could have an effect on vaginal lubrication and would need to be further investigated.
The present study had some methodological limitations and the findings should be interpreted with caution. Our study sample consisted mostly of peri- or postmenopausal women (70%). For this reason, representativeness of our study might be limited to the older female population, especially when considering that sexual dysfunction is more common in peri- and postmenopausal than in the non-climacteric period. However, prevalence rates of FSD in our sample are comparable to estimates found in other, younger populations [3]. Ideally, although similar populations are hard to come by, it would be important to replicate our GWAS findings in larger and independent samples before pursuing research into the underlying biological disease pathways. Our sample size may be one reason why our analysis did not reach conventional levels of GWA significance. Unfortunately, as is common in this field, there are no additional genotyped cohorts available with matching phenotypic data that could have been used to replicate our findings. Common diseases are typically influenced by multiple environmental as well as genetic factors. Our case and control participants may differ systematically for several of these environmental characteristics (e.g. education, anxiety levels, personality) which in turn could theoretically be related to genetic variation and to the disorder itself. Future studies in much larger sample sizes may be able to partition effects of known psychological predictors of FSD (such as sexual distress and anxiety levels) and if family-based differentiate genetic architecture of these co-morbid traits. Current approaches to perform GWAS are most successful if the common disease/common variant (CDCV) assumption holds [43]. Currently, exome sequencing has proven to be a powerful tool to identify rare coding variants and has the potential to overcome certain GWAS limitations by focusing on the identification of functional genomic structural variants rather than markers. Gene-environment interaction is also likely to have an influence on the development and maintenance of FSD [44]. In this regard, high throughput sequencing approaches would be again very useful as it can be used to interrogate functional aspects of the genome to identify epigenetic modifications such as DNA methylation, DNA-protein interaction, chromatin accessibility, etc.
In summary, we report the first GWAS of female sexual dysfunction symptoms in humans. This has pointed to several “risk alleles” and the implication of the serotonin and GABA pathways which we hope encourages further replication in large and independent population-based cohorts and then biological investigation to elucidate possible mechanisms. Ultimately, understanding key mechanisms via this research may lead to new FSD treatments and inform clinical practice and developments in psychiatric nosology.
Acknowledgments
We thank the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute led by Leena Peltonen and Panos Deloukas, for sample preparation, quality control, and genotyping; Le Centre National de Genotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors acknowledge financial support from the Wellcome Trust; the National Eye Institute via an National Institutes of Health/Center for Inherited Disease Research (NIH/CIDR) genotyping project; the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ National Health Service (NHS) Foundation Trust in partnership with King’s College London; and the Chronic Disease Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Laumann EO, Paik A, Rosen RC. Sexual Dysfunction in the United States. Prevalence and predictors. JAMA. 1999;281:537–44. doi: 10.1001/jama.281.6.537. [DOI] [PubMed] [Google Scholar]
- 2.Rosen RC, Taylor JF, Leiblum SR, Bachmann GA. Prevalence of sexual dysfunction in women: Results of a survey study of 329 women in an outpatient gynecological clinic. J Sex Marital Ther. 1993;19:171–88. doi: 10.1080/00926239308404902. [DOI] [PubMed] [Google Scholar]
- 3.Burri A, Spector T. Recent and lifelong sexual dysfunctions in a female UK population sample: prevalence and risk factors. J Sex Med. 2011;8:2420–2430. doi: 10.1111/j.1743-6109.2011.02341.x. [DOI] [PubMed] [Google Scholar]
- 4.Derogatis LR, Burnett AL. The epidemiology of sexual dysfunctions. J Sex Med. 2008;5:289–300. doi: 10.1111/j.1743-6109.2007.00668.x. [DOI] [PubMed] [Google Scholar]
- 5.Hawton DM, Gath D, Day A. Sexual Function in a Community Sample of Middle-Aged Women with Partners: Effects of Age, Marital, Socioeconomic, Psychiatric, Gynecological, and Menopausal Factors. Arch Sex Behav. 2000;23:375–395. doi: 10.1007/BF01541404. [DOI] [PubMed] [Google Scholar]
- 6.Basson R, Berman J, Burnett A, Derogatis L, Ferguson D, et al. Report of the international consensus development conference on female sexual dysfunction: Definitions and classifications. J Urol. 2000;163:888–893. [PubMed] [Google Scholar]
- 7.Basson R, Althof S, David S, Fugl-Meyer K, Goldstein I. Summary of the recommendations on sexual dysfunctions in women. J Sex Med. 2004;1:24–34. doi: 10.1111/j.1743-6109.2004.10105.x. [DOI] [PubMed] [Google Scholar]
- 8.Witting K, Santilla P, Rijsdijk F, Varjonen M, Stern P, et al. Correlated genetic and non-shared environmental influences account for the comorbidity between female sexual dysfunctions. Psychol Med. 2008;26:1–13. doi: 10.1017/S0033291708003206. [DOI] [PubMed] [Google Scholar]
- 9.Dawood K, Kirk KM, Bailey JM, Andrews PW, Martin NG. Genetic and environmental influences on the frequency of orgasm in women. Twin Res Hum Genet. 2008;8:27–33. doi: 10.1375/1832427053435427. [DOI] [PubMed] [Google Scholar]
- 10.Dunn KM, Cherkas LF, Spector TD. Genetic on variation in female orgasmic function: A twin study. Biol Lett. 2005;22:260–26. doi: 10.1098/rsbl.2005.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatr. 2001;158:1568–15. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatr. 2000;157:1552–15. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 13.Jang K, Wesley WJ, Vemon PA. Heritability of the Big Five Personality Dimensions and Their Facets: A Twin Study. J of Pers. 1996;64:577–591. doi: 10.1111/j.1467-6494.1996.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 14.Bishop JR, Moline J, Ellingrod VL, Schultz SK, Clayton AH. Serotonin 2A-1438 G/A and G-protein Beta3 subunit C825T polymorphismms in patients with depression and SSRI-associated sexual side-effects. Neuropsychopharmacol. 2006;31:2281–2288. doi: 10.1038/sj.npp.1301090. [DOI] [PubMed] [Google Scholar]
- 15.Ben Zion IZ, Tessler R, Cohen L, Lerer E, Raz Y, Bachner- Melman R, et al. Polymorphisms in the dopamine D4 receptor gene (DRD4) contribute to individual differences in human sexual behavior: desire, arousal and sexual function. Mol Psychiatry. 2006;11:782–786. doi: 10.1038/sj.mp.4001832. [DOI] [PubMed] [Google Scholar]
- 16.Gerber S, Bongiovanni AM, Ledger WJ, Witkin SS. Interleukin- 1beta gene polymorphism in women with vulvar vestibulitis syndrome. Eur J Obstet Gynecol Reprod Biol. 2003;107:74–7. doi: 10.1016/s0301-2115(02)00276-2. [DOI] [PubMed] [Google Scholar]
- 17.Spector T, Williams F. The UK Adult Twin Registry (TwinsUK). Twin Res Hum Genet. 2006;9:899–906. doi: 10.1375/183242706779462462. [DOI] [PubMed] [Google Scholar]
- 18.Andrews T, Hart DJ, Snieder H, de Lange M, Spector TD, et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics. Twin Res Hum Genet. 2001;4:464–77. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 19.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman’s reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83:1875–1880. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 20.Rosen R, Brown C, Heiman J, Leiblum S, Meston C, et al. The Female Sexual Function Index (FSFI): a multidimensional self-report instrument for the assessment of female sexual function. J Sex Mar Ther. 2000;26:191–208. doi: 10.1080/009262300278597. [DOI] [PubMed] [Google Scholar]
- 21.Burri A, Cherkas L, Spector T. Replication of psychometric properties of the FSFI and validation of a modified version (FSFI-LL) assessing lifelong sexual function in an unselected sample of females. J Sex Med. 2010;7:3929–39. doi: 10.1111/j.1743-6109.2010.01970.x. [DOI] [PubMed] [Google Scholar]
- 22.Marcini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nat Genet. 2009;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 23.Chen WM, Abecasis R. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nat Genet. 2009;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 25.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;49:986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UK IBD Genetics Consortium, Barrett JC, Lee JC, Lees CW, Prescott NJ, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genetic Analysis of Psoriasis Consortium, the Wellcome Trust Case Control Consortium 2, Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, et al. : Collaborative Association Study of Psoriasis. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–7. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barone P, Jordan D, Atger F, Kopp N, Fillion G. Quantitative autoradiography of 5-HT1D and 5-HT1E binding sites labelled by [3H]5-HT, in frontal cortex and the hippocampal region of the human brain. Brain Res. 1994;638:85–94. doi: 10.1016/0006-8993(94)90636-x. [DOI] [PubMed] [Google Scholar]
- 34.Shimron-Abarbanell D, Nothen MM, Erdmann J, Propping P. : Lack of genetically determined structural variants of the human serotonin-1E (5-HT1E) receptor protein points to its evolutionary conservation. Mol Brain Res. 1995;29:387–390. doi: 10.1016/0169-328x(95)00003-b. [DOI] [PubMed] [Google Scholar]
- 35.Frohlich PF, Meston CM. Evidence that serotonin affects female sexual functioning via peripheral mechanisms. Physiol Behav. 2000;71:383–93. doi: 10.1016/s0031-9384(00)00344-9. [DOI] [PubMed] [Google Scholar]
- 36.Zajecka J. Strategies for the treatment of antidepressant-related sexual dysfunction. J Clin Psychiatry. 2001;62:35–43. [PubMed] [Google Scholar]
- 37.Frazer A, Hensler JG. Serotonin. In: GJ InSiegel, BW Agranoff, RW Albers, FisherSK, editors. Uhler MD (eds.) Basic Neurochemistry. Lippincott-Raven: Philadelphia, PA; 1999. [Google Scholar]
- 38.Wada S, Yamada S, Yamanouchi K. Additive inhibition of lordosis by simultaneous treatments with GABA(A) and GABA(B) receptor agonists, muscimol and baclofen, in female rats. Pharmacol Biochem Res. 2008;90:590–3. doi: 10.1016/j.pbb.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Frye CA, Paris JJ. Effects of neurosteroid actions at N-methyl-D-aspartate and GABA A receptors in the midbrain ventral tegmental area for anxiety-like and mating behavior of female rats. Psychopharmacology. 2011;213:93–103. doi: 10.1007/s00213-010-2016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kogan MI, Kalinchenko SI, Avadieva NE. Sexual dysfunction in Russia: risk factors for women. Urologiia. 2009;5:8–12. [PubMed] [Google Scholar]
- 41.Ponholzer A, Roehlich M, Racz U, Temml C, Madersbacher S. Female sexual dysfunction in a healthy Austrian cohort: prevalence and risk factors. Eur Urol. 2005;47:366–74. doi: 10.1016/j.eururo.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Doyon Y, Selleck W, Lane WS. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JR, Kee HJ, Kim JY, Joung H, Nam KI, et al. Enhancer of polycomb1 acts on serum response factor to regulate skeletal muscle differentiation. J Biol Chem. 2009;284:16308–16. doi: 10.1074/jbc.M807725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends in Genetics. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]