Abstract
E26 transformation-specific sequence (ETS)-2 is a transcriptional modulator located on chromosome 21, alterations in its expression have been implicated with a reduced incidence of solid tumors in Down syndrome patients. MicroRNAs (miRNAs) are thought to participate in diverse biological functions; however, the regulation of miRNAs is not well characterized. Recently, we reported that miR-196b is highly expressed in gastric cancers. Herein, we demonstrate that miR-196b expression was significantly repressed by ETS2 during gastric cancer oncogenesis. We demonstrate that knockdown of endogenous ETS2 expression increases miR-196b expression. A genomic region between −751 and −824 bp upstream of the miR-196b transcriptional start site was found to be critical for the repression activity. This putative regulatory promoter region contains three potential ETS2-binding motifs. Mutations within the ETS2 binding sites blocked the repression activity of ETS2. Furthermore, knockdown of ETS2 or overexpression of miR-196b significantly induced migration and invasion in gastric cancer cells. In addition, alterations in ETS2 and miR-196b expression in gastric cancer cell lines affected the expression of epithelial–mesenchymal transition-related genes. The levels of vimentin, matrix metalloproteinase (MMP)-2 and MMP9 were drastically induced, but levels of E-cadherin were decreased in shETS2- or miR-196b-transfected cells. Our data indicate that ETS2 plays a key role in controlling the expression of miR-196b, and miR-196b may mediate the tumor suppressor effects of ETS2. We demonstrated that miR-196b was transcriptionally regulated by ETS2 and there was an inverse expression profile between miR-196b and ETS2 in clinical samples. This finding could be beneficial for the development of effective cancer diagnostic and alternative therapeutic strategies.
Introduction
Gastric cancer is a common cancer type and especially prevalent in the Andean region of South America and in the far east (1) and is the second leading cause of cancer-related deaths in the world (2). The development and progression of gastric cancer have been characterized by multiple genetic mutations and the dysregulation of both coding and non-coding genes including microRNAs (miRNAs) (3,4).
miRNAs are endogenous non-coding short RNAs of 21–23 nucleotides in length. MiRNAs were initially discovered in Caenorhabditis elegans and thousands have since been identified in many organisms, including humans, mammals, invertebrates, insects, plants and viruses (5). In humans, miRNAs play important roles in cellular physiology, development and disease by negatively regulating gene expression through translational repression or post-transcriptional degradation (6,7). MiRNAs regulate their target genes through the 3′untranslated region of the gene. Depending on the target gene, miRNAs act as tumor suppressor genes or have an oncogenic role in cancer formation. Tumor-suppressive miRNAs usually repress growth-promoting genes, and oncogenic miRNAs often target growth-stimulatory genes.
The miR-196s family of miRNAs is encoded at three paralogous loci in the mammalian HOX clusters and several HOX genes are regulated by miR-196s (8,9). The miR-196 family has been shown to be critical transcriptional regulators involved in embryo development (10–13), diseases (14–16) and tumorigenesis (17–20). Dysregulation of miR-196b has been reported in a variety of human cancers, such as colorectal cancer (21), leukemia (22–26), glioblastoma (27) and breast cancer (28), suggesting that miR-196b may be important in multiple tumor types. Our previous study also demonstrated that abnormal DNA hypomethylation induces the overexpression of miR-196b in human gastric cancers (29).
There is a significant amount of information on aberrantly expressed miRNAs and their tumorigenic effects in human cancers, but the detailed transcriptional regulations of these miRNAs remain poorly understood. Recent studies have suggested possible mechanisms, including epigenetic alterations and deregulated transcription processes. Most miRNAs are transcribed by RNA polymerase II. Therefore, similar to most protein-coding genes, the up or downregulation of miRNAs can be further controlled by transcription factors targeting their promoters. The mixed lineage leukemia gene directly upregulates miR-196b transcription in leukemia (22). Velu et al. (16) found that miR-196b is directly regulated by the transcription factor zinc finger protein growth factor independent-1 during myelopoiesis. In this study, we found that E26 transformation-specific sequence (ETS)-2 can regulate miR-196b expression in gastric cancers, and there is an inverse expression between ETS2 and miR-196b in clinical samples.
ETS2 is a member of a highly conserved transcription factor ETS family known to have a characteristic winged helix-turn-helix DNA-binding domain that interacts with a core GGAA/T consensus sequence found within promoter regions of target genes (30–32). ETS2 has been shown to play important roles in embryo development (33–35), senescence (36), vasculogenesis (34), immunity (37), osteogenesis (38) and tumorigenesis. The gene that encodes ETS2 is located on human chromosome 21q22.3 and has been reported to be overexpressed in the brain and fibroblasts of subjects with Down syndrome (DS) (39,40). Sussan et al. (41) demonstrated that enhanced ETS2 activity in a DS animal model induced significant inhibition of intestinal tumors formation. Therefore, ETS2 could have a tumor suppressor role. Whether the deregulated expression of ETS2 leads to a similar oncogenic activity in human gastric cancers is not known. In this study, we report that loss of ETS2 expression and increased miR-196b levels are observed in gastric cancer samples. We applied a bioinformatics approach to investigate the ETS2-binding potential to miR-196b promoter regions. Finally, we performed verification experiments using real-time quantitative PCR (RT–qPCR), a chromatin immunoprecipitation (ChIP) assay and a promoter reporter assay to demonstrate that downregulation of ETS2 could directly regulate miR-196b expression and affect migration and invasion activity in gastric cancer cells.
Materials and methods
Cell lines and human gastric tissues
The human gastric cancer cell lines, AGS, HR and the embryonic kidney cell, 293T were cultured as described previously (29). Paired samples of tumor and adjacent normal gastric tissues (63 pairs) were obtained from patients who had undergone curative gastric resection for primary gastric cancer at Taipei Veterans General Hospital, Taiwan. The study was approved by the Institutional Review Board, and informed consent was provided by all patients. Immediately after surgical resection, tissues were snap-frozen in liquid nitrogen and stored at −80°C until use.
Analysis of gene expression
Gene expression of ETS2 and epithelial–mesenchymal transition (EMT)-related genes in cell lines and tissues were detected with RT–qPCR using the SYBR Green I protocol (Bio-Rad, Hercules, CA). Expression analysis of miR-196b was performed by a quantitative stem-loop PCR, with U6 RNA as an internal control. All values were normalized against glyceraldehyde 3-phosphate dehydrogenase messenger RNA (mRNA) or U6 RNA. The primer sequences are listed in Supplementary Table 1, available at Carcinogenesis Online.
Plasmid constructs
The full-length human ETS2 (NM_005239) gene was subcloned into the pIRES-hrGFP-1a vector (Clontech, Mountain View, CA) and designated as Flag-ETS2. Luciferase reporter constructs containing the promoter regions of pre-miR-196b (nucleotides +36 to −996 with the ETS2 binding site GGA A/T at position −851 to −683 bp and −533 to −159 bp) were subcloned into the pGL3-basic vector (Promega, Madison, WI) and designated as different truncated wild-type pGL3-pre-miR-196b promoters: FL, N, C and P. ETS2 core-binding site mutations (GGA A/T) within the promoter regions of pre-miR-196b were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The pre-miR-196b promoter has three ETS2 binding sites at the core binding site (−851 to −683 bp) and the GGA A/T of the core binding site was mutated at the −824 to −821 site (M1), at the −767 to −764 sites (M2) or at the −755 to −752 sites (M3). The RT–qPCR primers used in the reporter and mutagenesis assay are listed in Supplementary Tables 2 and 3, available at Carcinogenesis Online.
A partial human pri-miR-196b gene (a 270 bp complementary DNA fragment encompassing 27 209 270 bp to 27 209 001 bp of chromosome 7) was subcloned into the lentiviral expression vector, pLKO.1-shLuc (National RNAi Core Facility, Academia Sinica, Taiwan) in place of shLuc and renamed pLKO.1-196b. The pLKO.1-196b was used to induce expression of miR-196b in cells.
RNA interference with shRNA and ectopic expression of miR-196b
Stable AGS cells were plated and infected with lentiviruses expressing shETS2, miR-196b or shLuciferase (shLuc) in the presence of 8 μg/ml polybrene for 24 h followed by puromycin selection (2 μg/ml; 48 h). RT–qPCR and/or western blotting were performed to validate the knockdown and the expression of mature miR-196b. The shRNA and miRNA constructs are described in the Supplementary Materials and Methods section, available at Carcinogenesis Online.
Antagomirs assays
The sequence complementary to miR-196b 5′-CCCAACAACAGGAAACUACCUA-3′ and the control sequence, 5′-GUGUAACACGUCUAUACGCCCA-3′ were synthesized with 2′-OMe modified bases, phosphothioate on the first two and last four bases, and a three cholesterol modification through a hydroxyprolinol linkage (Dharmacon, Lafayette, CO). The cells were incubated with 100 nM antagomir for 1 day to suppress miR-196b expression.
Antibodies, immunoblotting, immunostaining and staining for F-actin
Immunoblotting was performed as described previously (42). Protein lysates (30 μg) were electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA). The membranes were incubated with primary antibodies overnight at 4°C and then with the appropriate horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences, Piscataway, NJ).
The primary antibodies against Flag or actin were purchased from Sigma (St Louis, MO), and the antibody against ETS2 was obtained from Santa Cruz (Santa Cruz, CA). Protein signals were detected by an enhanced chemiluminescence kit (PerkinElmer, Waltham, MA). The levels of protein expression were normalized against actin.
Immunostaining was performed as described previously (42). The incubation with rhodamine–phalloidin (1 U/ml; Invitrogen, Carlsbad, CA) was performed at room temperature for 15 min followed by counterstaining with 4′,6-diamidino-2-phenylindole. The fluorescent images were captured using a Bio-Rad Radance 2100 confocal microscope (×60 oil immersion lens).
Cell migration/invasion assay
The cells were examined for migration and invasion abilities in vitro in a transwell assay The lower side or the upper side of polycarbonate membranes (8 μm pores) of a transwell (Costar, Lowell, MA) were coated with type I collagen (50 μg/ml) or Matrigel (80 μg/well) for the migration and invasion assays, respectively. The cells were added to the upper chamber of a transwell; after incubation for 16 h at 37°C, the cells on the lower side were stained with Giemsa stain. The level of migration or invasion was determined using a microscope at ×200 magnification. All experiments were repeated in triplicate.
Chromatin immunoprecipitation reactions
The ChIP assays were performed as described previously (42). The cells were fixed with 1% formaldehyde, harvested and lysed in sodium dodecyl sulfate buffer containing 50 mM Tris–HCl (pH 8.1), 0.5% sodium dodecyl sulfate, 100 mM NaCl, 5 mM ethylenediaminetetraacetic acid and protease inhibitors. The pellet was sonicated using 1 s pulses separated by 5 s for 4 min at output level 6 using a Sonicator XL2020 (Misonix, Farmingdale, NY). The sheared chromatin was precleared with protein A beads (30 μl; Amersham Biosciences), followed by incubation with 5 μg of anti-Flag mAb, anti-ETS2 or mouse IgG Ab (PerkinElmer). Untreated sonicated chromatin was used as input. Purified DNA was subjected to PCR reactions using the primers described in Supplementary Table 4, available at Carcinogenesis Online.
Promoter reporter assay
HEK293T cells (5 × 104 cells per well) were seeded in 24-well plate and cotransfected with 0.25 μg of either control vector or Flag-ETS2 construct in conjunction with 0.25 μg of either pLuc-FL (wild-type or mutant), the varied truncated forms and 0.05 μg of pRL-TK (Promega) using TurboFect reagent (Fermentas, MD). After 48 h, luciferase activity was measured using the Dual-Glo Luciferase assay System Kit (Promega).
Statistical analysis
All data are expressed as the mean ± SD; groups were compared using Student’s t-test. Categorical variables were compared using the Wilcoxon rank test. A P value <0.05 was considered statistically significant.
Results
Prediction of potential ETS2 binding sites in the miR-196b promoter
In previous studies, we investigated the expression and epigenetic regulation of miRNAs in human gastric cancers (29,43,44), including miR-196b, which has been implicated in many human cancers. Therefore, we investigated about the specific transcription control of this important miRNA gene in human cancer cells.
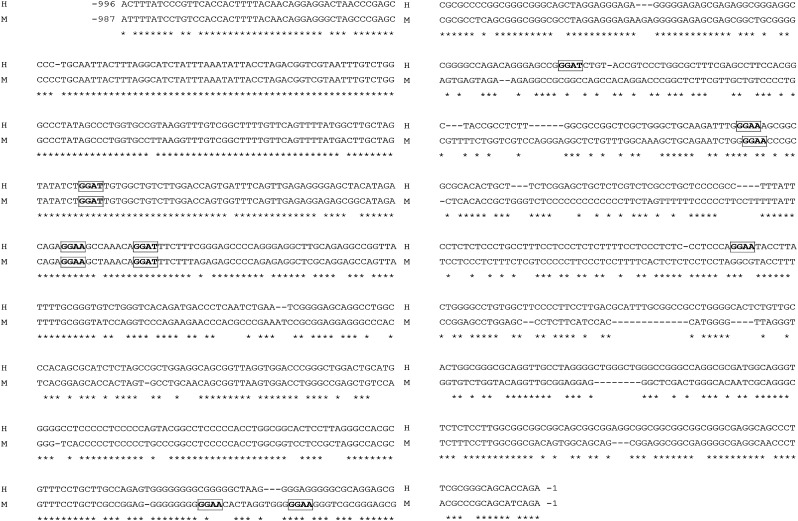
In preliminary examinations, bioinformatic tools (such as the TRANFAC public database) were utilized to interrogate the putative promoter region of miR-196b. Several transcription factor binding sites were implicated in the miR-196b promoter region, including ETS2. A consensus ETS2-binding motif, GGA A/T (30–32), was used to scan the upstream promoter region of miR-196b. In silico analyses revealed that six ETS2-binding motifs were located within upstream 1 kb region from the transcription start site of miR-196b, three (between −824 and −752) were located in the conserved non-coding block (CNB) and three additional motifs (between −399 and −205) were in non-conserved regions (Figure 1). It is known that the binding motifs of the evolutionarily conserved transcription factors tend to reside in CNBs within the promoter regions of their putative target genes (28,45).
Fig. 1.
ETS2 binding sites in miR-196b promoters. Human and mouse promoter regions of miR-196b contain six putative binding sites for ETS2 (GGA A/T), three of them located in a human–mouse conserved region (between −824 and −752). H, human; M, mouse.
Validation of the predicted ETS2-binding motifs using ChIP assays
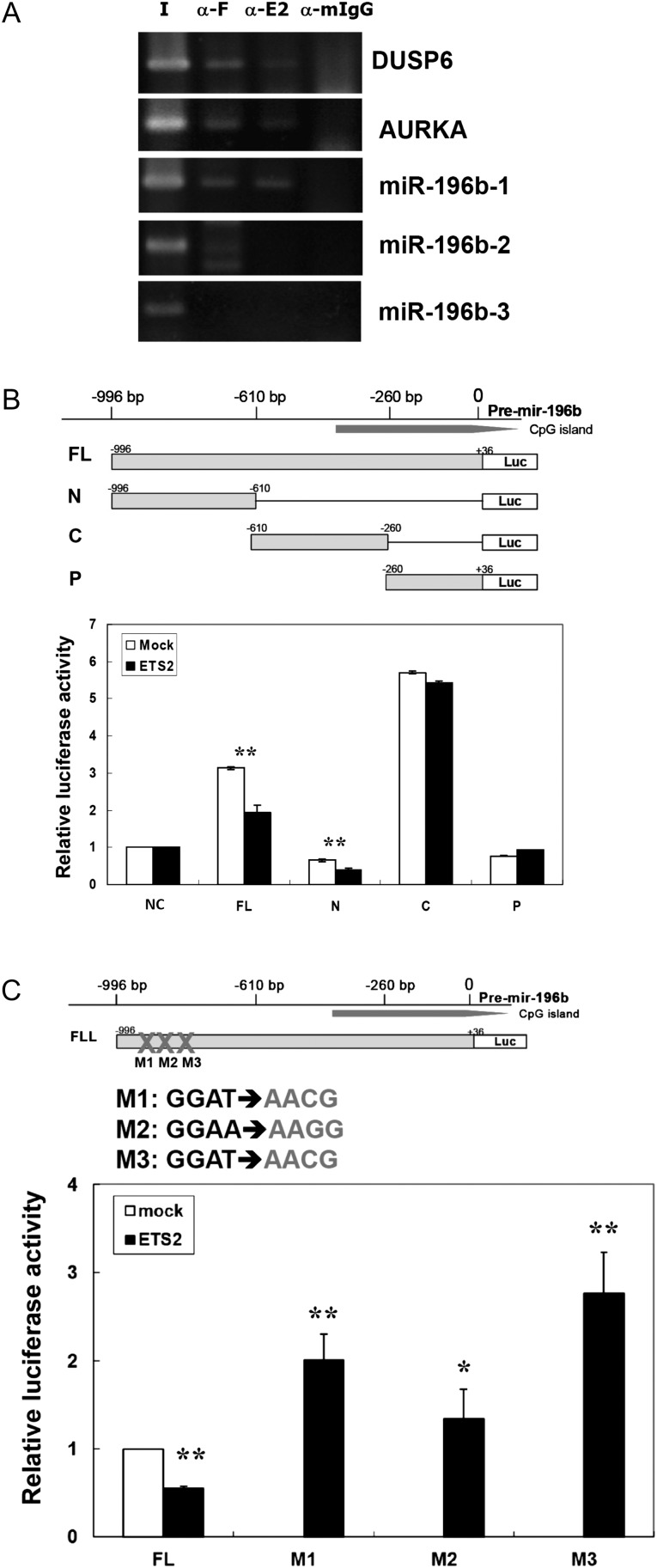
ChIP and reporter assays were performed to discover whether these motifs are functional binding sites for ETS2. ChIP is a powerful technique for analyzing transcription factor binding sites in living cells. We performed ChIP assays with AGS gastric cancer cells transfected with a Flag-ETS2 fusion protein construct using anti-ETS2 and monoclonal anti-Flag antibodies. In parallel, control ChIP assays experiments were performed using an anti-IgG antibody.
Briefly, we used formaldehyde to cross-link transcription factors to chromatin. The chromatin was sheared into small fragments and antibodies against Flag-ETS2 or control IgG were used to immunoprecipitate the nucleoprotein complexes. The DNA fragments were analyzed by PCR for three regions, including the three ETS2 binding sites in the CNBs (miR-196b-1: between −851 and −683 bp), the three ETS2 binding sites in the non-conserved regions (miR-196b-2: between −533 and −159 bp) and the lack of ETS2 binding sites in the nearby promoter region (miR-196b-3: between −154 and −3 bp). There are two target genes of ETS2 that have been previously empirically verified, DUSP6 and AURKA, which were used as our positive controls (46,47). As shown in Figure 2A, only the primers of miR-196b-1 produced strong PCR products, which suggested that ETS2 protein formed complexes with the predicted motifs between −851 and −683 bp of the miR-196b promoter. In addition, we have performed additional ChIP assays using the parental AGS cells and quantitative PCR to measure endogenous ETS2 binding to the miR-196b promoter region. As shown in Supplementary Figure 1, available at Carcinogenesis Online, the data were similar to the result in Figure 2A using the Flag-tagged ETS2 fusion protein. Only the miR-196b-1 region had the strong binding affinity. This result demonstrates that ETS2 protein specifically interacts with the miR-196b promoter region.
Fig. 2.
ETS2 directly regulates the expression of miR-196b. (A) ChIP assays with anti-Flag and anti-ETS2 antibodies showed binding of Flag-ETS2 to the promoters of miR-196b in AGS cells transfected with Flag-ETS2. Dual specificity phosphatase 6 (DUSP6) and aurora kinase A (AURKA) are confirmed ETS2 targets genes (46,47) and used as positive controls. The regions amplified by ChIP are: miR-196b-1: −851 to −683, miR-196b-2: −533 to −159 and miR-3: −154 to −3. I, input DNA; α-F, immunoprecipitation with anti-Flag antibody from cells expressing Flag-ETS2; α-E, immunoprecipitation with anti-ETS2 antibody from cells expressing Flag-ETS2; α-mIgG, immunoprecipitation with mouse IgG antibody was used as a negative control. (B) The promoter of miR-196b was repressed by ETS2. 293T cells were transiently cotransfected with Flag-ETS2 or mock vector and the wild-type miR-196b promoter (FL) or the various truncated miR-196b promoter constructs: N, C and P. These promoter sequences included in each were indicated by the numbers at the 5′ and 3′ termini. Normalized luciferase activity from triplicate samples is presented relative to that of cells transfected with the pGL3-basic construct. (C) The mutation of ETS2-binding motifs induced the promoter activities of miR-196b. 293T cells were transiently cotransfected with Flag-ETS2 or mock vector and wild-type miR-196b promoter (FL) or miR-196b promoter ETS2 binding site mutants (M1, M2 and M3). The miR-196b promoter constructs with the mutated core binding sequences were designated as M1, M2 or M3. Normalized luciferase activity from triplicate samples is presented relative to that of cells transfected with the FL construct. Luciferase activity was measured 48 h after transfection. The transfection efficiency was normalized against pRL-TK activity. The experiment was repeated twice with similar results. *P < 0.05, **P < 0.01.
Deletion analysis highlights a critical −996 to −610 bp fragment in the miR-196b promoter region
To evaluate if the negative regulation of miR-196b by ETS2 was a direct transcriptional modulation, we subcloned a −996 to +36 bp fragment of the promoter region of miR-196b into the luciferase expression pGL3-basic plasmid (Figure 2B). This construct was designated as FL construct and included the predicted ETS2 recognition sites. It was then transfected into 293T cells for a reporter assay. The expression levels of the FL construct were significantly increased compared with the pGL3-basic construct. These data indicated that the 1 kb upstream region of miR-196b gene contained a strong promoter region. Subsequently, when the FL reporter construct was cotransfected with an ETS2 expression vector into 293T cells, it induced a >40% reduction in luciferase activity compared with the mock control without ETS2 overexpression (Figure 2B).
To determine which portion of the miR-196b promoter region might be critical for the observed transcriptional regulation by ETS2, we created different truncated forms of the miR-196b promoter for ETS2 reporter assay experiments. These truncated form constructs included: N: −996 to −610 bp,; C: −610 to −260 bp and P: −260 to +36 bp. These constructs are designated from the miR-196b transcriptional start sites. The promoter structures and expression patterns are shown in Figure 2B. The N-fragment construct contains three ETS2-binding motifs in the evolutionarily CNB region and minimal expression of luciferase was conferred by the N-fragment construct which could be due to endogenous ETS2 activity. Intriguingly, the largest alteration in luciferase expression occurred when the region from −996 to −610 was deleted (the C construct). The P fragment did not affect expression activity compared with the control vector (Figure 2B). These data suggest that there is a critical region between −996 and −610 bp that is necessary for transrepression of the promoter constructs.
We next tested the ability of the exogenous ETS2 transcription factor to repress the miR-196b promoter. We cotransfected different miR-196b reporter constructs along with the ETS2 expression vector or an empty control vector into 293T cells. Overexpression of ETS2 led to a ∼50% decrease in the transactivation of the FL and N constructs, but the C and P constructs were not affected by ETS2 expression (Figure 2B, solid bars). Taken together with the ChIP data, these data indicated that the three ETS2-binding motifs in the CNB region (N: −996 to −610) are critical for the direct ETS2 repression of miR-196b promoter activity.
To clarify which ETS2 binding sites motifs are involved in the repression of miR-196b promoter activity, we generated an additional series of mutant constructs within the core binding site motif of ETS2. These constructs were as follows: M1 (−824 to −821, GGAT→AACG), M2 (−767 to −764, GGAA→AAGG) and M3 (−755 to −752, GGAT→AACG). In the presence of exogenous ETS2, the promoter activity of the wild-type FL construct was repressed by 50%, but mutations of the three ETS2-binding motifs resulted in significant enhancement in luciferase activity (1.5- to 3-fold; Figure 2C). These data imply that all three ETS2-binding motifs are involved in the repression of the miR-196b promoter region by ETS2.
In summary, these results indicate that ETS2 can directly repress miR-196b transcription activation through the three ETS2-binding motifs. This repression was strong within the CNB region. This finding provides evidence that miR-196b is indeed a target gene of ETS2.
An inverse correlation between ETS2 mRNA and miR-196b expression levels in gastric cancer tissues
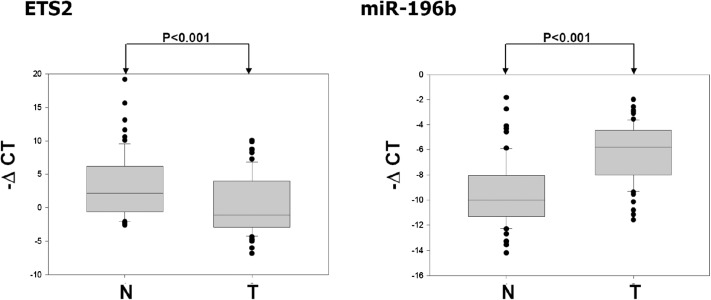
Previously, we showed that miR-196b is overexpressed in gastric cancers. Here, we investigated the expression of ETS2 and miR-196b in gastric cancers by RT–qPCR. Expression levels of ETS2 and miR-196b were analyzed in paired adjacent normal and gastric tumor tissues from 63 patients (Figure 3). An inverse correlation was observed between ETS2 mRNA and miR-196b level in the tumor specimens. ETS2 mRNA levels were significantly suppressed in the gastric tumors compared with the adjacent normal tissue (Figure 3, left panel, P < 0.001). In contrast, expression of miR-196b was remarkably higher in the tumor tissues (Figure 3, right panel, P < 0.001). Therefore, it is possible that in the course of gastric cancer progression, ETS2 was downregulated and miR-196b was subsequently upregulated.
Fig. 3.
Expression profiles of ETS2 and miR-196b in gastric tumors. Examination of ETS2 mRNA and miR-196b expression in 63 pairs of gastric cancer samples. The results were normalized against the expression level of glyceraldehyde 3-phosphate dehydrogenase mRNA or U6 RNA in each sample, respectively. The box plot shows the data distribution across the group classification and presents the 75th and 25th percentile (upper and lower quartile) with the median value in between. N indicates the average expression level of all 63 normal adjacent tissues; T indicates the average expression level of all 63 gastric tumor tissues. There was a significant difference between the level of ETS2 mRNA and miR-196b when tumors were compared with the adjacent tissue.
ETS2 regulates miR-196b expression in gastric cancer cell lines
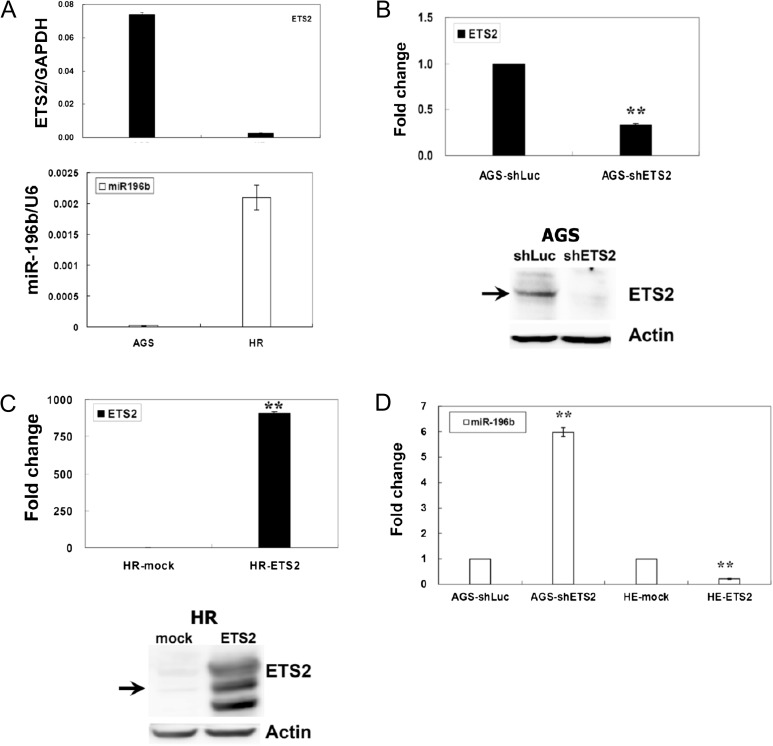
To determine the relevance of ETS2-miR-196b transcriptional regulation, the expression profiles of ETS2 and miR-196b were investigated in two gastric cancer cell lines by RT–qPCR. AGS and HR are the primary and metastatic gastric cancer cell lines, respectively (48,49). Primary gastric cancer cells (such as AGS) highly express ETS2, but metastatic gastric cancer cells (such as HR) poorly express ETS2 (Figure 4A). In contrast, AGS cells failed to show detectable levels of miR-196b, but HR cells do have high expression levels of miR-196b (Figure 4A). These data implied that ETS2 is a potential repressor of miR-196b and the dysregulation of ETS2 may be involved in gastric cancer progression.
Fig. 4.
Identification of ETS2 repressed miR-196b expression in gastric cell lines. (A) Expression profiles of ETS2 and miR-196b in gastric cancer cell lines. Expression levels of ETS2 mRNA and miR-196b in gastric cancer cell lines were detected using RT–qPCR by the SYBR Green I protocol. (B) Suppression of ETS2 in AGS cells was achieved with shETS2 lentiviral infection. A control shLuc lentivirus was used as a negative control. The effect of ETS2 knockdown was verified in the AGS–shETS2 cells by RT–qPCR and western blotting using an anti-ETS2 antibody. shETS2 knockdown reduced the level of endogenous ETS2 mRNA and protein by ≥60%. The expression of ETS2 and miR-196b were detected using RT–qPCR. shLuc was used as the negative control. (C) Overexpression of ETS2 in HR cells was achieved with Flag-ETS2 plasmids transfection. Overexpression was verified in the HR–ETS2 cells by RT–qPCR and western blotting using an anti-ETS2 antibody. HR cells transfected with Flag-ETS2 plasmids increased ETS2 expression in both mRNA and protein levels. Expression of ETS2 and miR-196b were detected using RT–qPCR. Mock transfections were used as negative controls. (D) After shETS2 knockdown of endogenous ETS2, expression of miR-196b in AGS cells was upregulated. After transfected with the Flag-ETS2 plasmid, the expression of miR-196b in HR cells was downregulated. In RT–qPCR, all values of ETS2 or miR-196b were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA or U6 RNA, respectively. For western blotting, total proteins (30 μg) were loaded and total actin was used as the internal control. The standard deviation is indicated. *P < 0.05, **P < 0.01.
To explore the ability of ETS2 to alter the expression of endogenous miR-196b in gastric cancer cells, the ETS2 shRNA vector was used to knockdown endogenous ETS2 expression in AGS cells. Transduction of shETS2 reduced ETS2 expression at both mRNA and protein levels in AGS cells (Figure 4B). In addition, we transfected HR cells with an ETS2 expression plasmid and found an increased expression of ETS2 mRNA and protein in HR cells (Figure 4C).
To further characterize the miR-196b gene as a direct ETS2 target, we analyzed the expression levels of miR-196b in AGS–shETS2 and HR–ETS2 cells (Figure 4D). The expression levels of the miR-196b gene were increased in the ETS2-knockdown AGS cells. In contrast, the expression levels of miR-196b were decreased in the ETS2-overexpressed HR cells.
Taken together, these data suggest that ETS2 suppressed miR-196b expression in gastric cancer cells.
Suppression of ETS2 or overexpression of miR-196b resulted in actin organization and increased invasion in AGS primary gastric cancer cells
Recently, miR-196b was found to be upregulated in human gastric tumors (29) and was considered a member of a metastasis-related miRNA family (28). However, the functional attributes of miR-196b associated with gastric tumor progression have not been experimentally demonstrated.
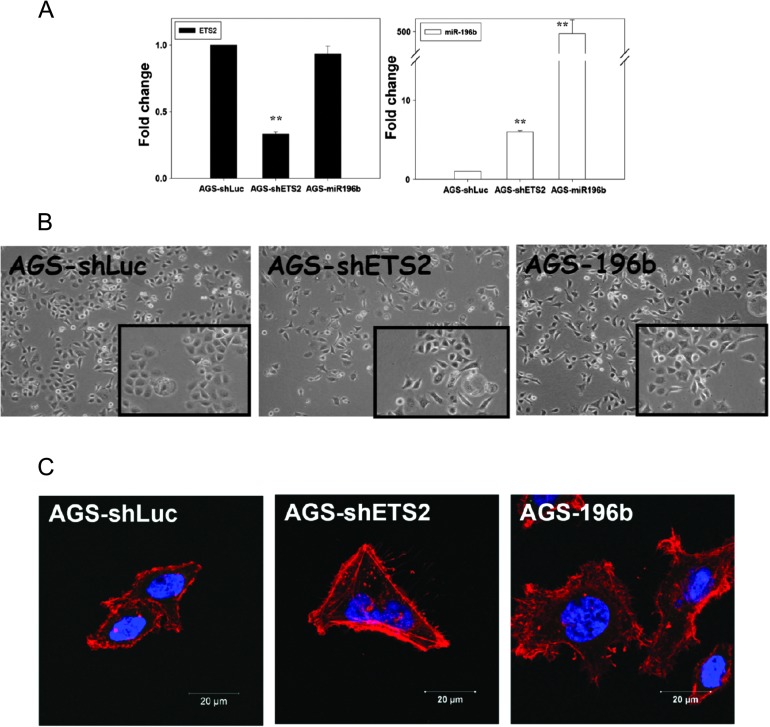
To explore the functional role of miR-196b in gastric cancers progression, ectopic expression of miR-196b was established using a lentiviral vector (lenti-196b; Figure 5A). Intriguingly, downregulation of ETS2 (using shETS2) and ectopic expression of miR-196b (using lenti-196b) significantly induced distinct morphological changes in the AGS primary gastric cancer cells; all AGS cells exhibited a transformed fibroblast-like cell arrangement (Figure 5B).
Fig. 5.
Knockdown of ETS2 or overexpression of miR-196b affected the morphological changes and actin organization of AGS cells. (A) Knockdown of ETS2 and overexpression of miR-196b in AGS cells were confirmed by reverse transcription–PCR. (B) The morphology of AGS cells after downregulation of ETS2 by shRNA or overexpression of miR-196b. Altered morphologies of AGS cells were revealed by phase contrast microscopy. Images were taken 5–7 days after infection. Original magnification: ×200. (C) Loss of actin organization in shETS2- and miR-196b-treated cells. Control shLuc-treated, shETS2- and miR-196b-treated AGS cells were fixed and stained with rhodamine–phalloidine and 4′,6-diamidino-2-phenylindole to detect F-actin and the nucleus, respectively. Bar, 20 μm.
The polymerization and depolymerization of F-actin are essential for cell motility. To assess whether the observed arrangements of the actin cytoskeleton could affect the migration of shETS2- or lenti-196b-treated AGS cells, we examined cellular actin structure with phalloidine staining. The F-actin positive membrane protrusions were significantly increased in addition to the F-actin organization enhancement in the shETS2- or lenti-196b-treated AGS cells (Figure 5C).
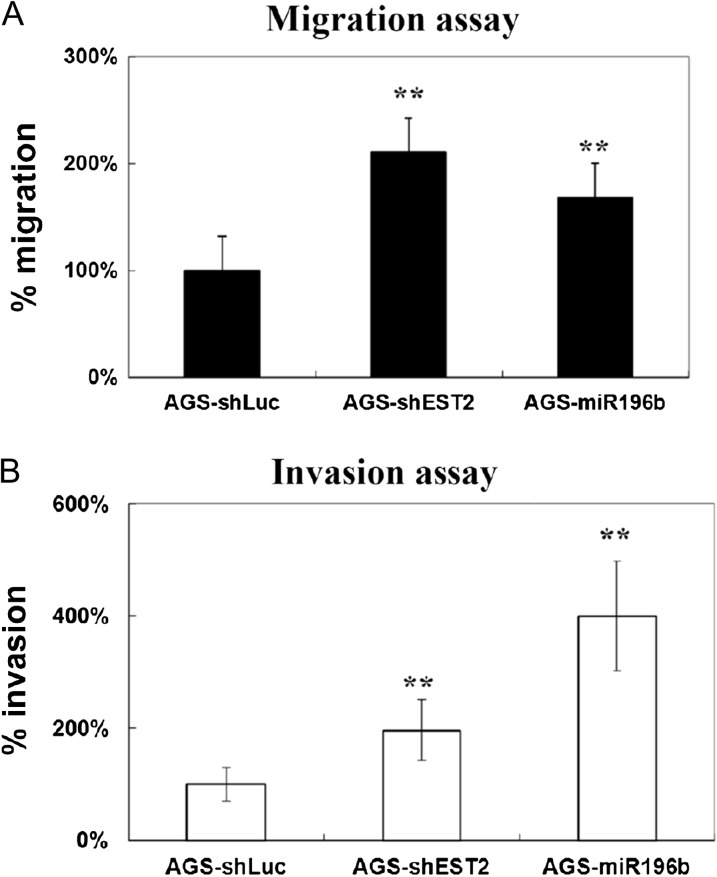
We examined the effects of silencing ETS2 or overexpressing miR-196b on cell migration and invasion. The knockdown of ETS2 drastically increased the migration and invasion ability of AGS cells (Figure 6). Furthermore, when miR-196b was overexpressed in AGS cells, a significant induction of cell migration and invasive activity was observed (Figure 6). In contrast, in the metastatic gastric cancer HR cells, overexpression of ETS2 or knockdown of miR-196b repressed cell migration and invasion (Supplementary Figure 2 is available at Carcinogenesis Online).
Fig. 6.
Knockdown of ETS2 or overexpression of miR-196b resulted in induction of migration and invasion. ETS2 knockdown and miR-196b overexpression resulted in a 30–50% induction in migration (A) and invasion (B) activity in AGS cells. AGS cells were infected with control shLuc, shETS2 or miR-196b for 16 h followed by a 2 day puromycin (2 μg/ml) selection before the cells were plated for the migration and invasion assay. The cell migration and invasion abilities were assessed as described in the Materials and methods. A total of 35 fields were counted for every filter. The data are an average of triplicates for each condition. *P < 0.05, **P < 0.01.
These results suggest that ETS2 affects cell migration partly via regulation of its downstream target genes, such as miR-196b. ETS2 may serve as an upstream participant in this particular miRNA regulation pathway for cytoskeleton migration.
Suppression of ETS2 expression and forced expression of miR-196b resulted in differential expression of EMT-related genes
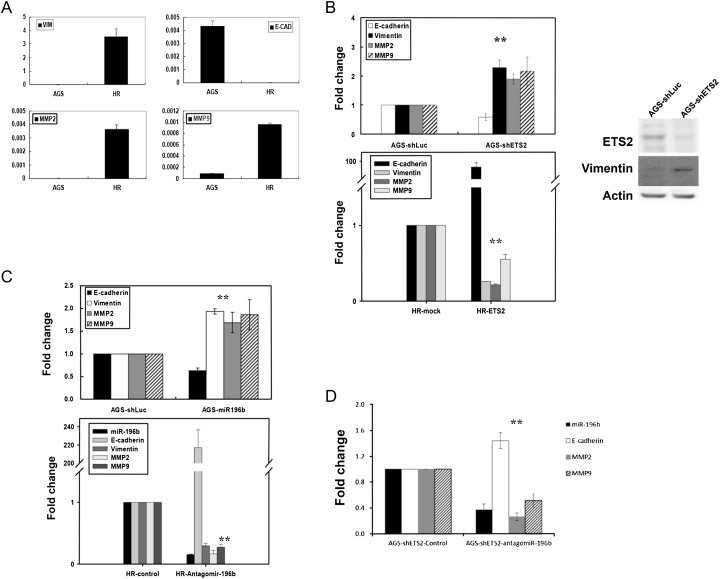
Due to the strong implication of this pathway in migration and invasion, we then investigated the possible involvement of ETS2 and miR-196b in the regulation of EMT-related genes. The process of EMT is a particular cellular program that converts polarized immotile epithelial cells into motile mesenchymal cells. During EMT, cells lose epithelial features through repression of E-cadherin expression and acquire mesenchymal characteristics such as increased vimentin expression. (50,51). The EMT processes enables cancer cells to increase metastatic behavior and promote malignant phenotypes (52). To explore the functional roles of ETS2 and miR-196b in gastric cancer metastasis, we investigated the expression profiles of EMT-related genes (E-cadherin, vimentin, matrix metalloproteinase (MMP)-2 and MMP9) with real-time reverse transcription–PCR. E-cadherin, a marker of the epithelial-type cells, was more highly expressed in parental AGS cells than in parental HR cells (Figure 7A). In contrast, the expression levels of vimentin, MMP2 and MMP9, markers for the mesenchymal-type cells, were elevated in parental HR cells (Figure 7A).
Fig. 7.
Knockdown of ETS2 or overexpression of miR-196b resulted in differential expression of EMT-related genes in gastric cancer cells. (A) HR cells expressed high levels of vimentin, MMP2 and MMP9, but AGS cells expressed high levels of E-cadherin. The expression patterns of genes were verified by RT–qPCR. (B) The gene expression patterns of E-cadherin, vimentin, MMP2 and MMP9 were detected by RT–qPCR in shETS2-treated AGS cells and ETS2-overexpressing HR cells. AGS cells expressed low levels of vimentin; increased expression of vimentin was detected after ETS2 knockdown in AGS cells (western blot). Antibodies to vimentin and actin (internal controls) were used. (C) The gene expression patterns of E-cadherin, vimentin, MMP2 and MMP9 were detected by RT–qPCR in miR-196b-treated AGS cells and antagomir-196b-treated HR cells. (D) The gene expression patterns of E-cadherin, MMP2 and MMP9 were detected by RT–qPCR in antagomir-196b-treated AGS–shETS2 cells. The expression levels of EMT-related genes were detected using RT–qPCR by the SYBR Green I protocol. All values were normalized against glyceraldehyde 3-phosphate dehydrogenase mRNA. shLuc and mock were used as negative controls. The data are an average of triplicates for each condition. *P < 0.05, **P < 0.01.
We then examined the effects of ETS2 expression on the levels of EMT-related gene. The levels of E-cadherin were drastically reduced in shETS2-treated AGS cells; however, the expression levels of vimentin, MMP2 and MMP9 were increased (Figure 7B, upper panel). In contrast, when ETS2 was overexpressed in HR cells, a significant reduction of vimentin, MMP2 and MMP9 and an increase in E-cadherin expression were observed (Figure 7B, lower panel).
Similar phenotypes were observed in miR-196b-treated cells. The levels of E-cadherin were significantly reduced in lenti-196b-treated AGS cells. In contrast, vimentin, MMP2 and MMP9 were upregulated (Figure 7C, upper panel). Antagomirs can efficiently and specifically silence endogenous miRs (53). Therefore, when endogenous miR-196b expression was suppressed by antagomir-196b in HR cells, the levels of vimentin, MMP2 and MMP9 were drastically reduced and E-cadherin was increased (Figure 7C, lower panel).
We further investigated whether the regulation of EMT-related genes by ETS2 was partly mediated through miR-196b. In AGS cells, the expression levels of miR-196b gene were increased by shETS2 treatment (Figure 4D). When we used the antagomir-196b to inhibit miR-196b expression in shETS2-treated AGS cells, the expression patterns of E-cadherin, MMP2 and MMP9 were reversed (Figure 7D). This supports our hypothesis and strongly suggests that a new ETS2-miR-196b regulatory cascade is operating during gastric cancer progression.
To explore the relevant roles between ETS2-miR-196b and EMT-related genes in clinical gastric cancer specimens, the expression profiles of ETS2, miR-196b, E-cadherin, vimentin, MMP2 and MMP9 in 59 gastric cancer sample pairs were further examined by real-time reverse transcription–PCR (Supplementary Figure 3 is available at Carcinogenesis Online). In most gastric cancer specimens, ETS2 and E-cadherin were downregulated. Expression of miR-196b was upregulated as expected, but the expression of vimentin, MMP2 and MMP9 were not significantly modulated in vivo. Unlike the in vitro two-dimensional cultured cells, these EMT-related genes might be modulated by many other signaling pathways in the tumor microenvironment. Nonetheless, our data strongly demonstrated the regulatory relationship between ETS2, miR-196b and E-cadherin in clinical gastric cancer tissues.
Discussion
MiRNAs have been shown to be a significant gene modulator during gastric cancer progression; therefore, global miRNA expression profiles have been performed using microarray, real-time PCR or next-generation sequencing approaches (3,4,54). There is a significant amount of information on aberrantly expressed miRNAs and their tumorigenic effects in gastric cancers, but the detailed control mechanisms of such miRNA dysregulation remain poorly understood. This is illustrated in our previous finding of a hypomethylated miR-196b promoter in cells (29).
In our previous study, miR-196b was dramatically overexpressed in gastric cancer tissue samples with a hypomethylated promoter. However, a hypomethylated miR-196b promoter was also observed in normal fibroblast cells that still expressed miR-196b at low levels. We then demonstrated that normal fibroblast cells did not restore the expression status of miR-196b following treatment with demethylation agents (29). This implies that the expression of miR-196b requires additional transcription regulatory factors. In this study, we demonstrated that ETS2 could be one of the key factors repressing the expression of miR-196b. These combined data indicate that the regulation of the expression of miR-196b required both cis elements (e.g. CpG island methylation) and trans-factors (e.g. ETS2) in gastric cancers.
The overexpression of ETS2 has been reported in a variety of human cancers (55–59), suggesting that ETS2 may be important in multiple tumors as an oncogene or a tumor repressor. Previous studies have revealed that certain tumor suppressor genes are regulated by ETS2. For example, ETS2 acts as a transcriptional repressor by downregulating the tumor suppressor gene BRCA1 in breast cancer cells (45).
The discovery of ETS2 downstream target genes could expand our knowledge regarding the ETS2 modulation of gastric cancer progression and would imply a range of novel ETS2 functions. Based on ChIP and reporter assay data, we demonstrate that ETS2 directly regulates miR-196b expression in gastric cancer cells. However, cumulative evidence suggests that ETS protein is not working alone but rather depends on requisite partners for target specificity and combinatorial control for gene regulation (60). In our study, ETS2 repressed expression of miR-196b but induced expression of AURKA and DUSP6. (46,47). A dichotomy of ETS2 transcriptional activity, whereby it acts as both an activator and a suppressor, was reported previously in various cell lines (60–62). Therefore, ETS2 has diverse activities across various human cell types; these activities are likely to be cell context-dependent involving different target genes (61,62).
DS patients present many clinical phenotypes including intellectual disability and an apparent decreased risk of certain cancers including gastrointestinal cancers (39,40,63). ETS2 is one of the key genes residing in the DS critical region on chromosome 21. Previous DS animal model studies demonstrated that ETS2 had a tumor suppression role in intestinal tumor oncogenesis (41). Our study is the first to report that ETS2 could serve as a transcriptional repressor to downregulate miR-196b expression for the inhibition of gastric cancer cell mobility. In addition to miR-196b, ETS2 has been reported to regulate the expression of miR-126 in endothelial cells and may mediate the angiogenesis process (34). Many miRNAs are dysregulated in gastric cancers (3,4,54). Whether ETS2 could modulate expression of other cancer-associated miRNAs in gastric cancers is unknown, but the implications of a novel transcription factor–miRNA regulatory circuitry are significant and deserve further elucidation.
Previous studies have demonstrated that ETS2 mediates cyclic adenosine 3′,5′-monophosphate-induced MMP2 expression activity during embryo development (33). In our study, we demonstrated that ETS2 regulated MMP2 and other EMT-related genes partially through miR-196b in vitro. We also observed a correlation between ETS2, miR-196b and E-cadherin expression alteration in clinical gastric cancer specimens. However, EMT is activated by many signaling pathways, such as the transforming growth factor β, Wnt and Notch pathways, in part through several intermediate critical transcription factors, including Snail1/2, bHLH, ZEB1/2 and Twist (52). In the in vivo three-dimensional microenvironment, EMT gene expression might be under additional regulatory signals in addition to ETS2-miR-196b. The investigation of downstream target genes is essential for understanding the true biological significance of miR-196b. MiR-196b cannot directly target the 3′untranslated region of vimentin, MMP2, MMP9 or E-cadherin as suggested by TargetScan predictions. Therefore, the miR-196b promotion of AGS cell migration may be mediated through other genes. More recently, miR-196b has been shown to regulate several metastasis-related genes, such as Annexin A1 and HOXB8, in various cancers (9,17,28). The detailed mechanism of the miR-196b promotion of gastric cancer migration and invasion is currently unclear and remains to be elucidated in future studies.
Our findings suggest that ETS2 and miR-196b play important roles in regulating the progression of gastric tumors. The potential of ETS2 and miR-196b as novel pathological staging markers and therapeutic targets for gastric cancer merits further investigation. The mechanisms by which miRNA participates in tumor promotion and progression are diverse and complicated. Gene profiling studies have demonstrated a number of significantly deregulated miRNAs and identified signatures of both diagnostic and prognostic value in gastric cancer. Our report provides new insights into the regulation of oncogenic miRNAs and their putative regulatory pathways.
Supplementary material
Supplementary Tables 1–4 and Figures 1–3 can be found at http://carcin.oxfordjournals.org/.
Funding
This work was supported in part by grants from the National Science Council (Taiwan NSC 100-2811-B-001-009, DOH100-TD-C-111-007), Center of Excellence for Cancer Research at Taipei Veterans General Hospital (DOH 99-TD-C-111-007) and Academia Sinica.
Supplementary Material
Acknowledgments
RNA interference reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- CNB
conserved non-coding block
- DS
Down syndrome
- EMT
epithelial–mesenchymal transition
- ETS
E26 transformation-specific sequence
- miRNA
microRNA
- mRNA
messenger RNA
- MMP
matrix metalloproteinase
- RT–qPCR
real-time quantitative PCR
References
- 1.Parkin DM, et al. Global cancer statistics. CA Cancer J. Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. , 1. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, et al. Aberrant DNA methylation and tumor suppressive activity of the EBF3 gene in gastric carcinoma. Int. J. Cancer. 2011;130((4)):817–826. doi: 10.1002/ijc.26038. [DOI] [PubMed] [Google Scholar]
- 3.Wu WK, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, et al. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett. 2010;297:137–143. doi: 10.1016/j.canlet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Liao Y-L, et al. In Lotfy,M. (ed) Gastric Carcinoma—Molecular Aspects and Current Advances. 2011. miRNAs in gastric cancer. ISBN: 978-953-307-412-2, InTech, Croatia. http://www.intechopen.com/articles/show/title/mirnas-in-gastric-cancer. [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 8.Yekta S. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 9.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki H, et al. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp. Ser., 2004 (48) 2004:211–212. doi: 10.1093/nass/48.1.211. [DOI] [PubMed] [Google Scholar]
- 11.Sehm T, et al. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev. Biol. 2009;334:468–480. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 12.McGlinn E, et al. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc. Natl Acad. Sci. USA. 2009;106:18610–18615. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asli NS, et al. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev. Biol. 2010;344:857–868. doi: 10.1016/j.ydbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 15.Guo X, et al. Down-regulation of VEZT gene expression in human gastric cancer involves promoter methylation and miR-43c. Biochem. Biophys. Res. Commun. 2011;404:622–627. doi: 10.1016/j.bbrc.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 16.Velu CS, et al. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luthra R, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol. Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou W, et al. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494–1504. doi: 10.1002/hep.23401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, et al. MicroRNA-196: critical roles and clinical applications in development and cancer. J. Cell. Mol. Med. 2011;15:14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YX, et al. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J. Dig. Dis. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 22.Popovic R, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schotte D, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1675–1682. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatia S, et al. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol. Cell. Biochem. 2010;340:97–106. doi: 10.1007/s11010-010-0406-9. [DOI] [PubMed] [Google Scholar]
- 25.Coskun E, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk. Res. 2011;35:208–213. doi: 10.1016/j.leukres.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Bhatia S, et al. Functional genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic leukemia. Mol. Cell. Biochem. 2011;346:103–116. doi: 10.1007/s11010-010-0597-0. [DOI] [PubMed] [Google Scholar]
- 27.Guan Y, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin. Cancer Res. 2010;16:4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70:7894–7904. doi: 10.1158/0008-5472.CAN-10-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai KW, et al. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 30.Yordy JS, et al. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- 31.Hsu T, et al. Ets proteins in biological control and cancer. J. Cell. Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei G-H, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staun-Ram E, et al. Ets-2 and p53 mediate cAMP-induced MMP-2 expression, activity and trophoblast invasion. Reprod. Biol. Endocrinol. 2009;7:135. doi: 10.1186/1477-7827-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris TA, et al. Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2010;30:1990–1997. doi: 10.1161/ATVBAHA.110.211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odiatis C, et al. New insights for Ets2 function in trophoblast using lentivirus-mediated gene knockdown in trophoblast stem cells. Placenta. 2010;31:630–640. doi: 10.1016/j.placenta.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Dwyer J, et al. Transcriptional regulation of telomerase activity: roles of the Ets transcription factor family. Ann. N. Y. Acad. Sci. 2007;1114:36–47. doi: 10.1196/annals.1396.022. [DOI] [PubMed] [Google Scholar]
- 37.Gallant S, et al. ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. (Warsz) 2006;54:149–163. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 38.Raouf A, et al. Ets transcription factors and targets in osteogenesis. Oncogene. 2000;19:6455–6463. doi: 10.1038/sj.onc.1204037. [DOI] [PubMed] [Google Scholar]
- 39.Pritchard M, et al. Down syndrome and the genes of human chromosome 21: current knowledge and future potentials. Cytogenet. Genome Res. 2008;121:67–77. doi: 10.1159/000124384. [DOI] [PubMed] [Google Scholar]
- 40.Patterson D. Molecular genetic analysis of Down syndrome. Hum. Genet. 2009;126:195–214. doi: 10.1007/s00439-009-0696-8. [DOI] [PubMed] [Google Scholar]
- 41.Sussan TE, et al. Trisomy represses ApcMin-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 42.Liao YL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 43.Tsai KW, et al. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 2009;4:587–592. doi: 10.4161/epi.4.8.10230. [DOI] [PubMed] [Google Scholar]
- 44.Tsai KW, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer. 2011;129((11)):2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 45.Baker KM, et al. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 2003;278:17876–17884. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- 46.Furukawa T, et al. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25:4831–4839. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 47.Furukawa T, et al. Feedback regulation of DUSP6 transcription responding to MAPK1 via ETS2 in human cells. Biochem. Biophys. Res. Commun. 2008;377:317–320. doi: 10.1016/j.bbrc.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Barranco SC, et al. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 1983;43:1703–1709. [PubMed] [Google Scholar]
- 49.Shimizu Y, et al. Characterization of human autotumor-reactive T-cell clones obtained from tumor-infiltrating lymphocytes in liver metastasis of gastric carcinoma. Cancer Res. 1991;51:6153–6162. [PubMed] [Google Scholar]
- 50.White NM, et al. Metastamirs: a stepping stone towards improved cancer management. Nat. Rev. Clin. Oncol. 2011;8:75–84. doi: 10.1038/nrclinonc.2010.173. [DOI] [PubMed] [Google Scholar]
- 51.Bracken CP, et al. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell. Mol. Life Sci. 2009;66:1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwatsuki M, et al. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 54.Kim YK, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dittmer J, et al. Ets transcription factors and human disease. Biochim. Biophys. Acta. 1998;1377:F1–F11. doi: 10.1016/s0304-419x(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 56.Stankiewicz MJ, et al. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge Y, et al. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia. 2007;22:521–529. doi: 10.1038/sj.leu.2405066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zabuawala T, et al. An Ets2-driven transcriptional program in tumor-associated macrophages promotes tumor metastasis. Cancer Res. 2010;70:1323–1333. doi: 10.1158/0008-5472.CAN-09-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munera J, et al. Ets2 regulates colonic stem cells and sensitivity to tumorigenesis. Stem Cells. 2011;29:430–439. doi: 10.1002/stem.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basuyaux JP, et al. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 1997;272:26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- 61.Trojanowska M. Ets factors and regulation of the extracellular matrix. Oncogene. 2000;19:6464–6471. doi: 10.1038/sj.onc.1204043. [DOI] [PubMed] [Google Scholar]
- 62.Mavrothalassitis G, et al. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- 63.Satge D, et al. Aspects of digestive tract tumors in Down syndrome: a literature review. Dig. Dis. Sci. 2006;51:2053–2061. doi: 10.1007/s10620-006-9131-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.