Abstract
Multiple breast cancer susceptibility loci have been identified in genome-wide association studies (GWAS) in populations of European and Asian ancestry using array chips optimized for populations of European ancestry. It is important to examine whether these loci are associated with breast cancer risk in women of African ancestry. We evaluated 25 single nucleotide polymorphisms (SNPs) at 19 loci in a pooled case–control study of breast cancer, which included 1509 cases and 1383 controls. Cases and controls were enrolled in Nigeria, Barbados and the USA; all women were of African ancestry. We found significant associations for three SNPs, which were in the same direction and of similar magnitude as those reported in previous fine-mapping studies in women of African ancestry. The allelic odds ratios were 1.24 [95% confidence interval (CI): 1.04–1.47; P = 0.018] for the rs2981578-G allele (10q26/FGFR2), 1.34 (95% CI: 1.10–1.63; P = 0.0035) for the rs9397435-G allele (6q25) and 1.12 (95% CI: 1.00–1.25; P = 0.04) for the rs3104793-C allele (16q12). Although a significant association was observed for an additional index SNP (rs3817198), it was in the opposite direction to prior GWAS studies. In conclusion, this study highlights the complexity of applying current GWAS findings across racial/ethnic groups, as none of GWAS-identified index SNPs could be replicated in women of African ancestry. Further fine-mapping studies in women of African ancestry will be needed to reveal additional and causal variants for breast cancer.
Introduction
While African American women have a 6% lower age-adjusted incidence rate of breast cancer than that of non-Hispanic White women in the USA, their mortality rate is 38% higher than the rate of non-Hispanic White women (1). In addition to their diagnoses at more advanced stages of disease, African American women are more likely to have young-onset breast cancer than their White counterparts (2). We have reported that 60% of the breast cancer cases were diagnosed <50 years in Nigeria, West Africa (3). The high proportion of young-onset breast cancer in women of African ancestry is probably due to a correspondingly high prevalence of pertinent genetic risk factors.
Multiple novel susceptibility loci for breast cancer have been identified in genome-wide association studies (GWAS) using several hundred thousand single nucleotide polymorphisms (SNPs) in commercial genotyping chips. These discoveries are poised to increase our understanding of breast cancer biology and also have the potential for cancer prediction in the clinical setting. However, these susceptibility loci were discovered primarily in women of White European ancestry and were validated in the same populations (4–13), with the exception being that a risk locus at chromosome 6q25.1 was identified in Chinese women (14). As GWAS are based on linkage disequilibrium (LD), which is quite different between Caucasians, Asians and Africans (15,16), the association of these novel SNPs with breast cancer risk will have to be replicated in other populations, including women of African ancestry. A few studies have evaluated these associations in African Americans, but only two recent studies have evaluated most published loci (17–22).
In the current study, we evaluated common genetic variants at 19 breast cancer susceptibility loci in a case–control study of women of African descent, which included 1509 breast cancer cases and 1383 controls. Specifically, we examined SNPs that showed the strongest statistical associations with breast cancer risk as reported in the initial GWAS (4–12,14). These variants are referred to as ‘index SNPs’ hereafter. In addition, we examined three SNPs that were revealed from fine-mapping studies conducted in women of African ancestry (23–25).
Materials and methods
Study subjects
We pooled samples from six epidemiologic studies of breast cancer among women of African ancestry, including 1509 cases and 1383 controls. All the studies have been approved in the corresponding institutional review boards of the participating institutions. Sample size and selected characteristics of these six study sites are presented in Table I. Below is a brief description of each study.
Table I.
Characteristics of breast cancer cases and controls
| Cases (n = 1509) | Controls (n = 1383) | P value | |
| Age in years | |||
| Mean ± SD | 48.0 ± 12.0 | 47.2 ± 17.2 | 0.08 |
| <50 years, No. (%) | 878 (58.4) | 799 (57.9) | 0.76 |
| Study site, No. | |||
| Nigeria | 681 | 282 | |
| Barbados | 93 | 244 | |
| Baltimore | 117 | 111 | |
| Pennsylvania | 151 | 272 | |
| Chicago | 268 | 261 | |
| Northern California | 199 | 213 | |
| Percentage of African ancestry, mean ± SD | |||
| Nigerian | 98.0 ± 1.2 | 98.1 ± 1.0 | 0.08 |
| Barbadian | 85.6 ± 10.4 | 85.7 ± 9.8 | 0.93 |
| African American | 77.6 ± 13.5 | 78.7 ± 12.0 | 0.10 |
| Baltimore | 80.7 ± 13.3 | 79.5 ± 12.2 | |
| Pennsylvania | 78.0 ± 13.4 | 79.3 ± 11.2 | |
| Chicago | 77.3 ± 14.1 | 79.6 ± 11.5 | |
| Northern California | 75.8 ± 12.5 | 76.3 ± 13.1 |
The Nigerian Breast Cancer Study.
The Nigerian Breast Cancer Study is an ongoing case–control study of breast cancer in Ibadan, Nigeria, initiated in 1998 (3,26). Breast cancer cases who were at least 20 years old were recruited at the University College Hospital, Ibadan, which is the oldest tertiary hospital in Nigerian with a catchment population of approximate 3 million. Controls were recruited from a randomly selected community adjoining the hospital. Names were then randomly selected from the community register and the individuals were invited to visit a clinic set up in the community for the study. The majority of the study subjects are Yoruban and Yoruban is one of the populations selected by the International HapMap Project to represent the African continent (16). Included in this study were 681 cases and 282 controls recruited between 1998 and 2009.
The Barbados National Cancer Study.
The Barbados National Cancer Study is a population-based case–control study designed to evaluate risk factors for incident breast and prostate cancer in the predominantly African population of Barbados, West Indies (27). Cases were identified through the only pathology department on the island, located at the Queen Elizabeth Hospital, and represented all histologically confirmed incident cases of breast cancers between July 2002 and March 2006. Controls were selected from a national database provided by the Barbados Statistical Services Department and were frequency matched to breast cancer cases at a 2:1 ratio and by 5 years age groups. Genotyping were conducted from 93 cases and 244 controls who have provided good quality of DNA.
The Northern California site of the Breast Cancer Family Registry.
The NC-BCFR is a population-based family study conducted in the Greater San Francisco Bay Area and one of six sites of the Breast Cancer Family Registry (28). African American breast cancer cases in NC-BCFR were diagnosed after 1 January 1995 and between the ages of 18 and 64 years. This study conducted genotyping for 199 invasive African American cases and 213 sister controls. The relatedness of cases and sibling controls requires special analysis (see below), but the family-based study design is immune from population admixture and thus can serve as a good validation set.
The Racial Variability in Genotypic Determinants of Breast Cancer Risk Study.
The Racial Variability in Genotypic Determinants of Breast Cancer Risk Study is a hospital-based genetic epidemiologic study conducted in Philadelphia and Detroit metropolitan areas from 1999 to 2003. Breast cancer cases were identified in the University of Pennsylvania Health System and Karmanos Cancer Institute. Local advertisement was also distributed to recruit breast cancer cases living in the Philadelphia and Detroit area. Controls were recruited in the same fashion as cases in these institutions except that they did not have breast cancer. Patients with invasive ductal breast cancer had to be recruited within 18 months of diagnosis. The study was designed to overrepresent women diagnosed <40 years. The Racial Variability in Genotypic Determinants of Breast Cancer Risk Study contributed 151 African American cases and 272 African American controls.
The Baltimore Breast Cancer Study.
The Baltimore Breast Cancer Study is a case–control study of breast cancer designed to identify and characterize markers of disease aggressiveness and poor outcome (29). Incident breast cancer cases and controls were recruited between February of 1993 and August of 2003 in six hospitals in the greater Baltimore area, including the University of Maryland Medical Center, the Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center and the Sinai Hospital. Controls were frequency-matched to cases by race and age. A total of 117 African Americans incident cases and 111 African Americans controls were included in this study.
The Chicago Cancer Prone Study.
The Chicago Cancer Prone Study is an ongoing hospital-based case–control study designed to investigate the genetics of young-onset breast cancer. Cases with histologically confirmed breast cancer were enrolled through the Cancer Risk Clinic at the University of Chicago. Young-onset cases and African Americans were oversampled. Controls were gender- and age-matched with cases and enrolled from patients who visited the same hospital and were willing to donate blood samples for genetic studies. The Chicago Cancer Prone Study contributed 268 cases and 261 controls to this study and most of the subjects were recruited between 1999 and 2008.
SNP selection and genotyping
The 22 SNPs that showed the strongest association with breast cancer in one or more GWAS were selected for genotyping, including SNPs on chromosomes 1p11, 2q35, 3p24, 5p12, 5q11, 6q22, 6q25, 8q24, 9p21, 10p15, 10q21, 10q22, 10q26, 11p15, 11q13, 14q24, 16q12, 17q23 and 19p13 (Table II) (4–12,14). In addition, three SNPs (rs9397435, rs2981578 and rs3104793) that were revealed from fine-mapping studies conducted in women of African ancestry were also selected (23–25). Detailed description of these 25 SNPs is presented in Supplementary Table 1, available at Carcinogenesis Online. To control for population stratification in African Americans and African Barbadians, we genotyped 30 ancestry-informative markers (AIMs). These 30 SNPs were selected from a set of 1373 AIMs with maximum allele frequency differences between European and African descendants, and these 30 AIMs gave ancestry estimates that were highly correlated (r = 0.89) with estimates using the entire set (30).
Table II.
Association of genetic loci from previous GWAS with breast cancer risk in women of African ancestry
| Locus (gene) | SNP | Allelesa (ref/risk) | Risk allele frequency |
Per-allele OR (95% CI)b | P for trend | Heterozygous OR (95% CI)b | Homozygous OR (95% CI)b | ||
| Case | Control | Two-sided | One-sided | ||||||
| 1p11.2 | rs11249433 | T/C | 0.102 | 0.101 | 1.07 (0.89–1.28) | 0.46 | 0.23 | 1.03 (0.84–1.26) | 1.41 (0.74–2.68) |
| 2q35 | rs13387042 | G/A | 0.768 | 0.749 | 0.99 (0.87–1.12) | 0.82 | 0.59 | 0.97 (0.69–1.36) | 0.96 (0.68–1.34) |
| 3p24 (SLC4A7) | rs4973768 | C/T | 0.355 | 0.350 | 1.06 (0.94–1.18) | 0.35 | 0.17 | 0.97 (0.82–1.14) | 1.20 (0.94–1.54) |
| 5p12 | rs10941679 | A/G | 0.178 | 0.189 | 0.94 (0.81–1.08) | 0.35 | 0.82 | 0.88 (0.74–1.04) | 1.07 (0.70–1.65) |
| rs4415084 | C/T | 0.649 | 0.655 | 0.95 (0.85–1.07) | 0.42 | 0.79 | 0.85 (0.66–1.09) | 0.87 (0.67–1.12) | |
| 5q11.2 (MAP3K1) | rs889312 | A/C | 0.321 | 0.340 | 0.93 (0.83–1.05) | 0.24 | 0.88 | 0.97 (0.82–1.14) | 0.84 (0.65–1.09) |
| 6q22 (RNF146) | rs2180341 | A/G | 0.341 | 0.316 | 1.09 (0.97–1.22) | 0.16 | 0.078 | 1.14 (0.97–1.35) | 1.12 (0.86–1.45) |
| 6q25.1 (ESR1/C6orf97) | rs2046210 | C/T | 0.646 | 0.627 | 1.02 (0.91–1.14) | 0.74 | 0.37 | 0.95 (0.75–1.21) | 1.01 (0.79–1.29) |
| rs9397435 | A/G | 0.093 | 0.074 | 1.34 (1.10–1.63) | 0.0035 | 0.0018 | 1.33 (1.07–1.65) | 1.83 (0.78–4.28) | |
| 8q24.21 | rs13281615 | A/G | 0.435 | 0.440 | 1.00 (0.89–1.11) | 0.95 | 0.52 | 1.08 (0.90–1.28) | 0.97 (0.78–1.21) |
| 9p21.3 (CDKN2BAS) | rs1011970 | G/T | 0.337 | 0.342 | 0.90 (0.80–1.01) | 0.076 | 0.96 | 0.90 (0.76–1.06) | 0.83 (0.64–1.06) |
| 10p15.1 | rs2380205 | T/C | 0.406 | 0.416 | 0.97 (0.86–1.09) | 0.60 | 0.70 | 1.00 (0.84–1.19) | 0.91 (0.73–1.15) |
| 10q21.2 (ZNF365) | rs10995190 | A/G | 0.808 | 0.832 | 0.88 (0.77–1.02) | 0.089 | 0.96 | 0.77 (0.48–1.23) | 0.70 (0.44–1.11) |
| 10q22.3 (ZMIZ1) | rs704010 | G/A | 0.061 | 0.076 | 1.04 (0.84–1.29) | 0.71 | 0.35 | 1.01 (0.80–1.28) | 1.39 (0.56–3.42) |
| 10q26 (FGFR2) | rs1219648 | A/G | 0.443 | 0.431 | 1.06 (0.95–1.19) | 0.26 | 0.13 | 1.06 (0.89–1.26) | 1.14 (0.91–1.42) |
| rs2981582 | C/T | 0.491 | 0.480 | 1.03 (0.92–1.15) | 0.59 | 0.29 | 0.99 (0.83–1.19) | 1.06 (0.86–1.32) | |
| rs2981578 | A/G | 0.905 | 0.871 | 1.24 (1.04–1.47) | 0.018 | 0.009 | 1.79 (0.95–3.37) | 2.08 (1.12–3.86) | |
| 11p15 (LSP1) | rs3817198 | T/C | 0.129 | 0.159 | 0.85 (0.73–0.99) | 0.040 | 0.98 | 0.84 (0.70–1.00) | 0.77 (0.44–1.34) |
| 11q13.2 | rs614367 | C/T | 0.139 | 0.128 | 1.09 (0.93–1.28) | 0.29 | 0.14 | 1.09 (0.91–1.31) | 1.21 (0.64–2.29) |
| 14q24.1 (RAD51B) | rs999737 | T/C | 0.974 | 0.967 | 0.93 (0.67–1.27) | 0.64 | 0.68 | 1.21 (0.16–8.96) | 1.11 (0.15–8.04) |
| 16q12 (TOX3) | rs3803662 | C/T | 0.510 | 0.516 | 0.96 (0.86–1.07) | 0.41 | 0.79 | 0.80 (0.66–0.97) | 0.91 (0.73–1.13) |
| rs3104793 | T/C | 0.625 | 0.585 | 1.12 (1.00–1.25) | 0.045 | 0.022 | 1.07 (0.86–1.33) | 1.23 (0.98–1.55) | |
| 17q23.2 (STXBP4) | rs6504950 | A/G | 0.635 | 0.654 | 0.90 (0.80–1.01) | 0.068 | 0.97 | 0.84 (0.66–1.07) | 0.79 (0.61–1.01) |
| 19p13 | rs2363956 | G/T | 0.515 | 0.501 | 1.08 (0.97–1.20) | 0.18 | 0.09 | 1.11 (0.92–1.35) | 1.16 (0.94–1.44) |
| rs8170 | C/T | 0.196 | 0.185 | 1.08 (0.94–1.24) | 0.27 | 0.13 | 1.05 (0.89–1.24) | 1.35 (0.86–2.14) | |
Bold represents ORs and P values of three SNP that were statistically significant and in the same direction as those reported in previous studies.
Reference/risk alleles in previous GWAS on the forward strand.
OR (95% CI) from logistic regressions adjusted for study site and African ancestry.
The SNPs in this study were genotyped using the Illumina GoldenGate platform (Illumina, San Diego, CA) as part of a larger panel of 1536 SNPs. Genotyping intensity and cluster data for all SNPs were reviewed individually. Of these, 122 (7.9%) SNPs failed as evidenced by low intensity or indistinguishable clustering. For successfully genotyped SNPs, the average call rate was 99.96%. Blind duplicates of 48 samples were included as between and within plate controls, dispersed in each 96-well plate. The reproducibility rate for duplicate samples was 99.95%. Four SNPs in this report (three index SNPs and one AIM) were not genotyped successfully. The remaining SNPs all had a call rate >99%. We imputed the three index SNPs using the software MACH v1.0 (31) with phased Yoruba in Ibadan, Nigeria and CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) data from HapMap Phased II (release 22) as the reference panel. The imputation quality was excellent because we included SNPs in LD with index SNPs as redundant markers (r2 = 1.00 for rs13387042, r2 = 0.98 for rs4415084 and r2 = 0.87 for rs2380205). Hardy–Weinberg equilibrium was assessed for each SNP using the stratified Hardy–Weinberg equilibrium test by Schaid et al. (32). One SNP (rs8170) was significant (P = 0.03) among the control samples while 1.25 was expected.
Statistical analysis
We estimated individual African ancestry from the 29 genotyped AIMs using the software Structure v.2.3.3 (33,34). We added Yoruba in Ibadan, Nigeria and CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) data from HapMap as benchmarks and our primary model assumed two subpopulations (African and European), although we also explored the possibility of three subpopulations. As the proportion of ancestry estimated in the third cluster was <0.03 in either Nigerians, African Americans or African Barbadians, and our study samples were best explained by the two subpopulation model, we only presented results under the two subpopulation model.
Case–control differences in age or African ancestry proportion were compared using t-tests. The association of each SNP with breast cancer risk was examined using unconditional logistic regression model adjusting for African ancestry proportion for each study site, except for the NC-BCFR site. Because of the relatedness between cases and controls in the NC-BCFR site, a conditional regression was used. Then, we used the Mantel–Haenszel method in meta-analysis to combine the estimated site-specific odds ratio (OR) and 95% confidence interval (CI). Alternatively, we fit unconditional logistic regression models adjusting for study site and African ancestry proportion. The two methods gave very similar results and thus, we presented the results from the second method to have a marginal or population average interpretation. Results from meta-analysis are presented in the Supplementary Material, available at Carcinogenesis Online. Both allele dosage effects (trend test) and genotypic effects were examined. The alleles associated with lower risk of breast cancer in previous studies were treated as the reference alleles. To emphasize the importance of the direction of association in genetic replication studies, we calculated one-sided P values in addition to two-sided P values. Note that a P value is two-sided unless otherwise specified. Furthermore, two composite risk models were built; one used the 19 risk variants reported in previous GWAS and the other used the 3 risk variants reported in fine-mapping studies in women of African ancestry. The composite risk score was calculated as the count of risk alleles and only one SNP per genetic locus was chosen. For individuals with missing data, the score was calculated as the average risk allele count multiplied by the number of total SNPs. As missing data were infrequent (<0.2%), missing data had no material impact on the composite score. Both continuous and categorical risk scores (grouped by quartiles) were examined in relation to breast cancer risk using logistic regression, adjusted for study site and genetic ancestry estimate. The statistical analysis was conducted using SAS 9.2 package (SAS Institute, Cary, NC). Additionally, we calculated LD measures (r2 and D′) in our study samples and the selected HapMap populations using Haploview v4.2 (35). A P value of <0.05 was considered statistically significant.
Results
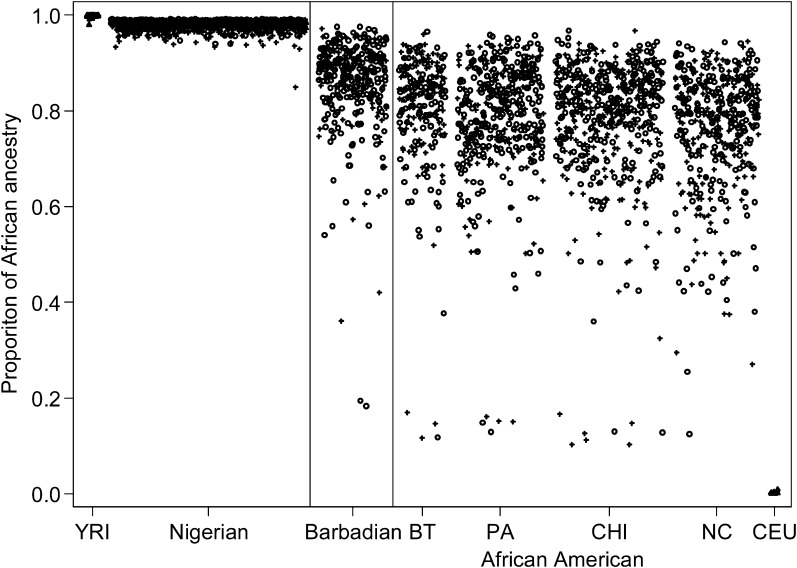
The mean age was 48.0 years in cases and 47.2 years in controls (Table I). The majority of cases and controls were <50 years. Figure 1 depicts the distribution of estimate of African ancestry proportion across the three populations. The mean (median) proportion of African ancestry was 0.857 (0.882) in Barbadians, 0.782 (0.808) in African Americans and close to 1 in Nigerians. There admixture estimates are in line with previous studies (36,37). There was no significant difference in the proportions of European admixture between cases and controls.
Fig. 1.
Distribution of estimate of African ancestry proportion by study populations and case/control status. African Americans were recruited at four sites (BT, Baltimore; PA, Pennsylvania; CHI, Chicago; NC, Northern California). Cases were presented in plus symbols and controls were presented in open circles. Two HapMap populations were also plotted as benchmarks (Yoruba in Ibadan, Nigeria, Yoruba in Ibadan, Nigeria; CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain collection).
We found three SNPs that were significantly associated with breast cancer risk (Table II). The allelic odds ratio was 1.24 (95% CI: 1.04–1.47; P = 0.018, one-side P = 0.009) for the rs2981578-G allele (10q26/FGFR2), 1.34 (95% CI: 1.10–1.63; P = 0.0035, one-sided P = 0.0018) for the rs9397435-G allele (6q25) and 1.12 (95% CI: 1.00–1.25; P = 0.045, one-sided P = 0.022) for the rs3104793-C allele (16q12). All the three SNPs were identified from previous fine-mapping studies in women of African ancestry and the associations were in the same direction and of similar magnitude as those originally reported. For each of the three SNPs, there was no statistically significant heterogeneity of effects across study sites (Table III). Positive associations existed in at least four of six study sites for all associated SNPs although there were minor differences in risk allele frequency across study sites. There was no significant heterogeneity of effects across studies for other SNPs (Supplementary Table 2 is available at Carcinogenesis Online).
Table III.
Association of three SNPs and breast cancer risk across studies
| SNP (ref/risk allele) | Study | Risk allele frequency |
Per-allele OR (95% CI)a | |
| Case | Control | |||
| rs2981578 (A/G) | Nigeria | 0.951 | 0.935 | 1.35 (0.88–2.05) |
| Barbados | 0.919 | 0.920 | 1.00 (0.53–1.87) | |
| Baltimore | 0.893 | 0.832 | 1.62 (0.93–2.83) | |
| Pennsylvania | 0.820 | 0.843 | 0.88 (0.60–1.28) | |
| Chicago | 0.872 | 0.837 | 1.44 (1.01–2.05) | |
| Northern California | 0.854 | 0.828 | 1.24 (0.85–1.82) | |
| P for heterogeneity | 0.34 | |||
| rs3104793 (T/C) | Nigeria | 0.664 | 0.622 | 1.20 (0.98–1.47) |
| Barbados | 0.591 | 0.602 | 0.95 (0.67–1.35) | |
| Baltimore | 0.625 | 0.595 | 1.13 (0.77–1.65) | |
| Pennsylvania | 0.573 | 0.568 | 1.03 (0.78–1.38) | |
| Chicago | 0.584 | 0.554 | 1.16 (0.91–1.48) | |
| Northern California | 0.601 | 0.568 | 1.14 (0.87–1.51) | |
| P for heterogeneity | 0.89 | |||
| rs9397435 (A/G) | Nigeria | 0.088 | 0.067 | 1.34 (0.92–1.96) |
| Barbados | 0.086 | 0.086 | 1.00 (0.55–1.83) | |
| Baltimore | 0.085 | 0.059 | 1.47 (0.71–3.04) | |
| Pennsylvania | 0.099 | 0.081 | 1.27 (0.78–2.06) | |
| Chicago | 0.091 | 0.073 | 1.29 (0.83–2.01) | |
| Northern California | 0.118 | 0.073 | 1.71 (1.06–2.74) | |
| P for heterogeneity | 0.48 | |||
OR (95% CI) from logistic regressions adjusted for African ancestry.
Four index SNPs (rs3817198 at 11p15, rs1011970 at 9p21, rs10995190 at 10q21 and rs6504950 at 17q23) were significant or marginally significant, but the associations reported here were in the opposite direction to the previous studies (9,10), as indicated by the one-sided P values, suggesting that these markers are not consistent with breast cancer risk in women of African ancestry. Although the allele frequencies of the four SNPs were not similar between African and European descents, there was no switch between minor and major alleles, suggesting that changes in association direction were not due to difference in minor alleles.
Table IV presents the LD measures between the significant SNPs and the index SNPs at the same locus. The index SNP at 6q25 (rs2046210) was strongly linked with the fine-mapping SNP (rs9397435) in the Chinese population, in which the locus was initially discovered. However, these two SNPs were only weakly linked in the present study samples of African descent. At the 10q26 locus, the two index SNPs (rs1219648 and rs2981582) were in tight LD with the fine-mapping SNP (rs2981578) in the European population, in which the locus was discovered, but the LD was moderate in the Chinese population, weak in African Americans and weakest in Africans. At the 16q12 locus, the LD pattern between rs3803662 and rs3104793 was also different across populations. These differences in LD correlations across populations are a possible reason why only the fine-mapping SNPs could be validated in the present study.
Table IV.
LD between significant SNPs and index SNPs in different populations
| Region | SNP pair | The present study |
The HapMap project |
||||||||||
| AA |
BB |
NG |
CEU |
CHB |
YRI |
||||||||
| r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | r2 | D′ | ||
| 6q25.1 | rs2046210–rs9397435 | 0.06 | 1.00 | 0.06 | 1.00 | 0.04 | 1.00 | 0.10 | 1.00 | 0.86 | 1.00 | 0.04 | 1.00 |
| 10q26 | rs2981578–rs1219648 | 0.13 | 0.98 | 0.08 | 1.00 | 0.05 | 1.00 | 0.85 | 1.00 | 0.32 | 0.83 | 0.06 | 1.00 |
| rs2981578–rs2981582 | 0.16 | 0.99 | 0.08 | 0.94 | 0.05 | 0.90 | 0.85 | 1.00 | 0.15 | 0.67 | 0.08 | 1.00 | |
| rs1219648–rs2981582 | 0.38 | 0.67 | 0.37 | 0.67 | 0.31 | 0.63 | 1.00 | 1.00 | 0.64 | 0.93 | 0.28 | 0.60 | |
| 16q12 | rs3803662–rs3104793 | 0.08 | 0.33 | 0.15 | 0.50 | 0.35 | 0.87 | 0.14 | 0.53 | 0.44 | 0.89 | 0.49 | 0.81 |
AA, African American; BB, Barbadian; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan; NG, Nigerian; YRI, Yoruba in Ibadan, Nigeria.
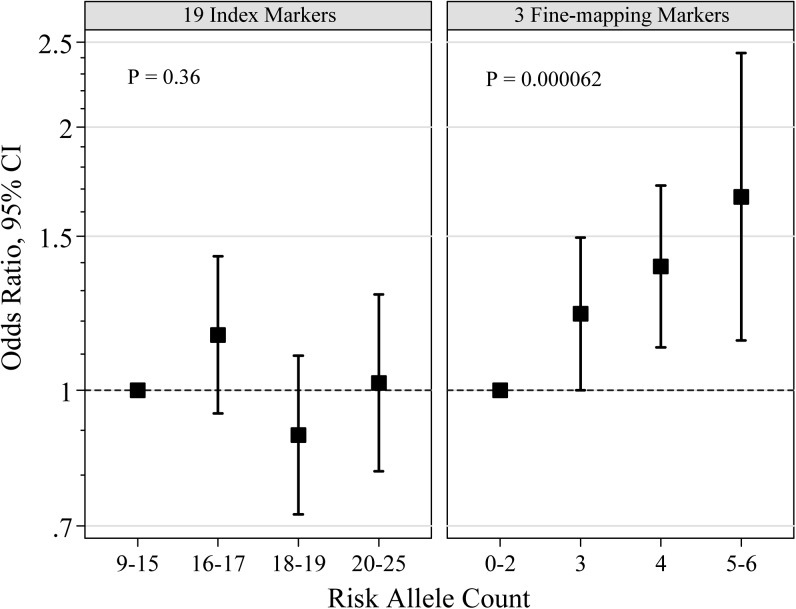
We further constructed composite risk score from unweighted risk allele counts using 19 index SNPs from previous GWAS or the three significant SNPs from previous fine-mapping studies (Figure 2 and Supplementary Table 3 is available at Carcinogenesis Online). The composite risk score from the 19 index SNPs was not associated with breast cancer risk in women of African ancestry (per-allele OR = 0.99; 95% CI: 0.96–1.02; P = 0.36). In contrast, the composite risk score from the three fine-mapping SNPs was significantly associated with breast cancer risk in women of African ancestry (per-allele OR = 1.19; 95% CI: 1.09–1.29; P = 6.2 × 10−5) and women with five or six risk alleles had a 66% increased risk compared with women with fewer than three risk alleles.
Fig. 2.
Relative risk of breast cancer in women of African ancestry, according to risk allele counts of 19 index markers from GWAS (left panel) or 3 markers from fine-mapping studies (right panel).
Discussion
We investigated the effects of 19 loci identified from breast cancer GWAS in women of African ancestry. We were not able to replicate any index SNPs reported in previous GWAS, which were conducted in populations of either European or Chinese ancestry. In contrast, we were able to validate three SNPs identified from fine-mapping studies that were conducted in women of African descent. The inability to replicate associations with any index SNPs is unlikely due to sample size. This present study was not powered to identify very small effects, but the study had sufficient power (>70%) for 7 of the 19 loci and our power to detect at least one locus approached 100%. Considering that four index SNPs showed associations in the opposite direction of the previous reports, it is possible that they are not causal variants or do not tag causal variants in women of African ancestry.
The 10q26 (FGFR2) locus was discovered in two GWAS among women of European descent (4,5), and the index SNPs rs2981582 and rs1219648 have been consistently replicated in European and Chinese populations (4,7,21,38,39). These index SNPs have been replicated among African Americans in some studies (17–19) but not in others (20,21). In one study, while replicated, the magnitude of association was smaller than that in initial GWAS (18). In a fine-mapping study using samples from African Americans, rs2981578 was found to have the strongest signal (23). In the present study, we failed to show that the two index SNPs at the FGFR2 locus were associated with breast cancer in women of African ancestry, but found that rs2981578 was significant with an allelic OR of 1.24, similar to that reported in the fine-mapping study. We believe this finding could be explained by the difference in LD pattern across populations as shown in Table IV. It is likely that the three SNPs tag the same causal variant(s), but the short LD pattern in women of African descent narrow the localization of the causal variant(s) in this region.
The 6q25 (ESR1/C6orf97) locus was discovered in a Chinese GWAS, with the index SNP rs2046210 (14), and this variant has been replicated in Chinese and Japanese populations (22). It was found to be significantly associated with breast cancer in women of European ancestry, but the effect was weaker (10,22). Several studies in African Americans have evaluated this SNP, but none found an association with breast cancer risk (18,20,22). A fine-mapping study using samples from women of European, African and Asian origin found that rs9397435 (2.9 kb away from rs2046210) carried the risk association in all three racial populations (24). Consistent with the fine-mapping study, we found strong association between rs9397435 and breast cancer (OR = 1.34; 95% CI: 1.10–1.63) but not rs2046210. After excluding the Nigerian samples, most of which had been included in the fine-mapping study, a statistically significant association was still observed for rs9397435 (OR = 1.36, 95% CI: 1.06–1.74; one-sided P = 0.008). Again, LD pattern difference across populations may explain these observations. In this locus, rs9397435 is strongly linked with rs2046210 only in Asians.
The SNP rs3803662 at 16q12 (TOX3) was identified as a breast cancer susceptibility variant in two GWAS, both conducted in European populations (4,6). This SNP remained the strongest signal for the 16q12 region in further studies in women of European ancestry (10,40). However, no studies have replicated this index SNP in African American populations (17–20,25,40), and Hutter et al. (20) found the association was in opposite direction to the initial GWAS findings. A fine-mapping study in African Americans found several SNPs were associated with breast cancer, with one SNP (rs3104793) being significant after permutation (25). In the present study, we did not find the index SNP rs3803662 to be associated with breast cancer in women of African ancestry but did validate that the association with rs3104793 was significant with an allelic OR of 1.12, similar to the prior fine-mapping study.
We have attempted to build a simple genetic risk prediction model and showed that the risk of breast cancer increased by 19% per one risk allele using the three fine-mapping SNPs. Additional risk variants in larger studies are necessary to generate a model that is useful in clinical settings. In a large pooled case–control study of breast cancer in African American, Chen et al. (18) found that the prediction model based on index SNPs (4% per one risk allele) has less predictive values than the model based on eight SNPs from their fine-mapping results (18% per one risk allele). One SNP (rs2981578) is shared between our study and the study by Chen et al., whereas other SNPs need cross-validation.
Several limitations need be considered when interpreting our study findings. This study utilized 29 AIMs to estimate European admixture proportion and the distribution of European ancestry proportion (1 − African ancestry proportion) was as we anticipated: no admixture for Nigerians, about 20% for African Americans and slightly less admixture (12%) for Barbadians. The SNP effect estimates with or without adjusting for ancestry proportions were in fact quite similar (difference by 1% on average and 5% the maximum). This is because (i) Nigerian is not an admixture population, (ii) African Americans from Northern California were sister pairs and (iii) the ancestry admixture was similar between cases and controls in the other study sites. These empirical data suggest that confounding due to population stratification is not large and unlikely to explain our study findings. Although this is a large study of women of African ancestry to investigate genetic risk factors for breast cancer, the statistical power for 12 of the 19 loci is <70%. Therefore, some of the null findings may be false negative. Another possible reason for lack of direct replication is disease heterogeneity because women of African Americans are more likely to have estrogen receptor-negative breast cancer (41,42), and genetic susceptibility SNPs are associated with specific breast cancer subtypes (39). In this replication study, we did not take into account multiple testing, which may lead to false-positive claims. Instead, we put our study findings in the context of previous studies to judge their validity.
In conclusion, this study serves to illustrate the complexity of applying current GWAS findings across racial/ethnic groups, as none of index SNPs from GWAS in non-African descent populations could be replicated in women of African ancestry. This lack of direct transferability of GWAS findings across populations has been observed for other cancer or non-cancer traits (43,44). Our successful validation of SNPs from fine-mapping studies suggests that fine-mapping studies and new GWAS in women of African ancestry promise to reveal additional and causal variants for breast cancer susceptibility. Although it is unlikely that there are major biological differences in breast cancer etiology among racial populations, interaction with environmental factors, allele frequencies and LD patterns may influence the genetic risk profiles in women of African ancestry.
Supplementary material
Supplementary Material and Tables 1–3 can be found at http://carcin.oxfordjournals.org/.
Funding
National Cancer Institute (R01CA141712 and P01CA82707). Support was also given by the Breast Cancer Research Foundation. The Northern California site of the Breast Cancer Family Registry (BCFR) was supported by the United States National Cancer Institute, National Institutes of Health under (RFA-CA-06-503) and through cooperative agreements with members of the BCFR and Principal Investigators, including the Northern California Cancer Center (U01 CA69417) and Georgetown University Medical Center Informatics Support Center (HHSN261200900010C).
Supplementary Material
Acknowledgments
We thank Drs Habibul Ahsan, James D. Fackenthal, Muhammad G. Kibriya and Jonathan Pritchard for their advice in study design. Samples from the Northern California site were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR) nor does mention of trade names, commercial products or organizations that imply endorsement by the United States Government or the BCFR.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AIM
ancestry-informative marker
- CI
confidence interval
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- OR
odds ratio
- SNP
single nucleotide polymorphism
References
- 1.Siegel R, et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance, Epidemiology, and End Results (SEER) Program. (2009) Limited-Use Data (1973-2006) National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch. www.seer.cancer.gov (1 September 2004, date last accessed) [Google Scholar]
- 3.Huo D, et al. Parity and breastfeeding are protective against breast cancer in Nigerian women. Br. J. Cancer. 2008;98:992–996. doi: 10.1038/sj.bjc.6604275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 7.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 8.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl Acad. Sci. USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher O, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 16.International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnholtz-Sloan JS, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417–1423. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum. Mol. Genet. 2011;20:4491–4503. doi: 10.1093/hmg/ddr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng W, et al. Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol. Biomarkers Prev. 2009;18:2761–2764. doi: 10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter CM, et al. Replication of breast cancer GWAS susceptibility loci in the Women's Health Initiative African American SHARe Study. Cancer Epidemiol. Biomarkers Prev. 2011;20:1950–1959. doi: 10.1158/1055-9965.EPI-11-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebbeck TR, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30:269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q, et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udler MS, et al. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 2009;18:1692–1703. doi: 10.1093/hmg/ddp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey SN, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Narvaez EA, et al. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol. Biomarkers Prev. 2010;19:1320–1327. doi: 10.1158/1055-9965.EPI-09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huo D, et al. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and breast cancer risk in Africans. Breast Cancer Res. Treat. 2008;110:367–376. doi: 10.1007/s10549-007-9720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemesure B, et al. Risk factors for breast cancer in a black population—the Barbados National Cancer Study. Int. J. Cancer. 2009;124:174–179. doi: 10.1002/ijc.23827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John EM, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boersma BJ, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J. Natl Cancer Inst. 2006;98:911–919. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Narvaez EA, et al. Validation of a small set of ancestral informative markers for control of population admixture in African Americans. Am. J. Epidemiol. 2011;173:587–592. doi: 10.1093/aje/kwq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaid DJ, et al. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. Am. J. Hum. Genet. 2006;79:1071–1080. doi: 10.1086/510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard JK, et al. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubisz MJ, et al. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Beroukhim R, et al. Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput. Biol. 2006;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zakharia F, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10:R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang J, et al. Genetic variants in fibroblast growth factor receptor 2 (FGFR2) contribute to susceptibility of breast cancer in Chinese women. Carcinogenesis. 2008;29:2341–2346. doi: 10.1093/carcin/bgn235. [DOI] [PubMed] [Google Scholar]
- 39.Broeks A, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2011;20:3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udler MS, et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet. 2010;19:2507–2515. doi: 10.1093/hmg/ddq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu KC, et al. Rates for breast cancer characteristics by estrogen and progesterone receptor status in the major racial/ethnic groups. Breast Cancer Res. Treat. 2002;74:199–211. doi: 10.1023/a:1016361932220. [DOI] [PubMed] [Google Scholar]
- 42.Huo D, et al. Population differences in breast cancer: survey in indigenous african women reveals over-representation of triple-negative breast cancer. J. Clin. Oncol. 2009;27:4515–4521. doi: 10.1200/JCO.2008.19.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang BL, et al. Validation of genome-wide prostate cancer associations in men of African descent. Cancer Epidemiol. Biomarkers Prev. 2011;20:23–32. doi: 10.1158/1055-9965.EPI-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shriner D, et al. Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS One. 2009;4:e8398. doi: 10.1371/journal.pone.0008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.