Abstract
Understanding the molecular pathways that contribute to the development of tamoxifen resistance is a critical research priority as acquired tamoxifen resistance is the principal cause of poor prognosis and death of patients with originally good prognosis hormone-responsive breast tumors. In this report, we provide evidence that Med1, an important subunit of mediator coactivator complex, is spontaneously upregulated during acquired tamoxifen-resistance development potentiating agonist activities of tamoxifen. Phosphorylated Med1 and estrogen receptor (ER) are abundant in tamoxifen-resistant breast cancer cells due to persistent activation of extracellular signal-regulated kinases. Mechanistically, phosphorylated Med1 exhibits nuclear accumulation, increased interaction with ER and higher tamoxifen-induced recruitment to ER-responsive promoters, which is abrogated by inhibition of Med1 phosphorylation. Stable knockdown of Med1 in tamoxifen-resistant cells not only reverses tamoxifen resistance in vitro but also in vivo. Finally, higher expression levels of Med1 in the tumor significantly correlated with tamoxifen resistance in ER-positive breast cancer patients on adjuvant tamoxifen monotherapy. In silico analysis of breast cancer, utilizing published profiling studies showed that Med1 is overexpressed in aggressive subsets. These findings provide what we believe is the first evidence for a critical role for Med1 in tamoxifen resistance and identify this coactivator protein as an essential effector of the tamoxifen-induced breast cancer growth.
Introduction
Estrogen is a key regulator for normal growth and differentiation of mammary glands as well as the malignant progression of breast cancer (1). Consequently, interruption of estrogen signaling by using endocrine therapies has been an effective therapeutic strategy. Tamoxifen is the most prolific therapeutic drug for the treatment of estrogen receptor (ER)-positive breast cancer, showing effective tumor growth inhibition, reduced mortality and prevention of disease recurrence (2). Despite excellent efficacy, the use of tamoxifen is limited by the development of acquired resistance as a majority of initially responsive tumors later develop resistance to tamoxifen (3). Several molecular mechanisms have been proposed for the development of acquired resistance, including expression of variant or mutant ER, intimate cross talk between the ER and HER2/growth factor signaling pathway, altered cytoplasmic and genomic signaling of ER, alternative proliferative and survival stimuli (3,4). Recent studies have revealed that distinct coregulatory complexes modulate transcriptional activity of ER. Ligand-bound ER recruits multiple coactivator/corepressor complexes, which serve as a bridge between ER and the general transcription machinery or associate with histone acetyltransferases or histone deacetylases (5). While the role of coregulatory complexes for ER is very well established in mediating genomic signaling of ER in response to different ligands during normal breast development and tumor progression, the importance of coregulators in development of tamoxifen resistance is still under investigation.
Coactivators that directly interact with liganded ER and facilitate transactivation include members of the p160/steroid receptor coactivator family (6). These proteins are thought to function in part by associating with potent histone acetyltransferases resulting in acetylation of nucleosomes and transactivation of ER-responsive genes (6). Another important, conserved multisubunit coactivator complex is a mediator complex that binds to liganded ER and plays a key regulatory role (7). Mediator complex positively influences ligand-dependent transactivation by interacting with basal transcription machinery, possibly by directly facilitating the recruitment of RNA polymerase II (8). Mediator complex is comprised of ∼30 subunits arranged in four subcomplexes known as the head, middle, tail and cdk8-containing modules (9,10). Mediator complex is directed to liganded ER by its anchor subunit, Med1, via two signature LXXLL motifs that serve as nuclear receptor-binding surfaces (11). Adding to its functional importance in regulating cell growth and development, Med1 also serves as a direct binding target for other regulatory transcription factors such as the GATA family of proteins, GABPα, BRCA-1 and p53 (12–17).
In this study, we present evidence that Med1 overexpression renders ER-positive breast cancer cells resistant to tamoxifen. Elevated expression of Med1, along with a significant increase in Med1 and ER phosphorylation due to persistent activation of extracellular signal-regulated kinase (ERK) in tamoxifen-resistant cells, leads to increased recruitment of ER-Med1 complex to ER-responsive genes upon tamoxifen treatment. Using isogenic Med1-knockdown tamoxifen-resistant cell line pair, we show that Med1 is required for tamoxifen-induced growth of tamoxifen-resistant cells in vitro and in vivo. Analysis of clinical samples showed that high Med1 expression is significantly linked to tamoxifen resistance in breast cancer. Also, in silico analyses of multiple independent microarray expression, datasets revealed Med1 overexpression in subsets of breast tumors correlated with increased aggressiveness, higher clinical risk and mortality. Therefore, Med1 could serve as a critical regulator altering ER-mediated transcription rendering cells nonresponsive to tamoxifen, thus mediating the development of tamoxifen-resistant phenotypes.
Materials and methods
Reagents, hormones and antibodies
17β-estradiol (E2), 4-hydroxytamoxifen (OHT), epidermal growth factor (EGF) and U0126 were from Sigma (St Louis, MO). Anti-Med1, anti-phospho-Med1 (Abcam), anti-ER (Santa Cruz), anti-phospho-ER (Serine-167), anti-phospho-ER (Serine-118), anti-phospho-ERK and anti-ERK (Cell Signaling) antibodies were used.
Cell culture
The human breast cancer cell line, MCF7, was obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gemini Bio-products, Woodland, CA) (18). Cell line authentication was done by analysis of known genetic markers or response (e.g. expression of ER and p53 and estrogen responsiveness). Cells were maintained in phenol red-free Dulbecco's modified Eagle's medium containing 10% charcoal-stripped dextran-treated serum (Gemini Bio-products) to reduce exogenous hormones and growth factors for 3 days prior to OHT and E2 treatment (18). EGF is known to activate ERK phosphorylation (19). U0126 (Sigma–Aldrich) is a highly selective inhibitor for both MEK1 and MEK2 that inhibit ERK phosphorylation. To determine the role of ERK phosphorylation in Med1 phosphorylation/function, cells were pretreated for 2 h with 100 ng/ml EGF (Sigma–Aldrich) or 30 μM U1026 (Sigma–Aldrich) followed by tamoxifen treatment for 2 h.
Generation of tamoxifen-resistant cell line
To generate a physiologically relevant model of acquired tamoxifen resistance, parent tamoxifen-sensitive MCF7 (MCF7-P) cells were grown in phenol red-free improved minimum essential medium supplemented with charcoal-stripped dextran-treated serum containing 1 μM 4-OHT for ∼12 months. Several tamoxifen-resistant (MCF7-OHT) clones of MCF7-P cells were selected and maintained in phenol red-free improved minimum essential medium supplemented with 10% charcoal-stripped dextran-treated serum. Representative data from multiple clones are presented in this report. To strengthen and generalize the findings of our study, we also used two different established models of tamoxifen-resistant breast cancer cells (LCC2 and MCF7-5/23) (20,21). LCC2 cells were a kind gift from Dr Robert Clarke, Georgetown University, USA (20). MCF7 5/21 and MCF7 5/23 were kindly provided by Dr Rob Sutherland, Garvan Institute of Medical Research, Australia and Dr John Zalcberg, University of Melbourne, Australia (21).
Clonogenicity assay
For colony-formation assay (22), cells were treated with 17β-estradiol or 4-OHT as indicated for 10 days; colonies were stained with crystal violet (0.1% in 20% methanol). Colonies containing >50 normal-appearing cells were counted.
Anchorage-independent growth assay
Anchorage-independent growth of cells was assayed by colony formation on soft agar (22). Colonies were stained with 0.005% crystal violet in phosphate-buffered saline for 1 h at room temperature and observed using Olympus IX50 inverted microscope. Colonies were counted in five randomly selected fields at ×10 magnification. Results are expressed as number of colonies counted.
Western blot
Whole cell lysate was prepared by scraping cells in 250 μl of ice-cold modified RIPA buffer (23,24). Western blot analysis was performed using the antibodies listed in the ‘Reagents, hormones and antibodies section’ above.
RNA isolation, reverse transcription–PCR, real-time PCR
Total cellular RNA was extracted using the TRIZOL Reagent kit (Life Technologies Inc., Rockville, MD). Reverse transcription–PCR was performed as described previously (18) using specific sense and antisense PCR primers. Taqman real-time quantitative PCR was performed using ABI 7900HT (Applied Biosystems). Details of primers are included in Supplementary Materials, available at Carcinogenesis Online.
Plasmids and luciferase assays
Construct pSilencer 2.1-U6 hygro Med1 and empty vector were kindly provided by Dr P.Lefebvre (Faculte de Medecine Henri Warembourg, France). Cells were transfected with vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Human ER promoter reporter construct used for luciferase assays was a kind gift from Dr Michael Wang (25). Cells were transfected with estrogen response element (ERE) luciferase construct using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and treated as indicated Luciferase activity was measured in a luminometer (BD Biosciences) after 48 h, and the resulting data were normalized for the background Renilla luciferase activity using the Dual Luciferase Reporter Assay system (Promega, Madison, WI).
Immunoprecipitation of Med1
Immunoprecipitation was performed essentially as described (26), whole cell lysates were incubated with specific antibodies for Med1 followed by incubation with protein A/G agarose beads. The precipitated protein-beads complexes were subjected to immunoblotting.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analyses were performed following our published procedure (18,27,28). Primers for ChIP assay are included in Supplementary Materials, available at Carcinogenesis Online. All ChIP assays were performed at least thrice with similar results.
Immunofluorescence and confocal imaging
To determine the role of ERK phosphorylation in Med1 localization, cells were pretreated for 2 h with 100 ng/ml EGF (Sigma–Aldrich) or 30 μM U1026 (Sigma–Aldrich) followed by tamoxifen treatment for 2 h. Cells (5 × 105 cells per well) were plated in four-well chamber slides (Nunc, Rochester, NY), treated with EGF or U0126 and subjected to immunofluorescence analysis as described earlier (22). Fixed and immunofluorescently stained cells were imaged using a Zeiss LSM510 Meta (Zeiss) laser scanning confocal system configured to a Zeiss Axioplan 2 upright microscope with a 63XO (NA 1.4) plan-apochromat objective. Cells were observed at five randomly selected fields in single focal plane for Med1 localization. All experiments were performed multiple times using independent biological replicates.
Med1 stable knockdown using lentiviral short hairpin RNA
Five premade lentiviral Med1 short hairpin RNA (shRNA) constructs and a negative control construct created in the same vector system (pLKO.1) were purchased from Open Biosystems (Huntville, AL). Paired Med1 stable knockdown cells were generated following our previously established protocol (22). Lentiviral helper plasmids (pCMV-dR8.2 dvpr and pCMV-VSV-G) were obtained from Addgene (Cambridge, MA). Transient lentivirus stocks were prepared following the manufacturer's protocol. One day before transfection, 1.5 × 106 293T cells were plated in 100 mm dishes. Cells were cotransfected with shRNA constructs (3 μg) together with 3 μg pCMV-dR8.2 dvpr and 0.3 μg pCMV-VSV-G helper constructs. Two days later, viral stocks were harvested from the culture medium and filtered to remove nonadherent 293T cells. To select for the MCF7-P and MCF7-OHT cells that were stably expressing shRNA constructs, cells were plated at subconfluent densities and infected with a cocktail of 1 ml of virus-containing medium, 3 ml of regular medium and 8 μg/ml polybrene. Selection with 0.5–2 μg/ml of puromycin was started 48 h after lentivirus infection. After 4 weeks of selection for MCF7-P and MCF7-OHT cells, monolayers of stably infected pooled clones were harvested for use and cryopreserved.
Athymic nude mice model for tamoxifen-resistant growth
Four- to six-week-old female athymic nude mice (Harlan Laboratories Inc., Indianapolis, IN) were given a subcutaneous injection of 5 × 106 cells (50% phosphate-buffered saline/50% Matrigel in a total volume of 50 μl). When tumors reached the size of 50 mm3, the animals were randomly allocated (n = 10 per group) to various groups. Vehicle and tamoxifen treatment (500 μg of tamoxifen citrate in peanut oil by subcutaneous injection daily Monday to Friday, for 6 weeks) were given for respective groups. Estrogen treatment was provided by a 60 d slow-release pellet containing 0.72 mg of 17β-estradiol (Innovative Research of America). Tumor growth was assessed and tumor volumes were measured as described previously (29). Tumors were harvested 6 weeks after the initiation of treatment and processed for protein (western blot analysis, RNA (reverse transcription–PCR analysis) and immunohistochemistry analysis).
Immunohistochemistry of clinical specimens
From 1986 until 1991, 564 premenopausal patients under 50 years with stage II (pT2 N0 M0, pT1 N1 M0 and pT2, N1 M0) invasive breast cancer were enrolled in a trial (SBII:2a) and randomized to either 2 years of adjuvant tamoxifen (n = 276) or no systemic adjuvant treatment (control) (n = 288). All patients received surgery followed by radiotherapy if indicated and only 2% (n = 9) of the patients received adjuvant polychemotherapy. The median follow-up time for patients with no recurrent breast cancer event was 13.9 years. Details of this trial have been described (30). This study was approved by the ethical committees at Lund and Linköping Universities.
Analysis/scoring of Med1.
Med1 expression was assessed immunohistochemically as absent = 0, very low = 1, low = 2, intermediate = 3, intermediate high = 4, high = 5 in combination with fraction of positive nuclei/cytoplasm where 0 = 0–1%, 1 = 1–10%, 2 = 11–25%, 3 = 26–50% and 4 = 51–100%. Med1 expression was scored using a combined analysis of nuclear/cytoplasmic intensity and fraction positive cells into 0–5 where 4–5 represented >50% cells showing staining of moderate or strong intensity. Since overexpression of Med1 in breast cancer was the scope of the examinations, we focused on the two highest subgroups of nuclear staining versus low expression. Cytoplasmic Med1 expression was scored into no/low expression or high. All immunohistochemical analyses were performed by a pathologist (L.J.).
Oncomine data
Supplementary figures include gene expression data and graphs from Oncomine (www.oncomine.org). Breast cancer profiling datasets (Supplementary Table 1, available at Carcinogenesis Online) were analyzed for Med1 expression levels.
Statistical analysis
All experiments were independently performed three times in triplicates. Statistical analysis was done using Microsoft Excel software. Significant differences were analyzed using student's t-test and two-tailed distribution. Data were considered to be statistically significant if P < 0.05. Data are expressed as means ± SE between triplicate experiments. The distribution of clinicopathological characteristics by Med1 expression was evaluated using Fisher's exact tests, Spearman's ρ or Pearson chi-square test. Continuous variables (age) were analyzed using Mann–Whitney U tests. Survival analyses were generated using the Kaplan–Meier method and compared using the log-rank test. Hazard ratios (HRs) were calculated using Cox proportional hazards regression. Statistical analyses were performed using SPSS software (version 15.0.1; Chicago, IL).
Results
ER function is modified in the acquisition of tamoxifen resistance
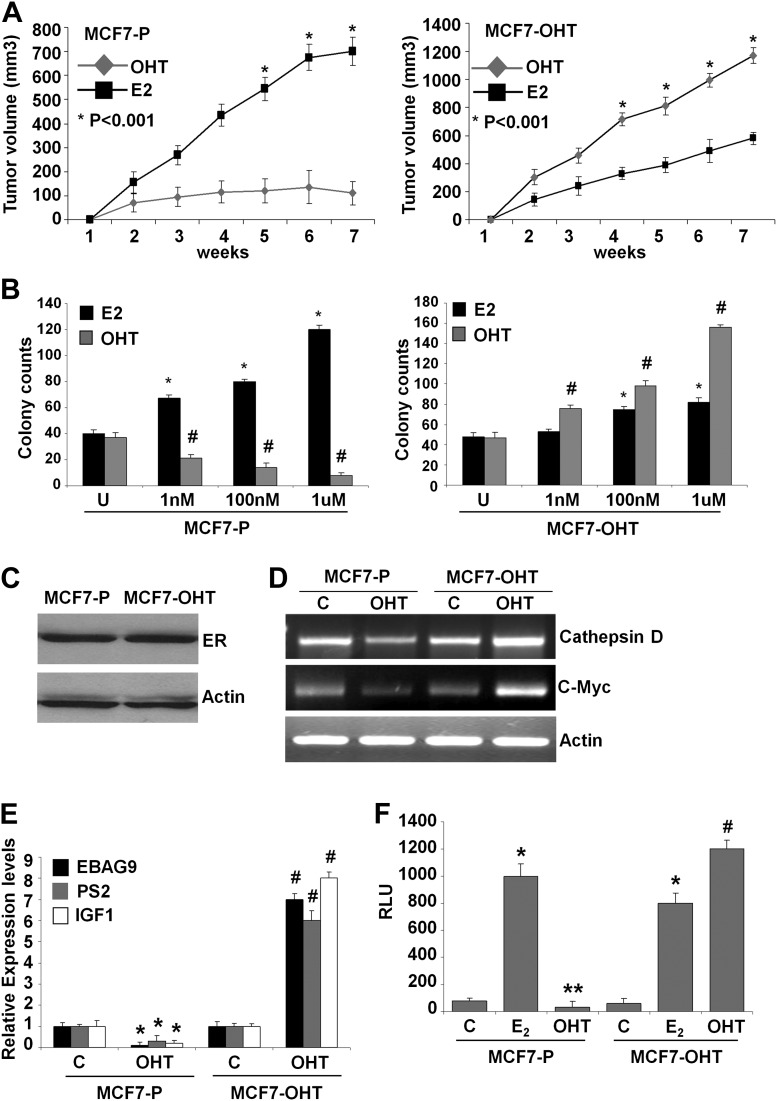
To understand how tamoxifen resistance develops in initially responsive ER-positive breast cancer cells, we developed isogenic cell line pairs (Supplementary Figure 1A, available at Carcinogenesis Online) exhibiting acquired tamoxifen resistance. In vivo, MCF7-OHT tumors grow rapidly in the presence of tamoxifen, whereas MCF7-P tumors get inhibited by tamoxifen treatment (Figure 1A). MCF7-P cells showed increased proliferation, clonogenicity and 3-D colony formation in response to estrogen treatment. MCF7-P cells form smaller colonies in the absence of estrogen while estrogen treatment enhances the number as well as the size of colonies. Tamoxifen treatment efficiently inhibited growth and clonogenicity of MCF7-P cells, whereas MCF7-OHT cells continued to grow at an increased rate (Figure 1B and Supplementary Figure 1B and C, available at Carcinogenesis Online). The resistant clones typically form similar number of colonies (containing >50 cells) as parent cells upon estrogen stimulation. However, the colonies of parent cells continue to grow in response to estrogen while resistant clones form smaller colonies displaying reduced response to estrogen stimulation. Change in ER level may modulate estrogen and tamoxifen response. However, immunoblot analysis showed similar levels of ER protein levels in both MCF7-P and MCF7-OHT cells (Figure 1C).
Fig. 1.
Tamoxifen promotes tumor growth, clonogenicity and augments ER-responsive gene expression and ER-mediated transactivation in tamoxifen-resistant cells. (A) Tumor growth curves of MCF7-P and MCF7-OHT cells implanted subcutaneous in athymic mice in the presence of estrogen and tamoxifen treatment. (B) Anchorage-dependent growth of MCF7-P and MCF7-OHT cells in the presence of various doses of 17β-estradiol (E2) or 4-OHT. Colonies containing >50 normal-appearing cells were counted. *, P < 0.001, compared with untreated controls (MCF7-P and MCF7-OHT); #, P < 0.001, compared with untreated controls (MCF-P and MCF7-OHT). (C) Immunoblot analysis of ER protein in MCF7-P and MCF7-OHT cells. (D) Semi-quantitative reverse transcription–PCR analysis of messenger RNA (mRNA) expression levels of cathepsin-D and c-Myc mRNA in MCF7-P and MCF7-OHT cells treated with 1 μM of 4-OHT for 2 h. (E) Real-time quantitative PCR analysis of EBAG9, pS2 and IGF1 expression in MCF7-P and MCF7-OHT cells treated with 1 μM of 4-OHT for 2 h. *, P < 0.005, compared with untreated controls (MCF7-P); #, P < 0.001, compared with untreated controls (MCF7-OHT). (F) Luciferase activity of pERE-Luc in MCF7-P and MCF7-OHT cells treated with either ethanol vehicle alone or 100 nM E2 or 1 μM OHT for 24 h. *, P < 0.001, compared with untreated controls (MCF7-P and MCF7-OHT); **, P < 0.02, compared with untreated controls (MCF7-P); #, P < 0.001, compared with untreated controls (MCF7-OHT).
Estrogen signaling regulates various biological processes such as cell cycle progression, proliferation, signaling pathways and apoptosis in normal as well as malignant growth via regulation of expression of various important cellular oncogenes and tumor suppressor genes. Overexpression/over-representation of E2-responsive genes has been associated with enhanced growth and aggressiveness of breast cancer (31, 32). Tamoxifen-mediated inhibition of these E2-responsive genes in MCF7 cells collectively leads to breast cancer growth inhibition. In contrast, tamoxifen activates the expression of these E2-responsive genes in tamoxifen-resistant cells altering the molecular network to enhance breast tumor growth. Steroid receptors, such as ER, regulate gene transcription either by binding directly to the promoter of target genes or by binding indirectly through other transcription factors (1, 33). Genes regulated through direct ER binding, such as EBAG9 (ER-binding fragment-associated antigen 9), cathepsin D and PS2, typically harbor a classic hormone-responsive element (34, 35). In contrast, genes, such as c-Myc and IGF1, are regulated by indirect binding of ER to non-classic HREs (36, 37). EBAG9, cathepsin D and PS2 were chosen to represent hormone-responsive element-containing target genes while c-Myc and IGF1 represented non-hormone-responsive element-containing target genes.
Examination of tamoxifen-mediated changes in expression pattern of endogenous ER-responsive genes (cathepsin D, c-myc, EBAG9, pS2 and IGF1) revealed inhibited expression upon tamoxifen treatment in MCF7-P cells, whereas MCF7-OHT cells showed increased expression in response to tamoxifen (Figure 1D and E). Furthermore, estrogen increased while tamoxifen inhibited ER-mediated transactivation in MCF7-P cells. MCF7-OHT cells showed significantly elevated levels of ER transactivation function in response to tamoxifen while showing an increased luciferase activity in response to estrogen albeit at a lower level in comparison with tamoxifen (Figure 1F). Collectively, these studies showed that the acquisition of tamoxifen resistance is associated with differential sensitivity to estrogen and tamoxifen, retention of ER protein while modifying its transactivation function in order to differentially alter expression of ER-responsive genes in response to tamoxifen.
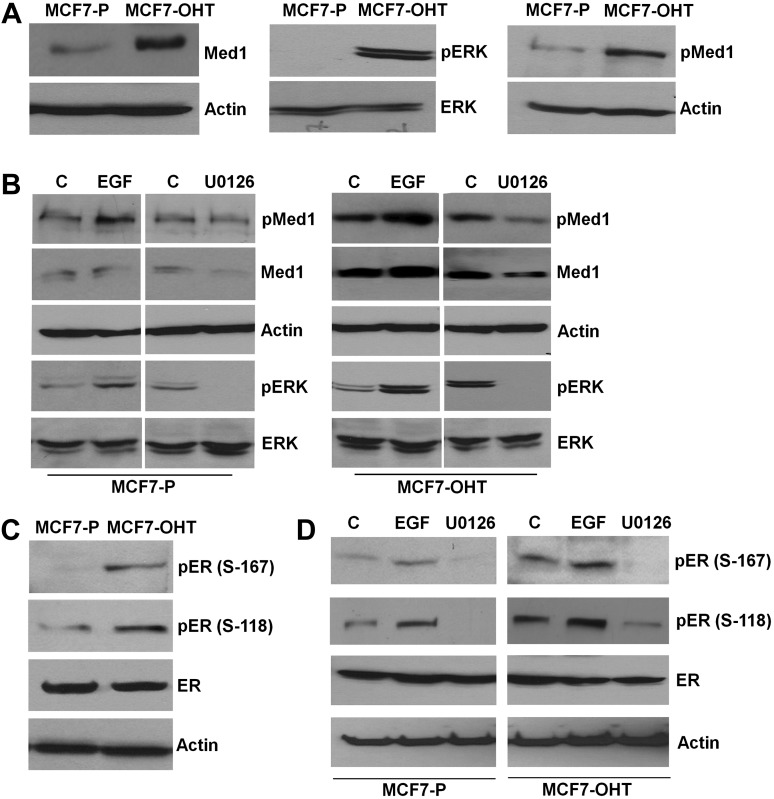
Alteration in expression and phosphorylation of Med1 and ER due to persistent activation of ERK modulates their functional interaction in acquired tamoxifen resistance
We discovered that Med1 was overexpressed in tamoxifen-resistant MCF7-OHT cells in comparison with parental tamoxifen-sensitive MCF7-P cells (Figure 2A). Med1 facilitates functional interactions between regulatory transcription factors and the general polymerase II initiation apparatus during ER-responsive gene regulation (9). Coregulatory functions of human Med1 may get modulated via altered phosphorylation. Interestingly, we observed persistent activation of ERK in MCF7-OHT cells indicating a possible role of ERK in tamoxifen resistance (Figure 2A). We hypothesized that Med1 phosphorylation may get altered by MAPK-ERK and acting as a regulatory mechanism may influence its association with the core mediator complex along with its coactivation functions. Increased accumulation of phosphorylated Med1 was observed in MCF7-OHT cells in comparison with MCF7-P cells (Figure 2A). EGF treatment is known to activate ERK (19). Stimulation with EGF (Figure 2B) or cotransfection with the MKK1 (NΔ4) or ERK2 (L73P/S151D) (38) expression constructs (data not shown) markedly enhanced Med1 phosphorylation, whereas addition of U0126, an inhibitor for ERK, inhibited Med1 phosphorylation (Figure 2B). Phosphorylation modifications of ER have been implicated in important regulatory function of ER. ER is phosphorylated at Ser-167 and Ser-118 in response to active ERK signaling (39). MCF7-OHT cells showed increased phosphorylation of ER at Ser-167 and Ser-118 sites (Figure 2C). ER phosphorylation at both Ser-167 and Ser-118 was further enhanced in response to EGF stimulation, whereas U0126 treatment led to inhibition of ER phosphorylation (Figure 2D).
Fig. 2.
Increased expression and phosphorylation of Med1 in tamoxifen-resistant cells due to persistent activation of ERK. (A) Immunoblot analysis of Med1, ERK, phospho-Med1 and phospho-ERK in MCF7-P and MCF7-OHT. (B) Immunoblot analysis of pMed1, pERK and ERK in MCF7-P and MCF7-OHT cells treated with EGF and U1026. (C) Immunoblot analysis of ER, p-ER (S-167) and p-ER (S-118) in MCF7-P and MCF7-OHT cells. (D) Immunoblot analysis of ER, p-ER (S-167) and p-ER (S-118) in MCF7-P and MCF7-OHT cells treated with EGF and of U1026.
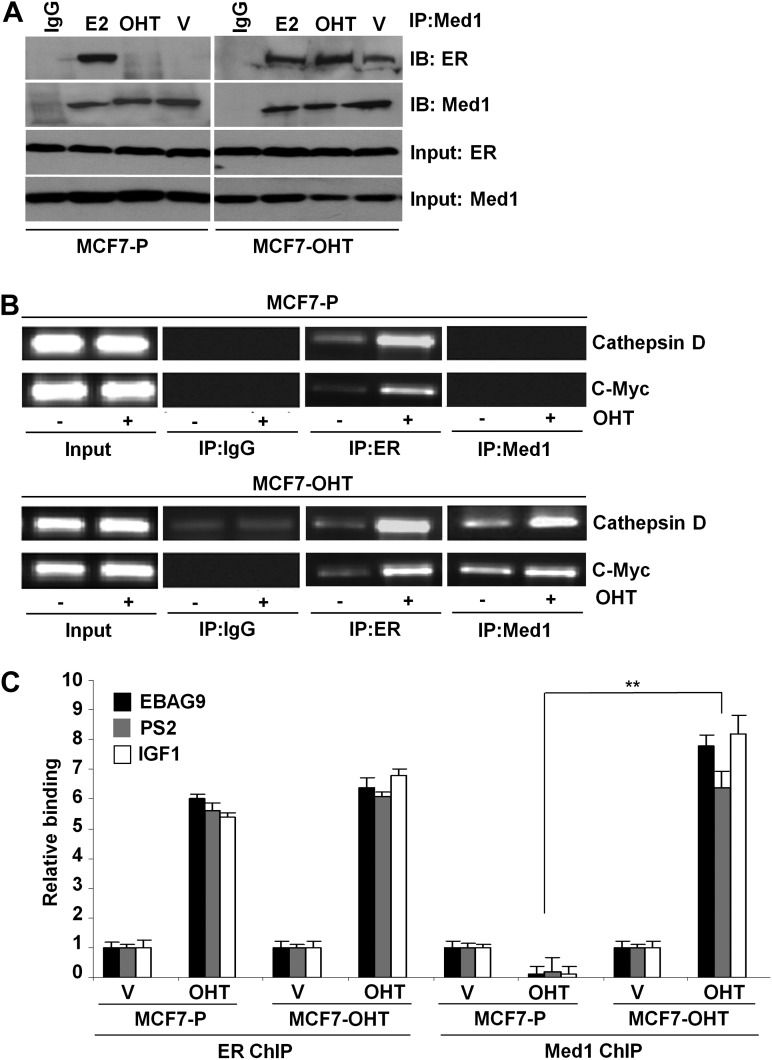
Activation of ERK pathway brings additional complexity because the ability of coactivators to interact with nuclear hormone receptors and influence transcription can be affected by their altered phosphorylation. In immunoprecipitation assay, MCF7-P cells exhibited Med1:ER interaction upon E2 treatment, whereas no interaction was observed in the presence of tamoxifen. In contrast, Med1 and ER showed a substantial interaction in a ligand-independent manner which was increased with E2 and tamoxifen treatment in MCF7-OHT cells (Figure 3A). In an effort to better understand the molecular events involved in acquired tamoxifen resistance, we used ChIP analyses to examine the recruitment of Med1 to specific ER-responsive gene promoters in response to tamoxifen treatment. ER got recruited to ER-responsive gene promoters in response to tamoxifen in MCF7-P and MCF7-OHT cells (Figure 3B, C). Coactivator Med1 did not bind to EREs in the presence of tamoxifen in MCF7-P cells. Intriguingly, tamoxifen treatment induced recruitment of Med1 to ER-responsive gene promoters in tamoxifen-resistant MCF7-OHT cells (Figure 3B, C). Tamoxifen inhibited the expression of ER-responsive genes, cathepsin D, c-myc, EBAG9, pS2 and IGF1 in MCF7-P cells, whereas MCF7-OHT cells showed increased expression upon tamoxifen treatment (Figure 5D and Supplementary Figure 3, available at Carcinogenesis Online).
Fig. 3.
Med1 associates with ER and tamoxifen increases recruitment of Med1 to ER-responsive gene promoters. (A) Immunoprecipitation of Med1 protein from MCF7-P and MCF7-OHT cells followed by immunoblot analysis of ER. (B) ChIP was performed using antibodies specific for ER and Med1 from MCF7-P and MCF7-OHT cells untreated and treated with 4-OHT (1 μM for 2 h). The purified DNA was analyzed by PCR using specific primers spanning the EREs of cathepsin D and c-myc gene promoters. (C) ChIP was performed as in (B). The purified DNA was analyzed by real-time quantitative PCR analysis using specific primers spanning the EREs of EBAG9, pS2 and IGF1 gene promoters. **, P < 0.001, compared with MCF-P treated with OHT.
Fig. 5.
Stable knockdown of Med1 reverses tamoxifen resistance in tamoxifen-resistant cells. (A) Immunoblot analysis of Med1 in stable pools of Med1-depleted (Med1shRNA) and vector control (pLKO.1) MCF7-P and MCF7-OHT cells. (B) Clonogenicity of MCF7-P-pLKO.1, MCF7-P-pMed1shRNA, MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA cells in the presence of 4-OHT (1 μM). *, P < 0.005, compared with untreated controls (MCF7-P-pLKO.1); **, P < 0.001, compared with untreated controls (MCF7-P-pMed1shRNA); #, P < 0.001, compared with untreated MCF7-OHT-pLKO.1; #*, P < 0.001, compared with untreated MCF7-OHT-pMed1shRNA. (C) Soft agar colony formation in MCF7-P-pLKO.1, MCF7-P-pMed1shRNA, MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA cells in the presence of 1 μM 4- OHT. *, P < 0.005, compared with untreated controls (MCF7-OHT-pLKO.1); #, P < 0.001, compared with tamoxifen-treated MCF7-OHT-pLKO.1. (D) Real-time quantitative PCR analysis of EBAG9, pS2 and IGF1 expression in MCF7-P-pLKO.1, MCF7-P-pMed1shRNA, MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA cells treated with 1 μM of 4-OHT for 2 h. *, P < 0.005, compared with untreated controls (MCF7-P-pLKO.1); #, P < 0.001, compared with untreated controls (MCF7-P-pMed1shRNA); **, P < 0.001, compared with untreated controls (MCF7-OHT-pLKO.1); ***, P < 0.005, compared with untreated controls (MCF7-OHT-pMed1shRNA).
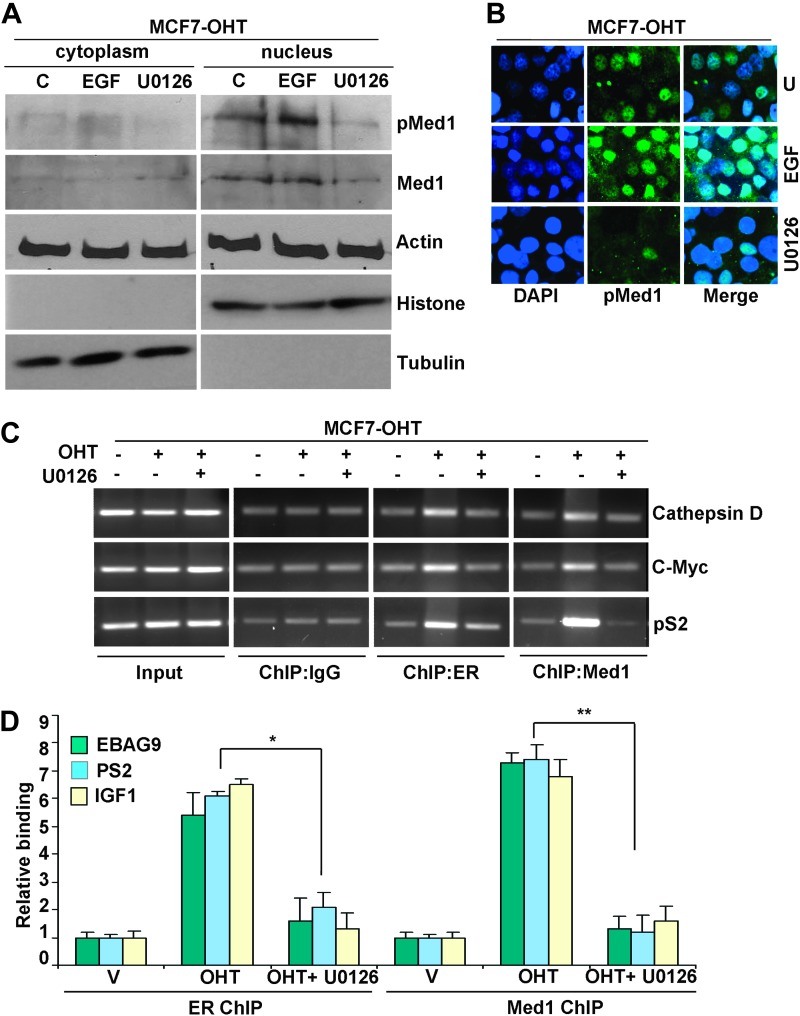
Next, we determined the impact of phosphorylation of Med1 on its nuclear translocation and tamoxifen-induced recruitment to ER-responsive promoters in tamoxifen-resistant cells. Med1 is majorly localized in nucleus in MCF7-OHT cells (Figure 4A and B). Treatment with U0126 reduces the total amount of Med1 indicating that the ERK pathway regulates Med1 protein expression (Figure 4A). Nuclear localization of Med1 is greatly enhanced in response to EGF stimulation (Figure 4A and B), which has been shown to increase its phosphorylation (Figure 2). ChIP analyses revealed that tamoxifen induced recruitment of ER and Med1 to ER-responsive genes which was abrogated by U0126 treatment (Figure 4C and D). Collectively, our findings suggest that persistent activation of ERK in tamoxifen-resistant cells leads to elevated phosphorylation levels of ER and Med1 that may act as a regulatory mechanism to promote Med1 association with ER in response to tamoxifen. Further mechanistic studies directly examining the role of phosphorylation in MED1-ER interaction may show that altered phosphorylation of Med1 and ER may facilitate a novel feed-forward action of tamoxifen during acquired tamoxifen resistance. These evidence support the notion that tamoxifen induced increased Med1-ER interaction and increased Med1 recruitment at ER-responsive gene promoters in tamoxifen-resistant but not in tamoxifen-sensitive cells may be due to altered phosphorylation of Med1 and ER in tamoxifen-resistant cells.
Fig. 4.
Med1 phosphorylation is important for its nuclear accumulation and tamoxifen-induced recruitment to ER-responsive gene promoters. (A) Immunoblot analysis of p-Med1 and Med1 in cytoplasmic and nuclear fractions of MCF7-OHT cells treated with EGF or U0126. (B) Immunofluorescence analysis of p-Med1 in MCF7-OHT cells treated as in A. (C) ChIP analysis of ER and Med1 in MCF7-OHT cells treated with 4-OHT alone or in combination with U0126. The purified DNA was analyzed by PCR using specific primers spanning the EREs of cathepsin D, c-myc and pS2 gene promoters. (D) ChIP was performed as in (C). The purified DNA was analyzed by real-time quantitative PCR analysis using specific primers spanning the EREs of EBAG9, pS2 and IGF1 gene promoters. *, P < 0.001, compared with cells treated with OHT alone (ER ChIP). **, P < 0.001, compared with cells treated with OHT alone (Med1 ChIP).
Targeted downregulation of Med1 in tamoxifen-resistant cells reverses acquired tamoxifen resistance
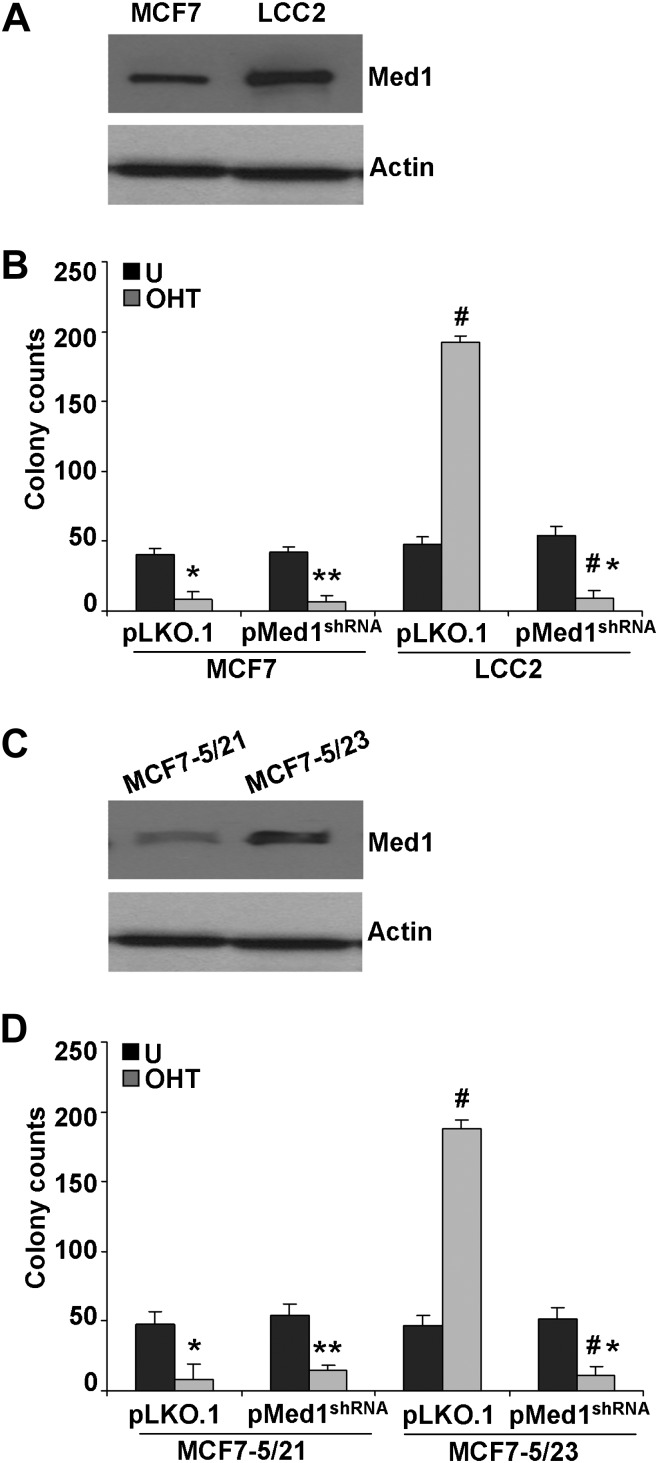
To determine the importance of Med1 in tamoxifen resistance, endogenous Med1 was transiently knocked down in MCF7-P and MCF7-OHT cells using psilencer-Med1 (Supplementary Figure 2A is available at Carcinogenesis Online). In MCF7-OHT cells transfected with psilencer-control, tamoxifen treatment induced significant growth. Interestingly, Med1 inhibition in MCF7-OHT cells significantly reduced tamoxifen-induced growth (Supplementary Figure 2B is available at Carcinogenesis Online) indicating the importance of Med1 in tamoxifen resistance while not affecting growth in the absence of tamoxifen. Next, we used Med1shRNA and pLKO.1 (vector control) lentivirus and puromycin to select for stable pools of MCF7-P and MCF7-OHT cells with stable Med1 depletion. Med1-depleted (Med1shRNA) MCF7-P and MCF7-OHT cells showed efficient knockdown of Med1 in comparison with vector control (pLKO.1) cells (Figure 5A). Examination of clonogenic potential and 3-D soft agar colony formation of MCF7-P-pLKO.1 and MCF7-P-pMed1shRNA cells in the presence of tamoxifen revealed that Med1 inhibition did not affect tamoxifen-mediated inhibition of MCF7-P cells. While knockdown of Med1 in MCF7-OHT (MCF7-OHT-pMed1shRNA) cells significantly inhibited tamoxifen-induced clonogenicity and 3-D soft agar colony formation in comparison with vector control (MCF7-OHT-pLKO.1) cells (Figure 5B and C). We also examined the effect of Med1 depletion on ER-responsive gene expression. Expression of ER-responsive genes was inhibited in response to tamoxifen treatment in MCF7-P-pLKO.1 and MCF7-P-pMed1shRNA cells while MCF7-OHT-pLKO.1 cells showed increased expression of cathepsin D, c-myc, EBAG9, pS2 and IGF1 upon tamoxifen treatment. Inhibition of Med1 in tamoxifen-resistant cells (MCF7-OHT-pMed1shRNA) reversed tamoxifen resistance leading to tamoxifen-mediated inhibition of expression of ER-responsive genes (Figure 5D and Supplementary Figure 3 is available at Carcinogenesis Online). Examination of four additional tamoxifen-resistant clones (Supplementary Figure 1 is available at Carcinogenesis Online) also showed higher Med1 expression along with increased ERK phosphorylation (data not shown). Silencing of Med1 in these tamoxifen-resistant clones inhibited tamoxifen-induced growth (data not shown). To strengthen and generalize these finding, we next investigated the impact of Med1 on tamoxifen resistance using two different established models of tamoxifen-resistant breast cancer cells (LCC2 and MCF7-5/23) (20,21). We found higher Med1 expression in tamoxifen-resistant LCC2 and MCF7-5/23 cells (Figure 6A and C) in comparison with their tamoxifen-sensitive counterparts. Also, stable Med1 depletion using Med1shRNA lentivirus in LCC2 and MCF7-5/23 cells led to reversal of tamoxifen resistance further corroborating the importance of Med1 in tamoxifen resistance (Figure 6B and D).
Fig. 6.
Stable knockdown of Med1 reverses tamoxifen resistance in LCC2 and MCF7-5/23, tamoxifen-resistant cells. (A) Immunoblot analysis of Med1 in MCF7 and LCC2 cells. (B) Clonogenicity of MCF7-pLKO.1, MCF7-pMed1shRNA, LCC2-pLKO.1 and LCC2-pMed1shRNA cells in the presence of 4-OHT (1 μM). *, P < 0.005, compared with untreated controls; **, P < 0.001, compared with untreated controls; #, P < 0.001, compared with untreated LCC2-pLKO.1; #*, P < 0.001, compared with tamoxifen-treated LCC2-pLKO.1. (C) Immunoblot analysis of MCF7-5/21 and MCF7-5/23 cells. (D) Clonogenicity of MCF7-5/21-pLKO.1, MCF7-5/21-pMed1shRNA, MCF7-5/23-pLKO.1 and MCF7-5/23- pMed1shRNA cells in the presence of 4-OHT (1 μM). *, P < 0.005, compared with untreated controls; **, P < 0.001, compared with untreated controls; #, P < 0.001, compared with untreated MCF7-5/23-pLKO.1; #*, P < 0.001, compared with tamoxifen-treated MCF7-5/23-pLKO.1.
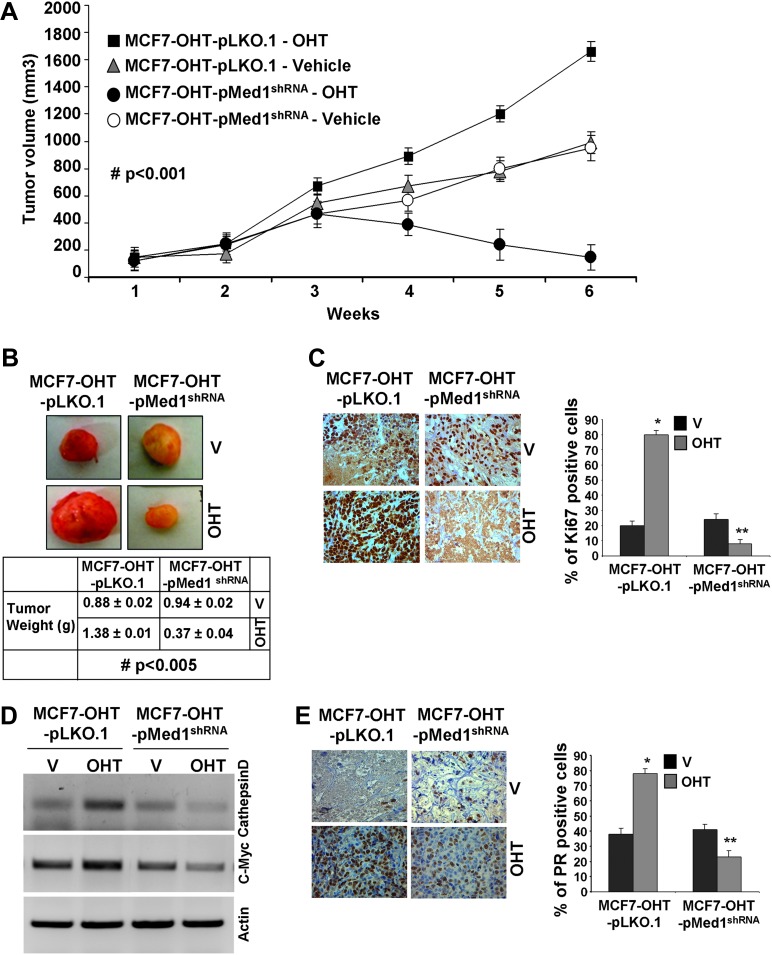
We next examined the importance of Med1 in tamoxifen resistance in vivo. MCF7-OHT-pLKO.1 tumors showed marked increase in tumor growth in the presence of tamoxifen treatment as compared with untreated (Figure 7A and B). MCF7-OHT cells also showed an increased tumor growth in response to tamoxifen treatment (data not shown). Importantly, MCF7-OHT-pMed1shRNA tumors exhibited significant inhibition of tumor growth in the presence of tamoxifen treatment showing that Med1 knockdown efficiently reversed tamoxifen resistance (Figure 7A and B). MCF7-OHT-pMed1shRNA tumors always appeared less vascularized. The Ki-67 antigen is a useful marker of cell proliferation (40). Upregulation of proliferation marker, Ki67, was observed in tamoxifen-treated MCF7-OHT-pLKO.1 tumors in comparison with tamoxifen-treated MCF7-OHT-pMed1shRNA tumors (Figure 7C). We further analyzed the alterations in ER-responsive gene expression in tumor samples and found overexpression of c-myc and cathepsin D in tamoxifen-treated MCF7-OHT-pLKO.1 tumors. Tamoxifen treatment did not induce expression of c-myc and cathepsin D in MCF7-OHT-pMed1shRNA tumors (Figure 7D). Immunohistochemistry analysis of tumor samples showed increased expression of ER-responsive gene, progesterone receptor (PR) in tamoxifen-treated MCF7-OHT-pLKO.1 tumors in comparison with tamoxifen-treated MCF7-OHT-pMed1shRNA tumors (Figure 7E).
Fig. 7.
Stable knockdown of Med1 reverses tamoxifen resistance in tamoxifen-resistant tumors in vivo. MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA cells derived tumors were developed in nude mice and treated with vehicle and 4-OHT. (A) Tumor growth #, P < 0.001, comparing tamoxifen-treated MCF7-OHT-pLKO.1 with tamoxifen-treated MCF7-OHT-pMed1shRNA. (B) Representative tumor images and tumor weight. (C) Immunohistochemical analysis using anti-Ki-67 antibody. Bar diagram shows quantitation of Ki-67 expression in tumors. Columns, mean (n = 6); *, P < 0.01, compared with vehicle controls; **, P < 0.001, compared with tamoxifen-treated MCF7-OHT-pLKO.1. (D) Semi-quantitative reverse transcription–PCR analysis of messenger RNA expression levels of c-Myc and cathepsin D in tamoxifen-treated and untreated MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA tumors. (E) Immunohistochemical analysis of PR expression in MCF7-OHT-pLKO.1 and MCF7-OHT-pMed1shRNA untreated and treated with tamoxifen. Bar diagram shows quantitation of PR expression in tumors. Columns, mean (n = 6); *, P < 0.005, compared with vehicle controls; **, P < 0.001, compared with tamoxifen-treated MCF7-OHT-pLKO.1.
Increased Med1 expression associates with higher tumor grade, increased clinical risk and recurrence after tamoxifen treatment
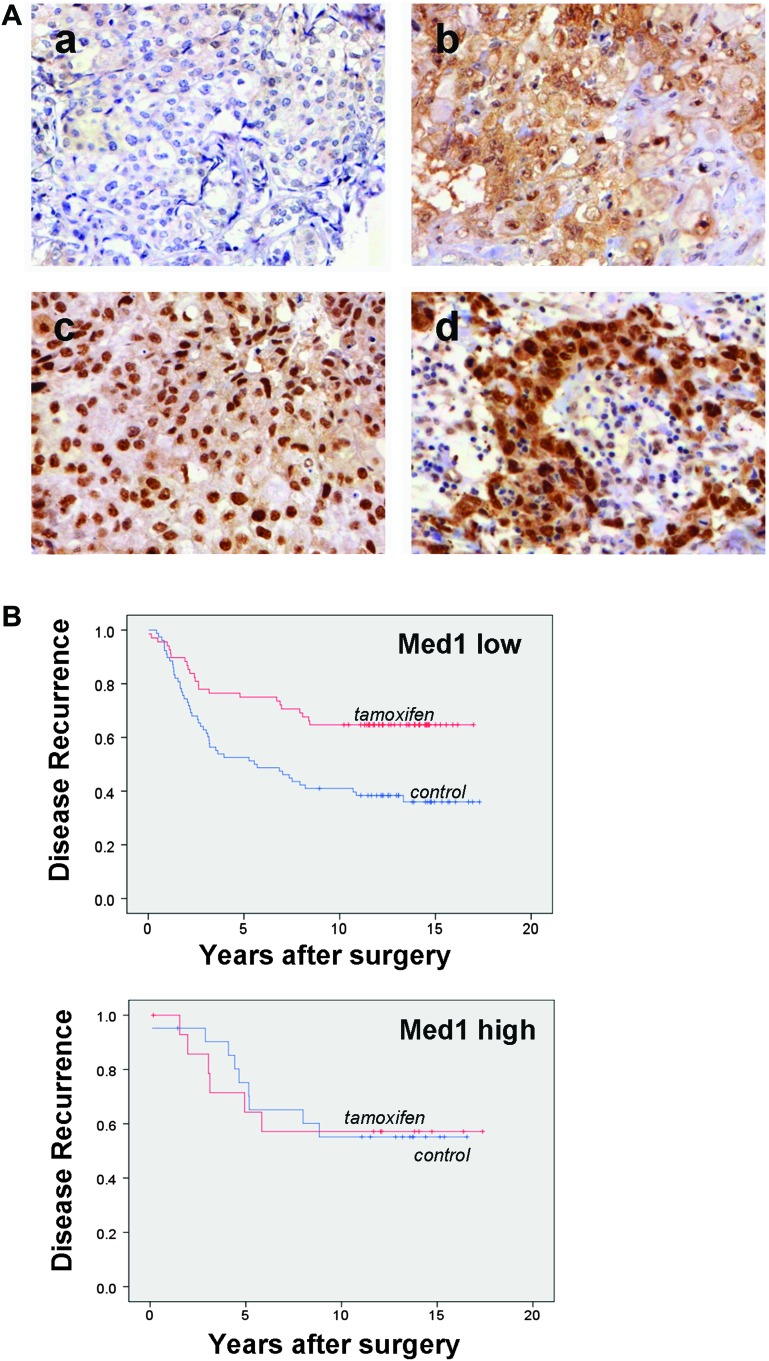
Our observation that tamoxifen-resistant cells exhibit increased Med1 expression and targeted downregulation of Med1 in tamoxifen-resistant cells reverses acquired tamoxifen resistance in vitro and in vivo raised the question whether Med1 might play an important role in the tamoxifen resistance in human breast cancer. If this were the case, we reasoned that women with breast cancers expressing higher levels of Med1 might relapse after tamoxifen treatment. We examined Med1 protein levels of hormone receptor-positive breast cancers in a cohort of 564 patients treated with adjuvant tamoxifen (n = 276) or no systemic treatment (n = 288) (30). Figure 8A shows representative immunohistochemical staining of Med1 in breast tumors indicating variations in positivity among tumor cells. In these discovery sets of tissues, statistically significant association between higher expression of Med1 and poorer recurrence-free survival was observed (HR = 0.65, 95% confidence interval 0.36–0.90, P = 0.005). In the Med1 low subgroup, patients treated with tamoxifen had a significantly better recurrence-free survival compared with control patients in the same subgroup (HR = 0.45, 95% confidence interval 0.28–0.74, P = 0.001) (Figure 8B and Supplementary Figure 4, available at Carcinogenesis Online). These data suggested that elevated expression levels of total Med1 (both nuclear and cytoplasmic) associate with the development of tamoxifen resistance in breast cancer patients. Although our in vitro data suggest that Med1 has to act in the nucleus to mediate tamoxifen resistance, further studies using larger set of tissues are required to examine if high nuclear or cytoplasmic Med1 expression levels alone associate with tamoxifen resistance. In addition, we performed in silico analysis of breast cancer utilizing published profiling studies for Med1 expression levels. Analysis of hormone receptor-positive, early-stage invasive breast cancers from 60 women uniformly treated with adjuvant tamoxifen alone (41) showed a distinct subset of tumors with tamoxifen recurrence exhibited higher Med1 expression (Supplementary Figure 5A, available at Carcinogenesis Online). We also found significant association between higher Med1 expression and low survival; P value = 0.02 and HR = 1.54. Patients with higher Med1 expression were at greater risk of mortality than patients with low Med1 expression (Supplementary Figure 5B, available at Carcinogenesis Online). Also, higher Med1 expression was associated with high tumor grade; P value = 0.029 (Supplementary Figure 5C, available at Carcinogenesis Online).
Fig. 8.
Increased Med1 expression associates with higher tumor grade and recurrence after tamoxifen treatment. (A) Examples of immunohistochemical Med1 staining of breast cancer. Breast cancer with a) low cytoplasmic and nuclear Med1 expression, b) high cytoplasmic but low nuclear Med1, c) high nuclear but low cytoplasmic and d) both high cytoplasmic and nuclear Med1. (B) Kaplan–Meier plots of breast cancer recurrences in relation to total Med1 status (high versus low) and treatment. Recurrence-free survival of patients was assessed among those who had been randomly assigned to tamoxifen or to no adjuvant systemic tamoxifen (control) treatment.
Collectively, these studies showed that Med1 overexpression is indispensable for acquired tamoxifen resistance as ablation of Med1 reverses tamoxifen resistance.
Discussion
Breast cancer progression in the presence of tamoxifen is a cardinal manifestation of the development of acquired tamoxifen resistance and represents the principal cause of poor prognosis and death in originally good prognosis hormone-responsive breast tumors. It is of great therapeutic interest to identify the molecular pathways that permit tumor cells to evade therapeutic effects of tamoxifen and continue malignant growth. In this report, we present in vitro and in vivo evidence for a molecular mechanism that contributes to the development of tamoxifen resistance. Our data demonstrate that Med1 is spontaneously upregulated during acquired tamoxifen-resistance development and that tamoxifen-resistant phenotype exhibits persistent activation of ERK and Med1 phosphorylation that has been linked to Med1 coactivation function. Tamoxifen-resistant MCF7-OHT clones exhibit slower growth in response to E2 in culture and in vivo in comparison to MCF7-P cells. Similar to MCF7-P cells, MCF7-OHT clones form ER:Med1 complex in response to E2. We hypothesize that differential response of MCF7-OHT clones to E2 stimulation is due to the difference between E2-responsive gene profiles among MCF7 and tamoxifen-resistant clones. Different groups of genes may be targeted by ER:Med1 complex in tamoxifen-resistant cells and parental cells in response to E2.
Consistent with a causal role for Med1 in acquired tamoxifen resistance, Med1 knockdown is sufficient to resensitize tamoxifen-resistant cells to tamoxifen-induced growth inhibition and gene expression modulation and to reverse acquired tamoxifen resistance in vivo.
Estrogen-bound ER stimulates transcription of ER-responsive genes utilizing various coactivator complexes (42), whereas tamoxifen-bound ER inhibits transcription via posing a steric impediment inhibiting coactivator recruitment while promoting binding of corepressor complexes (43–45). Importance of coregulators in ER function indicates that alteration in expression and/or activity of coregulatory molecules may potentially alter transcriptional response and downstream biological effects of ER during the development of tamoxifen resistance. Mediator is a conserved multisubunit complex that acts as a functional crossing point between DNA-bound regulatory transcription complexes and basal transcription machinery (46). Overexpression of Med1 in tamoxifen-resistant cells can potentially enhance mediator function leading to altered gene expression in response to tamoxifen. In humans, though Med1 is a critical component of the mediator complex but it is only variably associated with mediator (47,48). The mechanisms regulating its entry into the core mediator complex involves phosphorylation (49). A novel finding of this study is that tamoxifen-resistant cells have acquired overexpression of Med1 as well as increased phosphorylation of Med1 and ER via importunate ERK activation. Increased phosphorylation of Med1 increases its association with mediator complex hence transactivation functions. Further mechanistic studies will reveal if hyper-phosphorylated ER and Med1 exhibit increased interaction and significantly increased recruitment to ER-responsive gene promoters in response to tamoxifen. In light of the classical genomic action of ER including ligand binding, nuclear translocation, direct binding to ERE affecting gene expression (42), the functional significance of Med1 phosphorylation during tamoxifen resistance has to be considered. It is possible that Med1 phosphorylation facilitates a feed-forward action of ER in which coactivators as well as ER becomes activated in the presence of tamoxifen. Intrinsic to this model is the ERK-directed activation of ER, which has been shown to increase its nuclear translocation, interaction with coregulators, transcriptional activity and receptor turn over (39). Development of tamoxifen resistance attains ERK activation that may directly lead to phosphorylation of Med1 and hence promotes its association with mediator complex. Simultaneously, tamoxifen-bound hyperphosphorylated ER may bind to ERE and promote Med1 binding in tamoxifen-resistant cells. In this model, intrinsic abundance of phosphorylated Med1 and ER may potentiate subsequent ER-Med1 complex formation upon tamoxifen stimulus so that a maximal transcription response is rapidly achieved and sustained. Future studies should reveal whether other cellular signal transduction pathways target Med1 and ER for regulatory phosphorylation and affect their functional roles in acquired tamoxifen resistance.
Importantly, the association that we detected between high Med1 expression in a subset of breast tumors and recurrence after tamoxifen treatment in humans implies that Med1 overexpression may play a role in promoting tamoxifen resistance. Consistent with this, we have demonstrated that Med1 knockdown is sufficient to reverse tamoxifen resistance in mice tumor studies. We also found that Med1 overexpression was associated with increased aggressiveness and higher clinical risk in human tumor studies. It is worth noting that the association between Med1 overexpression and aggressiveness is consistent with our observation that mice tumors exhibiting intact Med1 overexpression levels showed higher Ki-67 staining, indicative of increased proliferation, in response to tamoxifen. Med1 overexpression also correlated with higher tumor grade in a subset of breast tumors across a panel of breast cancer profiling studies comprising multiple microarray experiments. To further validate our experimental findings, we analyzed Med1 in a large cohort of breast cancer patients from a randomized controlled trial for adjuvant tamoxifen treatment with long-term follow-up. The median follow-up time for patients was 13.9 years. Notably, in the Med1 low subgroup, patients treated with tamoxifen had a significantly better recurrence-free survival compared with control patients in the same subgroup. Multivariate interaction analyses showed that total Med1 was significantly associated with an altered tamoxifen response. As such, based upon our findings, we predict that a large-scale analysis of recurrent human breast cancers after tamoxifen treatment will reveal elevated levels of Med1 compared with tamoxifen-responsive tumors showing no recurrence.
ER can get phosphorylated at several amino acid residues encompassing all major structural domains affecting ER ligand binding, dimerization, nuclear localization, DNA binding, coactivator recruitment and transcriptional activation (50). Studies from several groups support the hypothesis that ligand-independent phosphorylation of ER may cause tamoxifen resistance. On one hand, kinases such as ERK and Akt have been shown to phosphorylate ER at S-118 and S-167 in a ligand-independent manner (51, 52); on the other hand, increased activation/accumulation of ERK and Akt have been associated with poor clinical outcome for breast cancer patients treated with tamoxifen (53, 54), providing indirect evidence for ER phosphorylation in tamoxifen resistance. Our studies found higher levels of phosphorylated ER (S-118 and S-167) in acquired tamoxifen-resistant cells. Multiple studies in recent years have analyzed phospho-specific sites in ER using human breast biopsy samples [summarized in (50)]. Increased levels of both phospho-ER (S-118) and phospho-ER (S-167) were found in secondary versus primary tumors suggesting that phospho-ER may be a useful marker for metastatic breast cancer (55). In contrast, phospho-ER (S-118) or phospho-ER (S-167) associated with less aggressive and more differentiated tumors (56, 57). Most of the studies to date have determined phospho-ER using primary breast tumors and shown associations with overall clinical prognosis and de novo endocrine resistance. Future studies utilizing clinical samples relevant for acquired tamoxifen resistance might show tighter associations between specific ER phosphorylation and development of acquired resistance.
For many years, oncologists have used ER/PR clinically to select treatment options. In spite of this, 25% of ER+/PR+ tumors, 66% of ER+/PR− tumors and 55% of ER−/PR+ tumors either present de novo resistance or develop acquired resistance (3). To date, there are no dependable markers to identify patients who are at risk of tumor recurrence in the setting of adjuvant tamoxifen. The ability to predict the development of tamoxifen resistance in hormone-positive tumors can be extremely useful in facilitating the use of alternative hormonal therapies such as the aromatase inhibitors letrozole and anastrozole (58–60), chemotherapeutic agents (61) or inhibitors of other signaling pathways (62–64) in this subset of patients hence improving their clinical outcome. Our results suggest the utility of Med1 expression levels in identifying subset of patients who are at risk for tumor recurrence in the setting of tamoxifen therapy and who may benefit from alternative therapeutic approaches. Beyond the ability to predict tamoxifen resistance, our findings raise the important possibility that acquired tamoxifen resistance might be prevented by therapeutic agents that inhibit Med1, either by preventing its upregulation or by inhibiting its function. For example, inhibition of Med1 phosphorylation by ERK inhibition inhibits transactivation function of Med1. Hence, development of cancer therapeutics that targets these upstream pathways may be useful. Future studies utilizing small molecule inhibitors for ERK activation are required to show if ERK inhibition is sufficient to reverse tamoxifen resistance in vivo. A definite causal role of Med1 cannot be yet confirmed in human breast tumor tamoxifen resistance since no specific drugs are available to specifically target Med1. Nonetheless, our findings may potentially open new avenues of research into the role of Med1 in the prediction and therapeutic targeting of acquired tamoxifen resistance.
Supplementary material
Supplementary Materials, Figures S1–S5 and Table 1 can be found at http://carcin.oxfordjournals.org/.
Funding
National Institute of Health (NIH) (K01DK076742 and R03DK089130) to NKS; NIH (R01CA131294) to DS; Komen for the cure (BCTR0503526) to DS; Swedish Cancer Society and Breakthrough Breast Cancer to GL.
Supplementary Material
Acknowledgments
The authors would like to thank Sooryanarayana Varambally for help with Oncomine data analysis, Elise Nilsson for technical assistance and the South Swedish and South-East Swedish Breast Cancer Groups.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- EGF
epidermal growth factor
- ER
estrogen receptor
- ERE
estrogen response element
- ERK
extracellular signal-regulated kinase
- OHT
hydroxytamoxifen
- shRNA
short hairpin RNA
References
- 1.Platet N, et al. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit. Rev. Oncol. Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Baumann CK, et al. Clinical use of selective estrogen receptor modulators and down regulators with the main focus on breast cancer. Minerva Ginecol. 2009;61:517–539. [PubMed] [Google Scholar]
- 3.Musgrove EA, et al. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 4.Arpino G, et al. Molecular mechanism and clinical implications of endocrine therapy resistance in breast cancer. Oncology. 2009;77(suppl. 1):23–37. doi: 10.1159/000258493. [DOI] [PubMed] [Google Scholar]
- 5.Welboren WJ, et al. Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated? Endocr. Relat. Cancer. 2009;16:1073–1089. doi: 10.1677/ERC-09-0086. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, et al. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang YK, et al. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl Acad. Sci. USA. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belakavadi M, et al. Role of the mediator complex in nuclear hormone receptor signaling. Rev. Physiol. Biochem. Pharmacol. 2006;156:23–43. doi: 10.1007/s10254-005-0002-0. [DOI] [PubMed] [Google Scholar]
- 9.Chadick JZ, et al. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Conaway JW, et al. The mammalian Mediator complex. FEBS Lett. 2005;579:904–908. doi: 10.1016/j.febslet.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Ren Y, et al. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 2000;20:5433–5446. doi: 10.1128/mcb.20.15.5433-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford SE, et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 13.Frade R, et al. RB18A, whose gene is localized on chromosome 17q12-q21.1, regulates in vivo p53 transactivating activity. Cancer Res. 2000;60:6585–6589. [PubMed] [Google Scholar]
- 14.Gordon DF, et al. MED220/thyroid receptor-associated protein 220 functions as a transcriptional coactivator with Pit-1 and GATA-2 on the thyrotropin-beta promoter in thyrotropes. Mol. Endocrinol. 2006;20:1073–1089. doi: 10.1210/me.2005-0115. [DOI] [PubMed] [Google Scholar]
- 15.Stumpf M, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl Acad. Sci. USA. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udayakumar TS, et al. Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J. Biol. Chem. 2006;281:14691–14699. doi: 10.1074/jbc.M600163200. [DOI] [PubMed] [Google Scholar]
- 17.Wada O, et al. BRCA1 function mediates a TRAP/DRIP complex through direct interaction with TRAP220. Oncogene. 2004;23:6000–6005. doi: 10.1038/sj.onc.1207786. [DOI] [PubMed] [Google Scholar]
- 18.Sharma D, et al. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66:6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duquesnes N, et al. The EGF receptor activates ERK but not JNK Ras-dependently in basal conditions but ERK and JNK activation pathways are predominantly Ras-independent during cardiomyocyte stretch. Int. J. Biochem. Cell Biol. 2009;41:1173–1181. doi: 10.1016/j.biocel.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Brunner N, et al. MCF7/LCC2: a 4-hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Res. 1993;53:3229–3232. [PubMed] [Google Scholar]
- 21.Hu XF, et al. Circumvention of tamoxifen resistance by the pure anti-estrogen ICI 182,780. Int. J. Cancer. 1993;55:873–876. doi: 10.1002/ijc.2910550529. [DOI] [PubMed] [Google Scholar]
- 22.Taliaferro-Smith L, et al. LKB1 is required for adiponectin-mediated modulation of AMPK-S6K axis and inhibition of migration and invasion of breast cancer cells. Oncogene. 2009;28:2621–2633. doi: 10.1038/onc.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saxena NK, et al. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena NK, et al. Leptin-induced growth stimulation of breast cancer cells involves recruitment of histone acetyltransferases and mediator complex to CYCLIN D1 promoter via activation of Stat3. J. Biol. Chem. 2007;282:13316–13325. doi: 10.1074/jbc.M609798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, et al. Neurite-localized estrogen receptor-alpha mediates rapid signaling by estrogen. J. Neurosci. Res. 2003;74:1–11. doi: 10.1002/jnr.10725. [DOI] [PubMed] [Google Scholar]
- 26.Sharma D, et al. Temporal formation of distinct thyroid hormone receptor coactivator complexes in HeLa cells. Mol. Endocrinol. 2000;14:2001–2009. doi: 10.1210/mend.14.12.0567. [DOI] [PubMed] [Google Scholar]
- 27.Sharma D, et al. Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol. Endocrinol. 2005;19:1740–1751. doi: 10.1210/me.2004-0011. [DOI] [PubMed] [Google Scholar]
- 28.Sharma D, et al. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl Acad. Sci. USA. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keen JC, et al. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2'-deoxycytidine. Breast Cancer Res. Treat. 2003;81:177–186. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 30.Ryden L, et al. Two years of adjuvant tamoxifen in premenopausal patients with breast cancer: a randomised, controlled trial with long-term follow-up. Eur. J. Cancer. 2005;41:256–264. doi: 10.1016/j.ejca.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 31.Tsuneizumi M, et al. Overrepresentation of the EBAG9 gene at 8q23 associated with early-stage breast cancers. Clin. Cancer Res. 2001;7:3526–3532. [PubMed] [Google Scholar]
- 32.Ellis MJ, et al. Insulin-like growth factors in human breast cancer. Breast Cancer Res. Treat. 1998;52:175–184. doi: 10.1023/a:1006127621512. [DOI] [PubMed] [Google Scholar]
- 33.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchiya F, et al. Molecular cloning and characterization of mouse EBAG9, homolog of a human cancer associated surface antigen: expression and regulation by estrogen. Biochem. Biophys. Res. Commun. 2001;284:2–10. doi: 10.1006/bbrc.2001.4892. [DOI] [PubMed] [Google Scholar]
- 35.Augereau P, et al. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol. Endocrinol. 1994;8:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 36.Dubik D, et al. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene. 1992;7:1587–1594. [PubMed] [Google Scholar]
- 37.Umayahara Y, et al. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J. Biol. Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- 38.Pandey PK, et al. Activation of TRAP/mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell. Biol. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- 40.Walker RA. Immunohistochemical markers as predictive tools for breast cancer. J. Clin. Pathol. 2008;61:689–696. doi: 10.1136/jcp.2006.041830. [DOI] [PubMed] [Google Scholar]
- 41.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Iwase H. Molecular action of the estrogen receptor and hormone dependency in breast cancer. Breast Cancer. 2003;10:89–96. doi: 10.1007/BF02967632. [DOI] [PubMed] [Google Scholar]
- 43.Hall JM, et al. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol. Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 44.Gao X, et al. The roles of sex steroid receptor coregulators in cancer. Mol. Cancer. 2002;1:7. doi: 10.1186/1476-4598-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hart SM. Modulation of nuclear receptor dependent transcription. Biol. Res. 2002;35:295–303. doi: 10.4067/s0716-97602002000200021. [DOI] [PubMed] [Google Scholar]
- 46.Yuan CX, et al. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik S, et al. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taatjes DJ, et al. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell. 2004;14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Belakavadi M, et al. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol. Cell. Biol. 2008;28:3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy LC, et al. Clinical significance of estrogen receptor phosphorylation. Endocr. Relat. Cancer. 2011;18:R1–R14. doi: 10.1677/ERC-10-0070. [DOI] [PubMed] [Google Scholar]
- 51.Kato S, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 52.Campbell RA, et al. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 53.Schiff R, et al. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin. Cancer Res. 2004;10:331S–336S. doi: 10.1158/1078-0432.ccr-031212. [DOI] [PubMed] [Google Scholar]
- 54.Kirkegaard T, et al. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J. Pathol. 2005;207:139–146. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita H, et al. Phosphorylation of estrogen receptor alpha serine 167 is predictive of response to endocrine therapy and increases postrelapse survival in metastatic breast cancer. Breast Cancer Res. 2005;7:R753–R764. doi: 10.1186/bcr1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy LC, et al. Phospho-serine-118 estrogen receptor-alpha expression is associated with better disease outcome in women treated with tamoxifen. Clin. Cancer Res. 2004;10:5902–5906. doi: 10.1158/1078-0432.CCR-04-0191. [DOI] [PubMed] [Google Scholar]
- 57.Jiang J, et al. Phosphorylation of estrogen receptor-alpha at Ser167 is indicative of longer disease-free and overall survival in breast cancer patients. Clin. Cancer Res. 2007;13:5769–5776. doi: 10.1158/1078-0432.CCR-07-0822. [DOI] [PubMed] [Google Scholar]
- 58.Buzdar AU. Anastrozole: a new addition to the armamentarium against advanced breast cancer. Am. J. Clin. Oncol. 1998;21:161–166. doi: 10.1097/00000421-199804000-00014. [DOI] [PubMed] [Google Scholar]
- 59.Ellis MJ, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1- and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: evidence from a phase III randomized trial. J. Clin. Oncol. 2001;19:3808–3816. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 60.Goss PE, et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N. Engl. J. Med. 2003;349:1793–1802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- 61.Colleoni M, et al. Preoperative and perioperative chemotherapy with 5-fluorouracil as continuous infusion in operable breast cancer expressing a high proliferation fraction: cytotoxic treatment during the surgical phase. Ann. Oncol. 2003;14:1477–1483. doi: 10.1093/annonc/mdg411. [DOI] [PubMed] [Google Scholar]
- 62.Wong ST. Emerging treatment combinations: integrating therapy into clinical practice. Am. J. Health Syst. Pharm. 2009;66:S9–S14. doi: 10.2146/ajhp090439. [DOI] [PubMed] [Google Scholar]
- 63.Adamo V, et al. Overview and new strategies in metastatic breast cancer (MBC) for treatment of tamoxifen-resistant patients. Ann. Oncol. 2007;18(suppl. 6):vi53–vi57. doi: 10.1093/annonc/mdm225. [DOI] [PubMed] [Google Scholar]
- 64.Arpino G, et al. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr. Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.