Abstract
It is known that an E146D site-directed variant of the Azotobacter vinelandii iron protein (Fe protein) is specifically defective in its ability to participate in iron-molybdenum cofactor (FeMoco) insertion. Molybdenum-iron protein (MoFe protein) from the strain expressing the E146D Fe protein is partially (≈45%) FeMoco deficient. The “free” FeMoco that is not inserted accumulates in the cell. We were able to insert this “free” FeMoco into the partially pure FeMoco-deficient MoFe protein. This insertion reaction required crude extract of the ΔnifHDK A. vinelandii strain CA12, Fe protein and MgATP. We used this as an assay to purify a required “insertion” protein. The purified protein was identified as GroEL, based on the molecular mass of its subunit (58.8 kDa), crossreaction with commercially available antibodies raised against E. coli GroEL, and its NH2-terminal polypeptide sequence. The NH2-terminal polypeptide sequence showed identity of up to 84% to GroEL from various organisms. Purified GroEL of A. vinelandii alone or in combination with MgATP and Fe protein did not support the FeMoco insertion into pure FeMoco-deficient MoFe protein, suggesting that there are still other proteins and/or factors missing. By using GroEL-containing extracts from a ΔnifHDK strain of A. vinelandii CA12 along with FeMoco, Fe protein, and MgATP, we were able to supply all required proteins and/or factors and obtained a fully active reconstituted E146D nifH MoFe protein. The involvement of the molecular chaperone GroEL in the insertion of a metal cluster into an apoprotein may have broad implications for the maturation of other metalloenzymes.
The biological reduction of dinitrogen to ammonia is catalyzed by the complex metalloenzyme nitrogenase (for recent reviews see refs. 1–6). This enzyme is composed of two proteins that are purified separately. The smaller of the two, the iron protein (Fe protein), is a 60,000 Mr dimer of two identical subunits encoded by the nifH gene. Each of the subunits has a binding site for MgATP, and the two subunits are bridged by a single [4Fe-4S] cluster. The molybdenum-iron protein (MoFe protein) is much more complex. It is a 230,000 Mr α2β2 tetramer with the α and β subunits encoded by the nifD and nifK genes, respectively. Each α subunit contains a [Mo-7Fe-9S-homocitrate] cluster designated FeMoco that serves as the site of dinitrogen binding and reduction by the enzyme. Bridged between each αβ subunit pair is another complex [8Fe-7S] cluster, designated the P-cluster, which is believed to mediate electron transfer from the Fe protein to FeMoco. This study concerns the assembly of the MoFe protein of nitrogenase and the role that the Fe protein plays in that process.
It is currently well established that the Fe protein has at least three separate functions in the cell. First, the Fe protein serves as a specific electron donor for the MoFe protein to support dinitrogen reduction (for reviews see refs. 1–6). To carry out this function, the reduced Fe protein first binds two molecules of MgATP and then undergoes a global conformational change before forming a very specific complex with the MoFe protein. Electrons are then transferred from the Fe protein to the MoFe protein in a reaction that is coupled to MgATP hydrolysis. The oxidized Fe protein then dissociates from the MoFe protein. Second, the Fe protein is somehow involved in the initial biosynthesis of FeMoco (for reviews see refs. 5 and 7–13). The complete pathway for FeMoco biosynthesis is not known, but it is well established that FeMoco is synthesized separately from the MoFe protein polypeptides. The role the Fe protein plays in the initial biosynthesis of FeMoco is not known, but mutant studies have established that to carry out this function the Fe protein does not need to undergo the MgATP-induced conformational change, it does not need to form the normal complex with the MoFe protein, it does not need to transfer electrons to any protein, and it does not need to hydrolyze MgATP (14–20).
The third Fe protein function, which is the subject of the current study, has to do with the final assembly of the MoFe protein, which includes the insertion of the separately synthesized iron-molybdenum cofactor (FeMoco) into the FeMoco-deficient MoFe protein. This final maturation of the MoFe protein occurs in a series of steps. First, a P-cluster-containing, but FeMoco-deficient, MoFe protein is synthesized that has the FeMoco site somehow inaccessible to FeMoco insertion (21–24). Strains that have deletions in the nifH gene accumulate this form of the MoFe protein. The FeMoco-deficient MoFe protein is then converted to a form with the FeMoco site accessible, and finally FeMoco is inserted. There may be a number of proteins and/or factors involved in these final assembly steps, but to date it has been established only that the reaction requires the Fe protein, MgATP, and, in Azotobacter vinelandii, a FeMoco chaperone-insertase protein designated gamma (25). Here, we show that the final assembly of the MoFe protein also requires the molecular chaperone GroEL. The requirement of GroEL for the insertion of a metal cluster into an apoprotein has not previously been observed and may represent a widespread mechanism for the maturation of metalloenzymes in general.
Materials and Methods
Unless otherwise noted, all chemicals and reagents were obtained from Fisher Scientific, Baxter Scientific Products (McGaw Park, IL), or Sigma.
Cell Growth and Protein Purification.
Wild-type and E146D nifH A. vinelandii strains were grown in 180-liter batches in a 200-liter New Brunswick Scientific fermentor under N2-fixing conditions on Burk's minimal media. The growth rate was measured by cell density at 436 nm by using a Spectronic 20 Genesys (Spectronic, Westbury, NY). The cells were harvested at the end of the exponential phase by using a flow-through centrifugal harvester (Cepa, Lahr/Schwarzwald, Germany). Published methods were used for the purification of wild-type Fe protein (26), wild-type MoFe protein (27), and the MoFe protein synthesized by the strain expressing the E146D Fe protein (28). Early in the purification, the E146D nifH MoFe protein is eluted with a linear 0.1–0.5 M NaCl gradient from a DEAE-cellulose (Whatman) ion-exchange column. This fraction was designated as partially pure E146D nifH MoFe protein.
A. vinelandii strain CA12 (ΔnifHDK) was grown on Burk's minimal medium supplemented with 2 mM ammonium acetate. After the consumption of the ammonia, the cells were derepressed for 3 hr, followed by harvesting as described above. The cell paste was washed with 50 mM Tris⋅HCl (pH 8.0) and kept on dry ice until needed.
GroEL was purified by using the following method. First, 500 g of cell paste of A. vinelandii strain CA12 (ΔnifHDK) was thawed in 1 liter of 50 mM Tris⋅HCl (pH 8.0), and degassed thoroughly. After adding 2 mM sodium dithionite and 10 μg/ml DNase, the cells were then broken by passing them through a Gaulin cell homogenizer two times at 5,000 p.s.i. to make a crude extract. The crude extract was degassed for an additional 2 hr, ultracentrifuged for 90 min at 35,000 rpm in a Beckman Ti 45 fixed-angle rotor, and then loaded onto a 5 × 25-cm DEAE-cellulose ion-exchange column. Throughout the purification procedure, the enzyme activity of 1–2 mg protein was measured as described later. GroEL did not bind to the DEAE-cellulose column. The GroEL-containing “run through” fraction was concentrated in an ultrafiltration cell (Amicon) by using a YM3 Membrane (Millipore). The protein was then loaded onto 2.5 × 100-cm Ultrogel AcA34 and subsequently Ultrogel AcA54 (BioSepra, Marlborough, MA) gel filtration columns. The elution buffer used was 50 mM Tris⋅HCl (pH 8.0) 0.1 M NaCl. The purified GroEL was analyzed by SDS/PAGE.
FeMoco Insertion Assays.
The assays designed to test for the presence of “free” or uninserted FeMoco in partially pure E146D nifH MoFe protein contained, in a 0.35 ml total volume, 25 mM Tris⋅HCl (pH 7.4), 20 mM Na2S2O4, and 0.15 mg purified cofactor-deficient MoFe protein from ΔnifB strain DJ1143 (29). The insertion was started by the addition of isolated FeMoco in N-methyl formamide (NMF) or partially pure E146D nifH MoFe protein. The samples were incubated at 30°C for 30 min, and, subsequently, the enzyme activity of 0.1 ml of the insertion mixture was determined as described previously (27). The product was analyzed as published elsewhere (14).
Experiments that monitored the insertion of “free” or uninserted FeMoco into partially pure E146D nifH MoFe protein were used as an activity assay to purify the protein needed for assembly that later turned out to be GroEL. Assays contained, in 0.35 ml total volume, 25 mM Tris⋅HCl (pH 7.4), 2.4 mM ATP, 4.8 mM MgCl2, 30 mM creatine phosphate, 24 units of creatine kinase, 20 mM Na2S2O4, 0.2 mg wild-type Fe protein, and 0.12 mg partially pure E146D nifH MoFe protein. The reactions were started by injecting 1–2 mg crude extract of A. vinelandii CA12 (ΔnifHDK) or 50 μg purified GroEL. After incubation at 30°C for 30 min, the insertion was stopped by addition of 40 nmol of (NH4)2MoS4. (NH4)2MoS4 is known to block FeMoco insertion into the FeMoco-deficient MoFe protein, probably by occupying the FeMoco site (30, 31). Subsequently, the enzyme activity was measured by C2H4 evolution. Those enzyme activity assays, containing 0.1 ml of the insertion mixture, were carried out as previously described (27). The product was analyzed as published elsewhere (14)
The assays designed to reconstitute pure E146D nifH MoFe protein contained, in a 0.7 ml total volume 25 mM Tris⋅HCl (pH 7.4), 1.2 mM ATP, 2.4 mM MgCl2, 15 mM creatine phosphate, 16 units of creatine kinase, 20 mM Na2S2O4, 0.12 mg wild-type Fe protein, 0.1 mg pure E146D nifH MoFe protein, and 2 mg crude extract of A. vinelandii CA12 (ΔnifHDK) or 50 μg purified GroEL. The insertion was started by the addition of isolated FeMoco in NMF. The samples were incubated at 30°C for 30 min, and, subsequently, the enzyme activity of 0.1 ml of the insertion mixture was determined as described previously (27). The product was analyzed as published elsewhere (14).
Results and Discussion
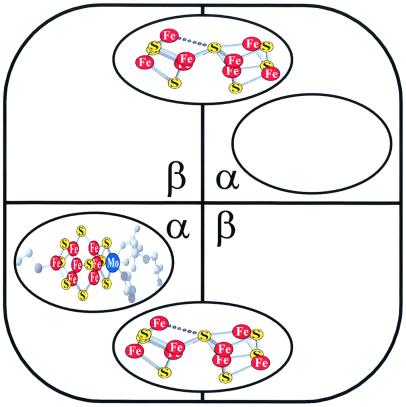
We have recently identified a region of the Fe protein around Glu146 that is specifically involved in the final assembly of the MoFe protein (28). An E146D Fe protein was shown to be fully functional as an electron donor to the MoFe protein and in the initial biosynthesis of FeMoco. This Fe protein variant, however, was partially defective in the final assembly process such that the A. vinelandii strain expressing the E146D Fe protein accumulated both uninserted FeMoco [probably bound to gamma (25)] and a FeMoco-deficient form of the MoFe protein. Analysis of the purified MoFe protein from that strain showed that it was ≈55% active because of the fact that only ≈55% of its FeMoco sites were occupied (28). Because FeMoco-deficent forms of the MoFe protein with all FeMoco sites vacant are notoriously unstable (24), whereas the E146D nifH MoFe protein is as stable as wild-type (28), it was concluded that the strain expressing the E146D Fe protein accumulated a small amount of fully active MoFe holoprotein whereas the bulk of the material is best represented as shown in Fig. 1, with one FeMoco site occupied and the other vacant.
Figure 1.
Schematic diagram of FeMoco-deficient MoFe protein expressed by E146D nifH A. vinelandii. Shown are the α and β subunits of the α2β2 tetramer. Each [8Fe-7S] P-cluster is bridged by one α and β subunit. The FeMoco has an overall stoichiometry of [Mo-7Fe-9S-homocitrate]. The FeMoco binding site is located in the α subunit. One binding site is occupied and the other one vacant. The MoFe protein shown represents ≈90% of the MoFe protein expressed by E146D nifH A. vinelandii.
FeMoco can be isolated from purified wild-type MoFe protein and studied as an independent entity in NMF, and it is this form of FeMoco that is generally used in in vitro MoFe protein assembly assays. With purified E146D nifH MoFe protein, however, the vacant FeMoco sites (Fig. 1) could not be filled in vitro simply by adding isolated FeMoco in NMF, the Fe protein, and MgATP (see below). This result was not surprising because we had previously shown that a FeMoco-deficient form of the MoFe protein synthesized by a nifH deletion strain could be activated in cell-free extracts on addition of FeMoco, Fe protein, and MgATP but could not be activated by using the same additions once it was purified (22–24). We also knew from the literature that gamma and possibly other proteins and/or factors were missing (24, 25).
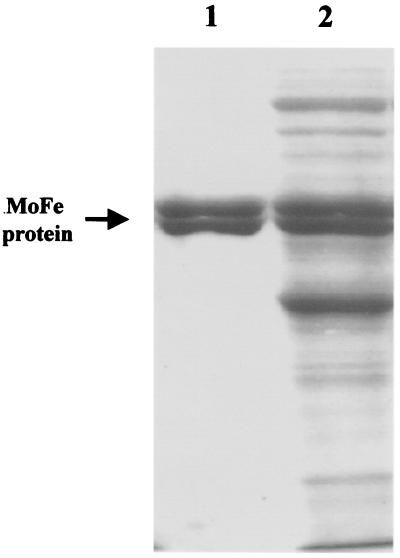
The specific activity of the purified, partially FeMoco-deficient, E146D nifH MoFe protein is 1110 ± 57 nmol C2H4 evolution/min/mg protein, which is ≈55% compared with wild-type MoFe protein (28). Our long-term goal was to fill the FeMoco sites in the E146D nifH MoFe protein to get 100% activity in vitro by using all purified proteins so that the function of the Fe protein and other proteins in this process could be established. Our strategy was to go back to an earlier stage of the purification where these other proteins might be present and to develop an assay for their purification. It was important to remove the defective E146D Fe protein so that wild-type Fe protein could be added in the in vitro assays. As described in Materials and Methods, the experiments described herein used a partially purified form of the E146D nifH MoFe protein that was completely separated from the E146D Fe protein. The specific activity of this partially purified MoFe protein is only ≈20%, compared with the purified E146D nifH MoFe protein (28). As shown in Fig. 2, the E146D nifH MoFe protein is at an early stage of the purification, and there are many proteins present, some of which, including gamma, may be required for the final assembly reaction. These proteins have not yet been identified, but they are present in all of the assays described below.
Figure 2.
Coomassie-stained 10% SDS/PAGE of partially purified E146D nifH MoFe protein of A. vinelandii. Lane 1 is 4 μg purified wild-type MoFe protein, and Lane 2 is 15 μg partially purified MoFe protein of E146D nifH A. vinelandii.
It was previously established that the A. vinelandii strain expressing the E146D Fe protein accumulates uninserted FeMoco [presumably on gamma (25)] that can be captured in vitro by using a purified His-tag form of a FeMoco-deficient MoFe protein (28). Using the same method, the data in Table 1 establish that this uninserted FeMoco is still present in solutions of the partially purified E146D nifH MoFe protein, so that the final assembly assays described below did not require the addition of isolated FeMoco in NMF.
Table 1.
Accumulation of FeMoco in the partially pure E146D nifH MoFe protein fraction
| Additions* | C2H4 evolution, nmol/min/mg ΔnifB MoFe protein |
|---|---|
| None | 0 ± 0 |
| Excess FeMoco† | 549 ± 59 |
| 0.6 mg partially pure E146D nifH MoFe protein | 263 ± 28‡ |
All assays contained 0.15 mg of purified FeMoco-deficient MoFe protein of ΔnifB A. vinelandii DJ1143.
Excess isolated FeMoco in NMF was added.
Background activity of partially pure E146D nifH MoFe protein alone has been subtracted.
As expected, the partially purified E146D nifH MoFe protein is ≈55% active, and we know from our study of the purified protein that this MoFe protein is missing about 45% of its FeMoco (28). This partially purified protein solution also contains sufficient uninserted FeMoco to fill all of the FeMoco sites and restore 100% activity. Nonetheless, the addition of wild-type Fe protein and MgATP did not result in the assembly of fully active MoFe protein. Therefore, some other protein(s) appeared to be required (Table 2). We decided to try to supply the missing protein(s) by using cell-free extracts from a ΔnifHDK strain of A. vinelandii, designated CA12 (32). As confirmed by activity assays and cross-reactivity with antibodies raised against the purified Fe protein and MoFe protein, these extracts do not contain any Fe protein or MoFe protein (data not shown). Because the Fe protein is required for FeMoco biosynthesis, the extracts also do not contain FeMoco (as confirmed by activity assays), but it was expected that they should still contain other proteins or factors that are needed for the insertion of isolated FeMoco into the FeMoco-deficient MoFe protein. As shown in Table 2, the addition of CA12 extracts to the partially purified E146D nifH MoFe protein was also necessary to allow the uninserted FeMoco to go into the vacant FeMoco sites (Fig. 1), increasing the specific activity by ≈45%.
Table 2.
Insertion of FeMoco into FeMoco-deficient partially pure MoFe protein of E146D nifH A. vinelandii
| Additions*
|
C2H4 evolution, nmol/min/mg protein | Activity, % | ||
|---|---|---|---|---|
| Fe protein | ATP | Other | ||
| − | − | − | 228 ± 32 | 56 |
| + | + | − | 215 ± 8 | 53 |
| + | + | Av CA12 extract† | 405 ± 54 | 100 |
| + | + | Pure GroEL | 399 ± 38 | 99 |
All insertion assays contained 0.12 mg partially pure MoFe protein of E146D nifH A. vinelandii. The insertion and activity portions of the assays were performed as described in Materials and Methods.
Av CA12, ΔnifHDK A. vinelandii strain CA12.
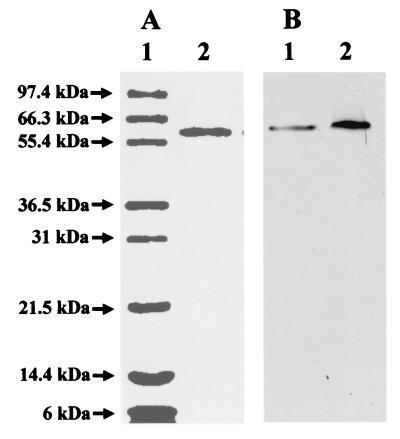
The results shown in Table 2 therefore established an assay for the purification of the protein from CA12 extracts that is required for the final assembly process. The purification procedure is described in detail in Materials and Methods. The required protein did not stick to DEAE cellulose, which is why it was not present in our partially purified E146D nifH MoFe protein solutions. Fig. 3A shows a one-dimensional Coomassie-stained gel of the purified protein that is required for the FeMoco insertion reaction. As shown, the protein has a subunit molecular weight of 58,800. Fig. 4 compares the NH2-terminal sequence of the protein to other proteins in the database. Its identification as A. vinelandii GroEL was also confirmed by crossreaction with commercially available antibodies raised against Escherichia coli GroEL (Fig. 3B). Purified GroEL of A. vinelandii was able to insert FeMoco into the partially purified E146D nifH MoFe protein, increasing the activity by ≈45% (Table 2).
Figure 3.
Coomassie-stained 12% SDS/PAGE (A). Lane 1 is 5 μg Mark12 wide range protein standard (NOVEX, San Diego). The molecular masses of the standard proteins are indicated. Lane 2 is 1 μg purified GroEL of A. vinelandii (molecular weight = 58,800). Western blot with commercially available antibodies raised against E. coli GroEL (B). Lane 1 is 20 μg of crude extract of A. vinelandii strain CA12 (ΔnifHDK), and lane 2 is 1 μg purified GroEL of A. vinelandii.
Figure 4.
Comparison of the NH2-terminal polypeptide sequence of GroEL of A. vinelandii with those of other organisms. Thirty-two amino acids of GroEL of A. vinelandii have been sequenced and compared with known sequences by using blast (NCBI, Bethesda, MD). NH2-terminal polypeptide sequences of the highest similarities are shown. P. aeruginosa = Pseudomonas aeruginosa, P. stutzeri = Pseudomonas stutzeri, P. putida = Pseudomonas putida, A. salmonicida = Aeromonas salmonicida, and P. multocida = Pasteurella multocida.
As mentioned earlier, the vacant FeMoco sites of purified E146D nifH MoFe protein (Fig. 1) could not be filled in vitro simply by adding isolated FeMoco in NMF, the Fe protein, and MgATP (Table 3). We assumed that gamma and possibly other proteins and/or factors were missing (24, 25). Having established that GroEL is one of the missing proteins, we tested whether the addition of purified GroEL is sufficient to insert FeMoco into purified E146D nifH MoFe protein. Purified GroEL of A. vinelandii alone or in combination with MgATP and Fe protein did not support the FeMoco insertion reaction (Table 3), demonstrating that there are still other proteins and/or factors missing. By using GroEL-containing extracts from a ΔnifHDK strain of A. vinelandii CA12, along with FeMoco, Fe protein, and MgATP, we were able to supply all required proteins and/or factors and obtained a fully active reconstituted E146D nifH MoFe protein (Table 3). The data in Table 3 supply an assay for the future purification of additional required proteins or factors.
Table 3.
Insertion of FeMoco into FeMoco-deficient pure MoFe protein of E146D nifH A. vinelandii
| Additions*
|
C2H4 evolution, nmol/min/mg protein | Activity, % | ||||
|---|---|---|---|---|---|---|
| Fe protein | ATP | FeMoco† | GroEL | Av CA12‡ extract | ||
| − | − | − | − | − | 1009 ± 123 | 51 |
| + | + | + | − | − | 1075 ± 88 | 54 |
| − | − | + | + | − | 1010 ± 83 | 51 |
| − | + | + | + | − | 1064 ± 88 | 53 |
| + | + | + | + | − | 1072 ± 13 | 54 |
| − | − | − | − | + | 993 ± 186 | 50 |
| + | + | + | − | + | 1992 ± 280 | 100 |
All insertion assays contained 0.1 mg pure MoFe protein of E146D nifH A. vinelandii. The insertion and activity portions of the assays were performed as described in Materials and Methods.
Excess isolated FeMoco in NMF was added.
Av CA12, ΔnifHDK A. vinelandii strain CA12.
E. coli GroEL is a well-characterized molecular chaperone whose role in the cell is to prevent aggregation and to facilitate proper protein folding. Protein folding in most cases is believed to occur in the central cavity of GroEL that is formed by two heptameric rings of 57-kDa subunits. In a MgATP- and cochaperone GroES-assisted mechanism, the cavity is able to accommodate polypeptides up to 70 kDa. During this process, GroES, a single heptameric ring of 10 kDa subunits, acts as a lid for the cavity forming a cis ternary complex in which the polypeptide is encapsulated within the GroEL-GroES structure (for detailed coverage see recent reviews in refs. 33–37). This study shows that GroEL is somehow required for the insertion of FeMoco into a form of the MoFe protein of nitrogenase that is already assembled into a 230,000 Mr, α2β2 tetramer (Fig. 1; ref. 28). Encapsulation in this case would not be possible. Recent reports in the literature also show that encapsulation is not the only mode of action for GroEL, that proteins larger than 70,000 daltons are able to bind to GroEL (38) and that GroEL can assist the folding of some protein without the GroES lid (39). It should be noted that we have not established whether or not GroES is required along with GroEL for the final assembly of the MoFe protein. If it is required, then it must be present in the extracts from the ΔnifHDK strain of A. vinelandii CA12 and in the partially purified E146D nifH MoFe protein solutions. Unfortunately, commercially available E. coli GroES antibodies showed extensive nonspecific crossreactivity with these solutions and were therefore not helpful in this regard.
Since the early 1990s, when GroEL research was in its infancy, several reports have appeared suggesting that GroEL might have something to do with nitrogenase regulation and assembly (40–44). For example, 35S pulse–chase experiments with Klebsiella pneumoniae suggested transient binding of newly synthesized Fe protein and MoFe protein polypeptides to GroEL. Recent work on other systems has shown that, in addition to helping to fold peptides into conformations that are capable of spontaneous assembly (45, 46), at least in the case of the mitochondrial branched-chain α-ketoacid dehydrogenase, GroEL/GroES interaction with an αβ heterodimeric intermediate is required for the final assembly of the α2β2 tetramer (47–50). Whether or not GroEL is required for the assembly of the tetrameric MoFe protein shown in Fig. 1, the data presented here show that it is definitely required for the final insertion of FeMoco into that protein.
At present it is not known which components of the system directly interact with GroEL, whose role might be to protect the vacant FeMoco site of the MoFe protein or to assist in the folding of that protein after insertion. The other known components of the system including gamma (25), FeMoco, the Fe protein, and MgATP (21, 22) are all small enough to be encapsulated by GroEL, which might serve to protect and hold them in the correct orientation during the insertion reaction.
It is interesting to note that, when the dual requirement for Fe protein and MgATP was first established, it was assumed that the MgATP was binding to the Fe protein and working with it in some way (22). Subsequently, numerous mutants were analyzed, showing that Fe protein variants that are altered in MgATP binding, ones that bind MgATP but do not undergo the MgATP-induced conformational change and ones that do not hydrolyze MgATP, all function fully in the final assembly of the MoFe protein (1, 4, 14, 15 and 19). These data, combined with the fact that GroEL is a protein that binds and hydrolyzes MgATP, suggest that the MgATP requirement may have nothing to do with the Fe protein but rather may be required for the function of GroEL in the final assembly reaction.
In this paper we report a function of GroEL that has not been described previously: its participation in the final insertion of a metal cluster into an apoprotein. This function of the well-known molecular chaperone GroEL might have broad implications for the assembly of other metalloproteins, including nickel-urease, nickel- and iron-hydrogenases, or molybdo-pterin cofactor-containing enzymes (for recent reviews, see refs. 51–54). Future studies should be directed toward purifying additional components of the nitrogenase system, by using the assay developed herein, so that the mechanistic details may be elucidated at the molecular level.
Acknowledgments
We wish to acknowledge Professor Agnes Henschen-Edman of University of California Irvine for the determination of the NH2-terminal polypeptide sequence of GroEL, Professor Dennis Dean of Virginia Polytechnic Institute and State University for kindly providing ΔnifB A. vinelandii strain DJ1143, and Dr. Hayley Angove for providing purified FeMoco-deficient MoFe protein from DJ1143. This work was supported by National Institutes of Health Grant GM-43144 (to B.K.B.).
Abbreviations
- MoFe protein
molybdenum-iron protein of nitrogenase
- Fe protein
iron protein of nitrogenase
- FeMoco
iron-molybdenum cofactor or [Mo-7Fe-9S-homocitrate] cluster of molybdenum-iron protein
- NMF
N-methyl formamide
References
- 1.Burgess B K, Lowe D J. Chem Rev. 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Howard J B, Rees D C. Chem Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- 3.Howard J B, Rees D C. Annu Rev Biochem. 1994;63:235–264. doi: 10.1146/annurev.bi.63.070194.001315. [DOI] [PubMed] [Google Scholar]
- 4.Seefeldt L C, Dean D R. Acc Chem Res. 1997;30:260–266. [Google Scholar]
- 5.Peters J W, Fischer K, Dean D R. Annu Rev Microbiol. 1995;49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 6.Thorneley R N F, Lowe D J. J Biol Inorg Chem. 1996;1:576–580. [Google Scholar]
- 7.Burgess B K. Chem Rev. 1990;90:1377–1406. [Google Scholar]
- 8.Burgess B K. In: Molybdenum Enzymes, Cofactors and Model Systems. Stiefel E I, Coucouvanis D, Newton W E, editors. Washington, DC: Am. Chem. Soc.; 1993. pp. 144–169. [Google Scholar]
- 9.Dean D R, Jacobson M R. In: Biological Nitrogen Fixation. Stacey G, Burris R H, Evans H J, editors. New York: Chapman & Hall; 1992. pp. 763–834. [Google Scholar]
- 10.Shah V K, Rangaraj P, Chatterjee R, Allen R M, Roll J T, Roberts G P, Ludden P W. In: Biological Nitrogen Fixation for the 21st Century. Elmerich C, Kondorosi A, Newton W E, editors. Boston: Kluwer; 1997. pp. 51–52. [Google Scholar]
- 11.Newton W E. In: Biological Nitrogen Fixation. Stacey G, Burris R H, Evans H J, editors. New York: Chapman & Hall; 1992. pp. 877–929. [Google Scholar]
- 12.Smith B E, Eady R R. Eur J Biochem. 1992;205:1–15. doi: 10.1111/j.1432-1033.1992.tb16746.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoover T R, Imperial J, Ludden P W, Shah V K. Biofactors. 1988;1:199–205. [PubMed] [Google Scholar]
- 14.Gavini N, Burgess B K. J Biol Chem. 1992;267:21179–21186. [PubMed] [Google Scholar]
- 15.Bursey E H, Burgess B K. J Biol Chem. 1998;273:29678–29685. doi: 10.1074/jbc.273.45.29678. [DOI] [PubMed] [Google Scholar]
- 16.Lowery R G, Chang C L, Davis L C, McKenna M C, Stephens P J, Ludden P W. Biochemistry. 1989;38:1206–1211. doi: 10.1021/bi00429a038. [DOI] [PubMed] [Google Scholar]
- 17.Wolle D, Kim C, Dean D R, Howard J B. J Biol Chem. 1992;267:3667–3673. [PubMed] [Google Scholar]
- 18.Wolle D, Dean D R, Howard J B. Science. 1992;258:992–995. doi: 10.1126/science.1359643. [DOI] [PubMed] [Google Scholar]
- 19.Rangaraj P, Ryle M J, Lanzilotta W N, Ludden P W, Shah V K. J Biol Chem. 1999;274:19778–19784. doi: 10.1074/jbc.274.28.19778. [DOI] [PubMed] [Google Scholar]
- 20.Rangaraj P, Shah V K, Ludden P W. Proc Natl Acad Sci USA. 1997;94:11250–11255. doi: 10.1073/pnas.94.21.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson A C, Dean D R, Burgess B K. J Biol Chem. 1987;262:14327–14332. [PubMed] [Google Scholar]
- 22.Robinson A C, Chun T W, Li J-G, Burgess B K. J Biol Chem. 1989;264:10088–10095. [PubMed] [Google Scholar]
- 23.Tal S, Chun T W, Gavini N, Burgess B K. J Biol Chem. 1991;266:10654–10657. [PubMed] [Google Scholar]
- 24.Gavini N, Ma L, Watt G, Burgess B K. Biochemistry. 1994;33:11842–11849. doi: 10.1021/bi00205a021. [DOI] [PubMed] [Google Scholar]
- 25.Homer M J, Dean D R, Roberts G P. J Biol Chem. 1995;270:24745–24752. doi: 10.1074/jbc.270.42.24745. [DOI] [PubMed] [Google Scholar]
- 26.Bursey E H, Burgess B K. J Biol Chem. 1998;273:16927–16934. doi: 10.1074/jbc.273.27.16927. [DOI] [PubMed] [Google Scholar]
- 27.Burgess B K, Jacobs D B, Stiefel E I. Biochim Biophys Acta. 1980;614:196–209. doi: 10.1016/0005-2744(80)90180-1. [DOI] [PubMed] [Google Scholar]
- 28.Ribbe M W, Bursey E H, Burgess B K. J Biol Chem. 2000;275:17631–17638. doi: 10.1074/jbc.275.23.17631. [DOI] [PubMed] [Google Scholar]
- 29.Christiansen J, Goodwin P J, Lanzilotta W N, Seefeldt L C, Dean D R. Biochemistry. 1998;37:12611–12623. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- 30.Shah V K, Ugalde R A, Imperial J, Brill W J. J Biol Chem. 1985;260:3891–3894. [PubMed] [Google Scholar]
- 31.Shah V K, Imperial J, Ugalde R A, Ludden P W, Brill W J. Proc Natl Acad Sci USA. 1986;83:1636–1640. doi: 10.1073/pnas.83.6.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop P E, Premakumar R, Dean D R, Jacobson M R, Chisnell J R, Rizzo T M, Kopezynski J. Science. 1986;232:92–94. doi: 10.1126/science.232.4746.92. [DOI] [PubMed] [Google Scholar]
- 33.Fenton W A, Horwich A L. Protein Sci. 1997;6:743–760. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin J, Hartl F U. Curr Opin Struct Biol. 1997;7:41–52. doi: 10.1016/s0959-440x(97)80006-1. [DOI] [PubMed] [Google Scholar]
- 35.Sigler P B, Xu Z, Rye H S, Burston S G, Fenton W A, Horwich A L. Annu Rev Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 36.Fink A L. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 37.Saibil H. Curr Opin Struct Biol. 2000;10:251–258. doi: 10.1016/s0959-440x(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y-S, Chuang D T. J Biol Chem. 1999;274:10405–10412. doi: 10.1074/jbc.274.15.10405. [DOI] [PubMed] [Google Scholar]
- 39.Wang J D, Weissman J S. Nat Struct Biol. 1999;6:597–600. doi: 10.1038/10636. [DOI] [PubMed] [Google Scholar]
- 40.Fisher H M, Babst M, Kaspar T, Acuna G, Arigoni F, Hennecke H. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer H-M, Schneider K, Babst M, Hennecke H. Arch Micobiol. 1999;171:279–289. [Google Scholar]
- 42.Govezensky D, Bochkareva E S, Zamir A, Girshovich A S. J Biol Chem. 1994;269:14003–14006. [PubMed] [Google Scholar]
- 43.Greener T, Govezensky D, Zamir A. EMBO J. 1993;12:889–896. doi: 10.1002/j.1460-2075.1993.tb05729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Govezensky D, Greener T, Segal G, Zamir A. J Bacteriol. 1991;173:6339–6346. doi: 10.1128/jb.173.20.6339-6346.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng X, Rosenberg L E, Kalousek F, Fenton W A. J Biol Chem. 1993;268:7489–7493. [PubMed] [Google Scholar]
- 46.Fedorov A V, Baldwin T O. J Mol Biol. 1997;268:712–723. doi: 10.1006/jmbi.1997.1007. [DOI] [PubMed] [Google Scholar]
- 47.Wynn R M, Davie J R, Song J L, Chuang J L, Chuang D T. Methods Enzymol. 2000;324:179–191. doi: 10.1016/s0076-6879(00)24230-4. [DOI] [PubMed] [Google Scholar]
- 48.Song J L, Wynn R M, Chuang D T. J Biol Chem. 2000;275:22305–22312. doi: 10.1074/jbc.M002038200. [DOI] [PubMed] [Google Scholar]
- 49.Wynn R M, Song J L, Chuang D T. J Biol Chem. 2000;275:2786–2794. doi: 10.1074/jbc.275.4.2786. [DOI] [PubMed] [Google Scholar]
- 50.Chuang J L, Wynn R M, Song J L, Chuang D T. J Biol Chem. 1999;274:10395–10404. doi: 10.1074/jbc.274.15.10395. [DOI] [PubMed] [Google Scholar]
- 51.Mobley H L, Island M D, Hausinger R P. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams M W, Stiefel E I. Curr Opin Chem Biol. 2000;4:214–220. doi: 10.1016/s1367-5931(99)00077-0. [DOI] [PubMed] [Google Scholar]
- 53.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer R K. Curr Opin Struct Biol. 1998;8:749–758. doi: 10.1016/s0959-440x(98)80095-x. [DOI] [PubMed] [Google Scholar]
- 54.Kisker C, Schindelin H, Baas D, Retey J, Meckenstock R U, Kroneck P M. FEMS Microbiol Rev. 1998;22:503–521. doi: 10.1111/j.1574-6976.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]