Abstract
Motivation: Computational characterization of ligand-binding sites in proteins provides preliminary information for functional annotation, protein design and ligand optimization. SiteComp implements binding site analysis for comparison of binding sites, evaluation of residue contribution to binding sites and identification of sub-sites with distinct molecular interaction properties.
Availability and implementation: The SiteComp server and tutorials are freely available at http://sitecomp.sanchezlab.org
Contact: roberto@sanchezlab.org; roberto.sanchez@mssm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The interaction of proteins with their ligands (metabolites, proteins, nucleic acids, lipids, etc.) is the most fundamental of all biological mechanisms. These interactions are often specific and are the consequence of distinct molecular interaction properties of the binding sites. Hence, the analysis and comparison of binding site properties can shed light on the basis of ligand affinity, selectivity and ultimately the molecular underpinnings of protein function. The most frequent questions that arise in binding site analysis are: (i) Does a binding site contain regions (sub-sites) with special molecular interaction properties? (ii) What residues contribute to the formation of a binding site? (iii) What are the differences between two similar binding sites? SiteComp is a webserver designed to answer these questions, hence facilitating the design of new experiments and the analysis of existing data in the context of elucidating molecular mechanisms and drug design.
While tools for the characterization of sub-sites within a ligand-binding region have been available since the development of the GRID approach (Goodford, 1985), no freely available webservers exist to carry out this type of analysis. Existing computational methods have also achieved success in the identification of ligand-binding sites (Ghersi and Sanchez, 2011), including detection of local similarity (Kellenberger et al., 2008), or comparison of interaction properties of complete proteins (Richter et al., 2008). However, these methods are not well-suited for identifying differences between similar binding sites, which can be exploited to improve ligand selectivity. Methods that address the question of residue contribution to a binding site can be divided into two groups: (i) computational alanine scanning methods (Chong et al., 2006; Kortemme et al., 2004; Kruger and Gohlke, 2010; Massova and Kollman, 1999); and (ii) energy decomposition methods (Benedix et al., 2009; Schymkowitz et al., 2005; Zoete and Michielin, 2007). The former have been developed exclusively for protein–protein interaction surfaces. While the latter, which are relatively accurate, require computationally expensive molecular dynamics or Monte Carlo simulations.
SiteComp complements the existing methods, bridging several of the current gaps, by providing a web-based interface for identification of differences between similar binding sites, discovery of sub-sites with different interaction properties and for fast (albeit more approximate) calculations of residue contribution to binding sites. It integrates these three modes of binding site analysis into an easy to use interactive interface with graphical input and output.
2 METHODS
2.1 Types of SiteComp analyses
SiteComp uses molecular interaction fields (MIFs) as descriptors of small-molecule ligand binding sites. MIFs describe the spatial variation of the interaction energy between a target molecule (e.g. a protein) and a probe, which represents a specific chemical group or atom (Ghersi and Sanchez, 2009). SiteComp provides three types of MIF-based analyses:
Binding site comparison identifies regions where two proteins exhibit differences in ligand-binding properties. After superposition of the two input proteins, a difference MIF is calculated and post-processed using the SiteHound algorithm (Ghersi and Sanchez, 2009) to identify difference clusters (see Supplementary Materials for details). These clusters identify regions with more favorable probe interactions with one protein than the other. The difference clusters can be used, for example, as guides to explain or design ligand selectivity between two proteins (Fig. 1).
Binding site decomposition evaluates the contribution of specific side chains to protein–ligand interaction regions. This is achieved by comparing the MIFs of the wild-type protein with that of the same protein with one or more residues mutated to alanine. Up to 10 residues can be selected in a user-defined region of the protein. A single protein is required as input and SiteComp produces the variants where alanine replaces the wild-type residue. This type of analysis can be used to identify key residues in a previously identified binding site and design mutations that disrupt binding.
Multi-probe characterization facilitates visual comparison of MIF clusters detected in a single protein with different chemical probes. It also facilitates the exploration of different parameters for MIF calculation (energy cutoff) and clustering (algorithm). Hence, this type of analysis enables an advanced characterization of the molecular interaction properties of a user-defined region in one protein. One application of this analysis is the identification of sub-sites with different interaction properties within a larger binding site (Fig. 2). Visualization of the output in the server facilitates comparison and combination of MIF clusters detected with different parameters and probes.
Fig. 1.
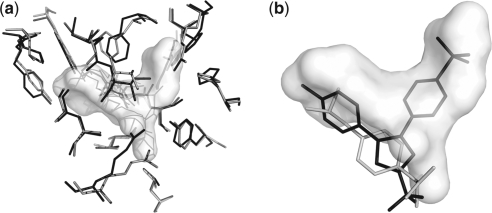
Example of binding site comparison. Comparison of the binding sites of two cyclooxygenase (COX) enzymes was carried out using SiteComp. COXs are targets for non-steroidal anti-inflammatory drugs. (a) SiteComp difference region (white surface) favorable for COX-2 (gray sidechains) over COX-1 (black sidechains). (b) The non-selective COX inhibitor Ibuprofen (gray) does not take advantage of the difference region, while whereas the selective COX-2 inhibitor Celecoxib (black) occupies most of the predicted selectivity region (Wang, et al., 2010).
Fig. 2.
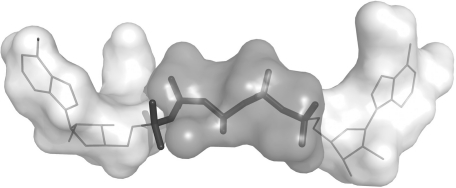
Example of multi-probe characterization. Sub-sites in the active site of adenylate kinase (ADK) were identified using SiteComp. ADK catalyzes the phosphate transfer from ATP to AMP. The figure shows AP5A, an ADK inhibitor (Abele and Schulz, 1995) that mimics the structure of the two substrates in the ADK active site. Sub-sites identified with the methyl carbon probe (white surfaces) highlight the regions of the active site that recognize the adenosine groups in the inhibitor and the substrates (thin lines), while sub-sites identified with the phosphate oxygen probe (gray surface) delineate the phosphate transfer region (thick lines).
2.2 Integration of analyses
The three types of SiteComp analyses can be integrated into a combined analysis. For example, a difference region identified in binding site comparison can be selected to be directly analyzed using binding site decomposition to identify residues that are important contributors to that region. Alternatively, it could be directed into multi-probe characterization to provide detailed information about the molecular interaction properties of the difference site. SiteComp is also integrated with the SiteHound-web binding site identification server (Hernandez et al., 2009), which enables seamless analysis of predicted binding sites using the SiteComp tools.
2.3 Usage and output
For each of the analyses, the user can upload PDB files or specify PDB codes for the proteins of interest. SiteComp processes the structures and prompts the user to select chains for calculation. In binding- site decomposition and multi-probe characterization, additional chains and ligands can be selected for display only. Next, a region of interest, the calculation box, is defined using a graphical user interface (GUI) based on the Jmol molecular structure viewer. The center of the calculation box can be defined interactively by selecting an atom in Jmol, entering a residue number or specifying coordinates. The box dimensions can also be modified interactively. Subsequently, parameters for MIF calculation and clustering are selected. Finally, the calculation is carried out and the output is presented in a Jmol-based GUI. Runtime is usually less than a few minutes, depending on the size of the calculation box.
The user can retrieve the results from the calculation at runtime or within 30 days after the calculation has completed using a unique and private URL generated at the time of job submission. After 30 days the results and input files are deleted from the server.
The SiteComp website includes step-by-step tutorials for each type of analysis. The server requires Java and Javascript to be enabled and has been tested on all major operating systems and web browsers.
Supplementary Material
ACKNOWLEDGEMENT
Dr Dario Ghersi for help with EasyMIFs and SiteHound usage.
Funding: National Institutes of Health (NIH) [HG004508, GM081713].
Conflict of Interest: none declared.
REFERENCES
- Abele U., Schulz G.E. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995;4:1262–1271. doi: 10.1002/pro.5560040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedix A., et al. Predicting free energy changes using structural ensembles. Nat. Methods. 2009;6:3–4. doi: 10.1038/nmeth0109-3. [DOI] [PubMed] [Google Scholar]
- Chong L.T. Kinetic computational alanine scanning: application to p53 oligomerization. J. Mol. Biol. 2006;357:1039–1049. doi: 10.1016/j.jmb.2005.12.083. [DOI] [PubMed] [Google Scholar]
- Ghersi D., Sanchez R. EasyMIFS and SiteHound: a toolkit for the identification of ligand-binding sites in protein structures. Bioinformatics. 2009;25:3185–3186. doi: 10.1093/bioinformatics/btp562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi D., Sanchez R. Beyond structural genomics: computational approaches for the identification of ligand binding sites in protein structures. J. Struct. Funct. Genomics. 2011;12:109–117. doi: 10.1007/s10969-011-9110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Hernandez M., et al. SITEHOUND-web: a server for ligand binding site identification in protein structures. Nucleic Acids Res. 2009;37:W413–W416. doi: 10.1093/nar/gkp281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E., et al. How to measure the similarity between protein ligand-binding sites. Curr. Comput.-Aid. Drug Des. 2008;4:209–220. [Google Scholar]
- Kortemme T., et al. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- Kruger D.M., Gohlke H. DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein-protein interactions. Nucleic Acids Res. 2010;38:W480–W486. doi: 10.1093/nar/gkq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massova I., Kollman P.A. Computational alanine scanning to probe protein-protein interactions: a novel approach to evaluate binding free energies. J. Am. Chem. Soc. 1999;121:8133–8143. [Google Scholar]
- Richter S., et al. webPIPSA: a web server for the comparison of protein interaction properties. Nucleic Acids Res. 2008;36:W276–W280. doi: 10.1093/nar/gkn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J., et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.L., et al. The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: the second clinical candidate having a shorter and favorable human half-life. Bioorg. Med. Chem. Lett. 2010;20:7159–7163. doi: 10.1016/j.bmcl.2010.07.054. [DOI] [PubMed] [Google Scholar]
- Zoete V., Michielin O. Comparison between computational alanine scanning and per-residue binding free energy decomposition for protein-protein association using MM-GBSA: application to the TCR-p-MHC complex. Proteins. 2007;67:1026–1047. doi: 10.1002/prot.21395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.