Abstract
Summary: Rapid identification of non-human sequences (RINS) is an intersection-based pathogen detection workflow that utilizes a user-provided custom reference genome set for identification of non-human sequences in deep sequencing datasets. In <2 h, RINS correctly identified the known virus in the dataset SRR73726 and is compatible with any computer capable of running the prerequisite alignment and assembly programs. RINS accurately identifies sequencing reads from intact or mutated non-human genomes in a dataset and robustly generates contigs with these non-human sequences (Supplementary Material).
Availability: RINS is available for free download at http://khavarilab.stanford.edu/resources.html
Contact: abhaduri@stanford.edu or kqu@stanford.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
The appeal of searching for pathogen sequences in high-throughput sequencing data has grown as massively parallel sequencing capabilities have developed (Shendure and Ji, 2008) and sequencing has identified pathogens such as the Merkel cell polyoma virus as a contributing factor in Merkel cell carcinoma (Feng et al., 2008). Algorithms such as PathSeq (Kostic et al., 2011) have emerged that apply computational subtraction to the task of pathogen detection. These algorithms are computationally intensive and thus still require cloud computing scale resources. Here, we present rapid identification of non-human sequences (RINS), an alternative to computational subtraction that can efficiently identify the presence of pathogens from a custom reference in high-throughput sequencing datasets. The speed and local computing-based nature of RINS makes it attractive for hypothesis-driven discovery of pathogens in large datasets. The accessibility of a workflow such as RINS opens the door for extensive pathogen discovery in a variety of contexts such as cancer and other difficult to treat diseases.
RINS employs intersection analysis with a user provided reference set, as opposed to computational subtraction. The latter is a process that maps first to the reference organism's genome and then attempts to assign all unmapped reads to a non-reference organism. Because mapping algorithms require intense RAM to store these unmapped reads, computational subtraction is slow and requires cloud scale resources. While both methods ultimately filter through a reference organism (e.g. human) and look for pathogenic sequences, RINS first maps to a query dataset, thereby lowering the computational requirements. It can use any non-human reference set, including genomes of viruses, bacteria or other pathogens. This set is used as the template for the RINS initial search. The reference genome set included with the RINS package contains viruses of all known classes that infect a variety of organisms in order to offer the broadest template to identify pathogens.
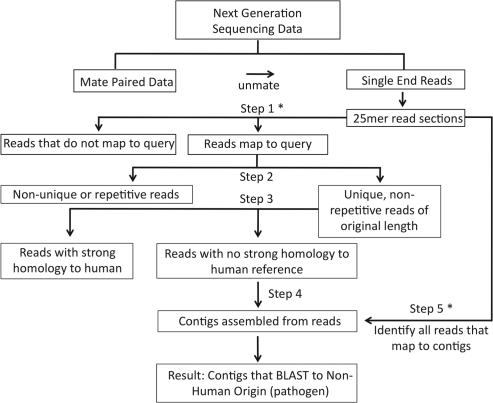
The workflow starts by generating non-overlapping 25 mers of each read that maximize the sensitivity of the alignment with Blat (Kent, 2002) against the reference genomes(s) provided, using an 80% match threshold (Fig. 1, RINS Step 1). This threshold was optimized for sensitivity and specificity performance on a randomly mutated test set. Step 2 of RINS removes duplicates that may have been generated by Step 1 alignment and filters the longest-associated read for each mapped 25 mer for complexity using a Lempel–Ziv–Welch (LZW) (Welch, 1984) compression ratio of 50% (RINS Step 2). The LZW compression method (Yozwiak et al., 2010) uses a dictionary-based approach to quantitate the complexity of a sequence by adding to the dictionary a new ‘word’ for every unique string of characters, eliminating repeat regions. These repeats are frequently found in both humans and microbes, making organismal origin difficult to pinpoint. This complexity ratio was optimized to minimize loss of non-human reads while further filtering the data for potentially confounding repetitive sequences. The filtered reads are then mapped against the human genome using Bowtie (Langmead et al., 2009) (RINS Step 3), and after this intersection, reads that mapped to the human genome are removed from the read set. Remaining reads, with their mate pairs (if the dataset is paired end) are assembled into contigs with the de novo assembler Trinity (Grabherr et al., 2011) (RINS Step 4). Using a local version of BLAST to classify contigs (RINS Step 5), those contigs with minimal homology to human sequences are then extended by mapping the original read set back to the contig. The process of identifying mate pairs and assembling the contig is repeated as before. This method of extension allows for reads that are part of the contig but were eliminated by other filtration methods to be reincorporated into the contig to increase the sensitivity and specificity of the results. RINS will then output a tab-delimited text file detailing the candidate contigs that have been generated, presented with the number of supporting reads and a BLAST e-value. Parameters used here are modifiable by the user if desired.
Fig. 1.
RINS uses intersection (marked by asterisks), not subtraction, to identify non-human reads. The workflow intersects the reads in the dataset with a reference of non-human genomes of interest using Blat to align non-overlapping 25 mers for each read. Reads with >80% homology are aligned to the human genome and reads with >97% homology are removed from the read set. Remaining reads are complexity filtered with an LZW compression ratio of 0.50 and mate pairs for sufficiently complex reads are identified. This read set is then assembled into pathogen sequence contigs.
Sensitivity of RINS was evaluated with a randomly mutated test set of viral genomes, which served as a stringent measuring system. In this test set, mutations occurred randomly throughout the genome without any conserved regions that are often found in nature. With this test set, it was shown that using the 25 mer reads promotes identification of mutated non-human genomes (Supplementary Fig. S1a). Specifically, at mutation rates >25%, mapping to the custom reference with these shorter read segments is significantly better at identifying genomes than with the full length reads (Supplementary Fig. S1b). This indicates known pathogens are identified with confidence, and genomes with >50% homology to the reference genomes can be extracted from the data using RINS.
A positive control was used to test RINS accuracy and speed (Supplementary Table S1). Sequencing data from the CA-HPV-10 prostate cancer cell line (SRR073726) (Prensner et al., 2011) was analyzed with RINS and accurately retrieved only HPV serotype 18 (the transforming virus) with a 570 bp contig (Supplementary Tables S1 and S2). RINS took <2 h to perform this analysis on a dual core machine with 8 GB of RAM and a 2.93 GHz processor. Accuracy of RINS was further tested in RNA sequencing data from Sézary syndrome, SRA046736 (Lee et al., manuscript in preparation) where a contig with homology to vector constructs and HIV was generated (Supplementary Table S2). Using PCR amplification and Sanger sequencing (data not shown), the existence of this laboratory contaminant in the cDNA of the relevant sample was confirmed.
Comparisons of RINS to the pathogen discovery algorithm PathSeq show similar performance, with better speed and lower cost for RINS. PathSeq sensitivity and specificity are derived from statistics presented by the authors in their work using an analogous test set of randomly mutated viral genomes. The sensitivity of PathSeq was 99.22% at a mutation rate of 0%, and drops to 0% at a mutation rate of 50% (Kostic et al., 2011, Table S3 and Figure S3a). RINS has a similar sensitivity of 99.78% based upon test set read recovery at a mutation rate of 0%. The RINS sensitivity drops to 0% at any mutation rate >50%, though at 50% there is a minimal sensitivity of 0.5–1% (Supplementary Fig. S1). Specificity for the test set is 100% for PathSeq. RINS also has no false positives for the test set and no false positives were identified from the CA-HPV-10 data or the Sezary syndrome data, giving RINS a specificity of 100%. The additional rigor of PathSeq would confidently allow identification of novel pathogens, though the ability of RINS to identify reads with up to 50% divergence from the reference genome suggests this could also be feasible with RINS if the novel pathogen has at least 50% homology to one or more of the custom reference genomes. PathSeq requires 13 h of cloud computing time and costs to process 10 million reads, whereas RINS takes <2 h for the 13 million reads in SRR073726. Importantly, RINS scales up well and is able to complete the six high-throughput datasets from SRA046736 with an average read depth of 112 million reads in <4 h each at no additional cost beyond access to a computer. The lower cost and faster speed are significant for accessibility to researchers interested in either hypothesis driven queries of datasets or queries of many different datasets in a reasonable timeframe.
RINS is optimized for mate-paired high-throughput sequencing data with reads at least 36 bp and up to 500 bp, and can be run on sequencing data from any species. Non-paired end sequencing data can also be used, though contig generation and extension will be less robust. As the read length and number of reads increases, the computational time required to complete RINS will increase as Blat and Trinity processing speeds will decrease. Included in the online package are 32 102 viral genomes of all classes curated by GenBank (Benson et al., 2008) and the International Committee on Taxonomy of Viruses (ICTV), retrieved through the National Center for Biotechnology Information (NCBI). Also provided are all scripts to run the processes, with options for user control of all default parameters. The user must provide other open source softwares referenced above.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Webster and A. Zehnder for helpful pre-submission review.
Funding: USVA Office of Research and Development, NIH/NIAMS AR49737.
Conflict of Interest: none declared.
REFERENCES
- Benson D.A., et al. GenBank. Nucl. Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., et al. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J.R., et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Welch T.A. A technique for high-performance data-compression. Computer. 1984;17:8–19. [Google Scholar]
- Yozwiak N.L., et al. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.