Abstract
Motivation: Public genomic databases, which are often used to guide genetic studies of human disease, are now being applied to genomic medicine through in silico integrative genomics. These databases, however, often lack tools for systematically determining the experimental origins of the data.
Results: We introduce a new data provenance model that we have implemented in a public web application, BioQ, for assessing the reliability of the data by systematically tracing its experimental origins to the original subjects and biologics. BioQ allows investigators to both visualize data provenance as well as explore individual elements of experimental process flow using precise tools for detailed data exploration and documentation. It includes a number of human genetic variation databases such as the HapMap and 1000 Genomes projects.
Availability and implementation: BioQ is freely available to the public at http://bioq.saclab.net
Contact: ssaccone@wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The rapid expansion of biotechnology continues to drive the public availability of massive, complex genomic databases. These databases are often used in applications of in silico integrative genomics to genetic studies of human disease (Hawkins et al., 2010; Hirschhorn, 2009) as well as genomic medicine (Green et al., 2011). It is therefore critical for investigators to have tools for systematically assessing the credibility of the data (Baggerly, 2010).
We have developed a straightforward model for tracing experimental process flow in genomic databases. The goal is to isolate the key entities in the database—the biologics, the experiments and the experimental results—and to express their relationships in terms of experimental process flow. We call this the Biologic-Experiment-Result (BERT) model and have implemented this model in our BioQ web application (http://bioq.saclab.net). BioQ builds on our dbSNP-Q application (Saccone et al., 2011) to allow investigators to visualize experimental process flow and retrieve process-related data with powerful query tools.
2 TRACING EXPERIMENTAL PROCESS FLOW
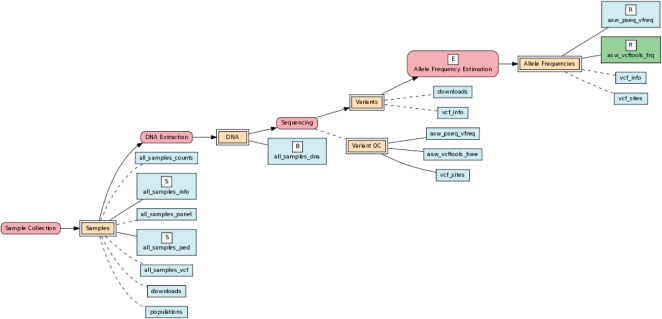
Figure 1 shows an application of the BERT model, as implemented in BioQ, to allele frequency data from the 1000 Genomes Project (Durbin et al., 2010) (see Supplementary Figs S1 and S2 for additional examples). These estimates, which are a key experimental result in the 1000 Genomes database, can be clearly traced back through specific processes, such as the methods and software applied to the original 1000 Genomes data files, to detailed information on the subjects and their biologics, including data on pedigree structure and DNA extraction from the Coriell Institute (http://ccr.coriell.org). In BioQ these process flow diagrams are fully interactive. Each node in Figure 1, which is taken directly from BioQ (http://tinyurl.com/75rlujj), is linked to detailed documentation and powerful data query tools (see Supplementary Figs S3 and S4).
Fig. 1.
How the BERT model is used to trace experimental process flow back to the original samples and biologics for allele frequency estimates in the ASW population (African Americans in the Southwestern USA) from the October 2011 Release of the 1000 Genomes Project. The double rectangles represent groups of tables that may be input or output for the various processes and experiments (represented by ovals) in the database; these are referred to as flow groups. Processes that yield key experimental results in the database are designated as Experiments and labeled ‘E’. Tables in the database are represented by simple rectangles. Tables containing data on subjects, biologics and key experimental results are indicated by the labels ‘S’, ‘B’ and ‘R’, respectively. The goal is to trace the results back to subjects and biologics. Some groups and individual tables contain auxiliary reference data for processes and flow groups, respectively; reference nodes are indicated by dashed lines. See the Supplementary Materials for addition information on the BERT model.
Process flow diagrams should ideally include information on diagnostics and quality control (QC). Our application to the HapMap and 1000 Genomes projects, for example, includes detailed results from our own QC analyses including sample- and marker-specific call rates, tests of Mendelian errors and Hardy–Weinberg equilibrium (see the Supplementary Materials for details). In particular, when developing tables related to Mendelian errors in the HapMap (Altshuler et al., 2010) Phase III Release III database we discovered a familial misspecification in the HapMap data files. The details are described in the BioQ documentation for the table summary_samples (http://tinyurl.com/7q5of5e) (see the Supplementary Materials for details).
Clearly there will be some genomic databases of interest to geneticists that lack data on experimental origins. BioQ, for example, includes the NHGRI genome-wide association study (GWAS) database (Hindorff et al., 2009). The NHGRI database contains the top association results from published GWAS. It does not, however, include detailed data on QC measures for individual association results, although links to publications are provided as indicated by the BioQ documentation for this database (http://tinyurl.com/7ztufdg). BioQ uses the BERT model to clearly and systematically show when experimental results are not traceable to subjects and biologics and when QC data is lacking (see Supplementary Figs S5 and S6).
3 DISCUSSION
The goal of the BERT data provenance model is to provide investigators with a practical means of systematically establishing a reasonable level of credibility when using genomic databases. Our BioQ web application implements this model in a highly transparent way with numerous tools for data retrieval. Other models, such as FuGE (Jones et al., 2007), are more appropriate for capturing the full spectrum of experimental detail. The relative simplicity of the BERT model will allow applications such as BioQ to adapt as the requirements of investigators evolve.
Very few tools allow investigators to trace the experimental source of genomic data and to assess QC in detail as in BioQ. Without tools such as BioQ these undertakings can take days, even weeks given the enormity of tests required to thoroughly assess QC, the sheer size of the datasets involved and the difficulty in locating provenance data. It is therefore likely that in many cases these assessments will not be done, and this may lead to the use of faulty data. A recent incident at Duke University (Samuel Reich, 2011), for example, involving the use of flawed data in a translational genomic study has led to the creation of a new framework at Duke on the quality of translational genomic medicine in which data provenance plays a minor role (http://tinyurl.com/6pkfdgd, accessed March 19, 2012).
BioQ is a direct extension of our previous web application dbSNP-Q (Saccone et al., 2011). While dbSNP-Q provides powerful query and documentation tools for the dbSNP database, it does not implement the BERT model. These methods and tools for systematically tracing experimental process flow are unique to BioQ. Resources such as the UCSC database (Fujita et al., 2011) focus on graphical representations of genomic data. These resources do not provide tools for systematically tracing experimental process flow and for querying the underlying relational databases in a web browser as in BioQ. The BERT model and the BioQ application are designed to complement these resources by providing tools for systematically determining the origins of and assessing the quality of the data used in other tools such as the UCSC genome browser.
The ability to establish the experimental origins of genomic data and to systematically assess QC at all stages of the experimental process is a systemic issue relevant to all genomic applications. It is therefore useful to have a separate resource dedicated to resolving this issue that can be used in conjunction with other applications. As new bioinformatics applications emerge that further bridge the gap between genomics and medicine, the availability of tools for systematically determining experimental origins will be crucial for maintaining reasonable levels of credibility.
The BioQ project has taken a number of measures to ensure that updates to external genomic databases are incorporated punctually and accurately. To each database there corresponds a custom command-line driven program, written in Perl, that downloads the data, processes it, checks for errors, performs QC analyses and creates the MySQL databases used in BioQ. Barring any major changes to the formats released by these databases, these programs allow the BioQ update process to be completely automated. All code is publicly available from our Subversion server (http://svn.saclab.net) and our code review resource (http://fisheye.saclab.net). These programs can be easily modified to accommodate any changes encountered in updates to external genomic databases and will be used to incorporate these updates as they become available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gaurang Mehta for his valuable advice in deploying the BioQ public web server.
Funding: National Institute on Drug Abuse (K01 DA024722); National Institute of Mental Health (U24 MH068457).
Conflict of Interest: none declared.
REFERENCES
- Altshuler D.M., et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly K. Disclose all data in publications. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- Durbin R.M., et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita P.A., et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E.D., et al. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- Hawkins R.D., et al. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L.A., et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J.N. Genomewide association studies—illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Jones A.R., et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat. Biotechnol. 2007;25:1127–1133. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- Saccone S.F., et al. New tools and methods for direct programmatic access to the dbSNP relational database. Nucleic Acids Res. 2011;39:D901–D907. doi: 10.1093/nar/gkq1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel Reich,E. Cancer trial errors revealed. Nature. 2011;469:139–140. doi: 10.1038/469139a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.