Abstract
Recently the concept of apoptotic cell elimination was expanded and programmed cell death is no longer viewed as an individual cellular event. The complete description of the apoptotic process now includes two phases: the self-driven cell disassembly and the externally-controlled elimination of apoptotic cell corpses by phagocytizing cells. The second, phagocytic phase is essential, highly conserved and is even more important than the internal phase of cell disassembly. This is because it ensures the complete degradation of the dying cell’s DNA, preventing release of pathological, viral and tumor DNA and self-immunization. In different cells and species from mammals to flies, a single conserved enzyme - DNase II is responsible for elimination of cellular DNA in the second, ‘mopping up’ phase, of apoptosis. Here we present an assay for selective detection of the phagocytic phase of apoptosis. The technology capitalizes on the fact that phagocytic DNase II produces identifiable signature DNA breaks, which can be labeled by vaccinia topoisomerase. The assay permits labeling of the previously underestimated phase of apoptotic execution and is a useful tool in the apoptosis detection arsenal.
Keywords: apoptosis labeling, DNA breaks, in situ detection, vaccinia topoisomerase I, DNase II-type breaks, blunt-ended DNA breaks, 5’OH DNA breaks, phagocytizing cells, apoptotic cell corpse elimination
1. INTRODUCTION
1.1. Cell-autonomous and lysosomal (phagocytic) nucleases in apoptosis
Based on their role in cell corpse elimination, all apoptotic nucleases are divided into two categories: cell-autonomous and waste-management nucleases (1). These two groups of nucleases have very different functions. Cell-autonomous nucleases cleave the DNA within a cell as it undergoes apoptosis. The lysosomal (phagocytic) nucleases take part in the engulfment-mediated DNA degradation — the 'cleaning up' of corpses of cells that have died by apoptosis. While cell-autonomous DNA degradation is important for carrying out an individual cell death program, the waste-control nucleases are essential for the life of other cells in the organism. The proper disposal of post-apoptotic corpses is critically important because their uncleaned debris pose danger to other cells (1).
Even though the cell-autonomous nucleases are important, they are dispensable in many cell types. This is because in the organism, after a cell has died, its corpse is destroyed and engulfed by other cells. Therefore even if a cell has not degraded its own DNA, its neighbors will do it instead. However, there is no such “plan B” in the case of waste-management nucleases. If they fail to clean the corpses, the organism would be poisoned with non-degraded DNA. Thus lysosomal nucleases are essential for life (1, 2).
Tissue section assays which can label activity of these two categories of enzymes permit comprehensive imaging of apoptotic degradation.
Here we present an assay which labels phagocytosis of DNA from apoptotic cell corpses by the 'mop up' cells. The assay addresses the challenges created by a very recent shift in the apoptotic paradigm which now views apoptosis not as a “private matter” of a dying cell, but as a broad reaction analogous to the immune response, continuing beyond the individual cell program and requiring participation of other specialized cells (1, 3). The assay takes into consideration this novel broad perspective of programmed cell death which is necessary for better understanding of apoptosis-related pathologies.
The technology capitalizes on the fact that phagocytic nuclease activities in apoptosis are highly conserved. They produce characteristic and highly specific DNA breaks. The assay uses the enzymatic action of vaccinia topoisomerase I to label signature DNA breaks produced by waste-management nucleases.
1.2. DNase I- and DNase II-types of DNA breaks
The ability to accurately label apoptotic cells and visualize the progression of apoptosis in the tissue section format is critical for many branches of biomedical science. However the task of choosing an appropriate apoptotic marker is complicated by the fact that apoptotic pathways are remarkably complex. They not only vary in different cell types and tissues, but can change within the same cell. Nucleolytic activities are critical for apoptotic disassembly as they degrade cellular DNA, preventing release of pathological, viral and tumor DNA and self-immunization (1, 3).
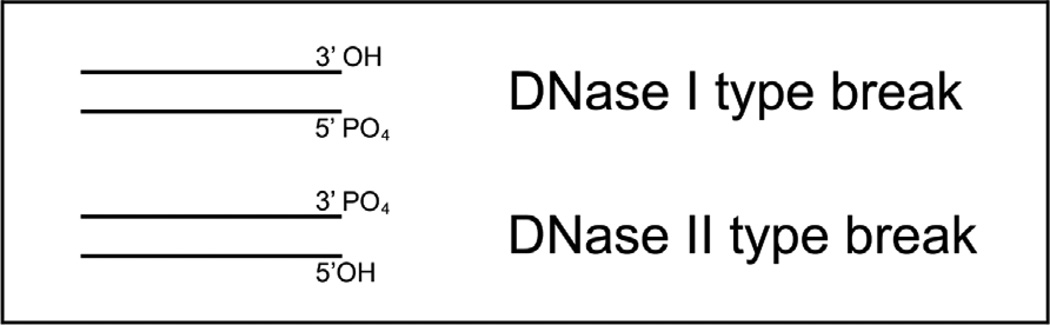
Every apoptotic cell goes through sequential steps ensuring its complete disassembly and disappearance. Massive and systematic DNA fragmentation is one of the characteristic features of this process. Not surprisingly, it is often used as specific marker in apoptosis detection. When apoptotic DNA is cut, the result is an abundant amount of DNA fragments. These DNA fragments are not random. Mainly they possess blunt ends and some have ends with short, single-nucleotide staggers with one of the strands protruding slightly (4-6). Different functional groups can be present at those ends. A double-strand break forms when a DNA duplex is cut through. It exposes the 3’ and 5’ ends of the two DNA strands. These ends can carry either a phosphate (PO4) group or a hydroxyl (OH) group (Fig. 1). The distribution of these groups at the ends provides important information about the enzyme which did the cutting.
Figure 1.
DNase I and DNase II type DNA breaks
Based on the distribution of functional groups, the two types of cuts are identified. The cuts with 3’OH/5’PO4 configuration are DNase I type cuts. The cuts with the inverse distribution of functional groups - 3’PO4/5’OH, are of DNase II type (7,8). (Fig.1)
The cuts received their names because they match the cleavage patterns of the two major nucleases - DNase I and DNase II. The terms are used for convenience and do not imply any specific relationship between these enzymes and the apoptotic cutters. The actual apoptotic nuclease might or might not be related to DNase I and II.
So far in the majority of apoptotic cells in mammals DNase I type fragmentation was detected (3’OH/5’PO4 at the ends) (1, 9, 10).
DNase II type was detected in many important cases too (2, 3, 11, 12). Although in general, this type of cleavage is less prevalent than the self-inflicted apoptotic DNA fragmentation. The underdevelopment of techniques for selective labeling of DNase II type cleavage might have contributed to this situation. In fact, the enzymatic assays for labeling apoptosis in tissue sections focus exclusively on DNase I type cleavage.
Different features of DNase I cleavage are used by various apoptosis assays. For example, the TUNEL assay specifically detects one marker of DNase I cleavage and labels the 3’OH groups with help of the enzyme Terminal Deoxyribonucleotidyl Transferase (TdT) (13). The other assay, in situ nick translation, labels nicks or single-stranded DNA breaks with 3’OH using DNA polymerase I (14). Yet another technique, in situ ligation, selectively detects double-stranded DNA breaks (10, 15). It relies on attachment of double-stranded DNA probes with blunt ends, or short 3’ overhangs, to the ends of such breaks. The ligation reaction is carried out by the enzyme T4 DNA ligase, which needs terminal 5’PO4 in the breaks to attach the probe. Consequently, the assay detects exclusively 5’ phosphorylated double-strand breaks and does not label DNase II type breaks with 5’OH. Therefore in all of these assays, the cells with DNase II type cleavage go undetected.
In the meantime, important changes occurred in the very concept of apoptotic cell elimination which significantly increased the value of detecting DNase II type breaks. It was demonstrated that DNase II plays a critical role in apoptosis at a level different from executioner nucleases.
1.3. Role of DNase II in elimination of apoptotic cell corpses
The apoptotic paradigm has recently changed to incorporate the notion that the apoptotic process is not an internal cellular event (1, 3). Instead, it continues beyond the individual cell reaction and requires participation of surrounding cells. The complete apoptotic process now includes two phases: the self-driven cell disassembly and the externally-controlled elimination of apoptotic cell corpses by phagocytizing waste-management cells (1). This externalized waste-control phase is essential, highly conserved and is considered to be even more important than the internal phase of cell disassembly (1, 2, 12). This is because it ensures complete degradation of the dying cell’s DNA, preventing release of pathological, viral and tumor DNA and self-immunization (see Note 1).
DNase II plays a fundamental role in engulfment-mediated DNA degradation during the waste-management phase of apoptosis (1, 3). DNase II is present in lysosomes, the sac-like organelles inside cells that contain digestive enzymes that break down cellular components. The enzyme destroys DNA of apoptotic cells after their corpses are engulfed by waste-management cells, such as tissue macrophages and many other tissue cells capable of phagocytosis.
Consequently, novel approaches for apoptosis detection are needed which will take this paradigm change into account. Several attempts were made to accomplish this task.
Initially to resolve this problem, the T4 DNA kinase-based technique was introduced for the detection of 5’OH bearing DNA breaks (8). However in this case, the 5’PO4 breaks must be labeled first so that in the subsequent reaction with T4 DNA kinase all remaining 5’OH breaks are converted to the detectable 5’ phosphate format. As a result, the approach requires two successive overnight labeling reactions with multiple controls, making it complicated and time consuming.
We have subsequently developed a new approach for labeling of 5’OH double-strand DNA breaks. The approach was an offshoot of our work in bio-nanotechnology developing an oscillating nano-size device, which contained a vaccinia virus encoded protein linked with a dual labeled DNA part (16) (also featured in (17). The construct exemplified a practical approach to the design of molecular devices and machines, and illustrated our notion that nano-size constructs that use mechanisms developed in the evolution of biological molecules are simpler and uniquely suitable for nanoscale environments. To make it into a useful assay, we adapted this development for practical usage as an oscillating double-hairpin oligonucleotide probe which uses vaccinia DNA topoisomerase I for simultaneous detection of two specific types of DNA damage in situ (16, 18).
However the later work permitted us to significantly simplify the assay and make it faster and more cost-effective. The improvements resulted in the development of a new much shorter probe of a different configuration, which substitutes for the previously used oscillating double-hairpin. This significantly increased the speed of detection which now takes only minutes for completion and makes the assay significantly more robust and more sensitive.
Here we describe this new and improved technique. The assay selectively detects blunt ended DNA breaks with terminal 5’OH groups. It labels the waste-management phase of apoptotic DNA degradation in tissue sections. This technique takes into account recent changes in the apoptotic paradigm that views apoptosis as a broad reaction continuing beyond the individual cell program and requiring participation of other cells.
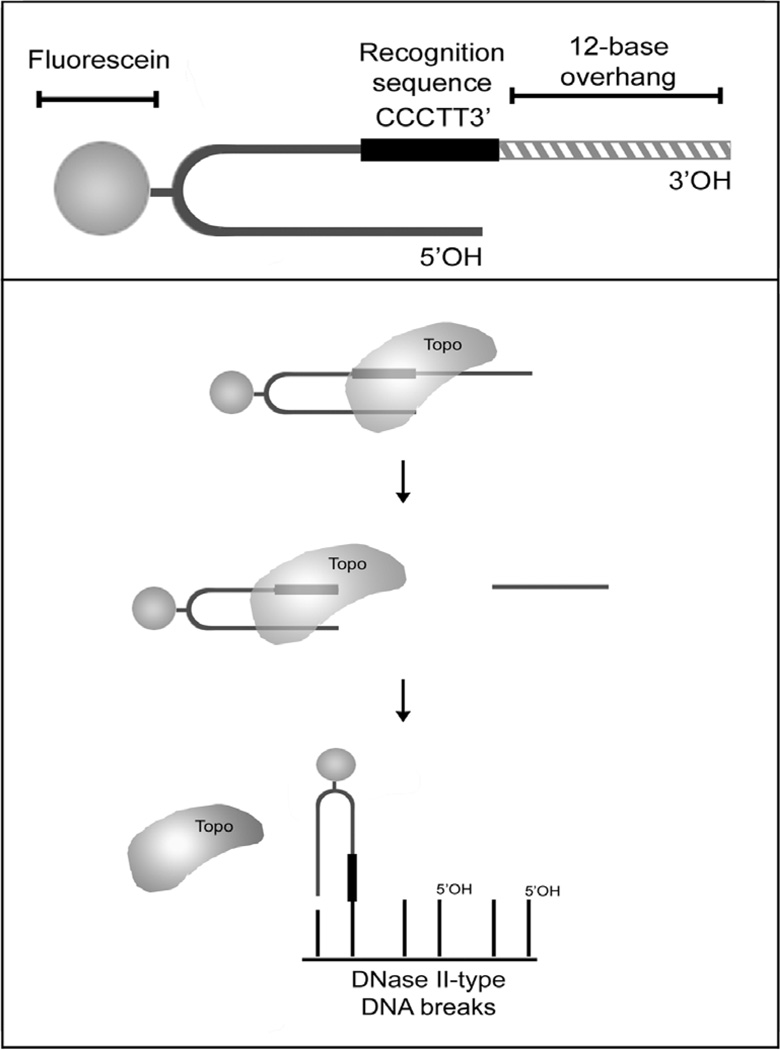
1.5. Principle of the assay (Fig.2)
Figure 2. Principle of the assay for DNA damage detection in situ using vaccinia topoisomerase I.
Vaccinia topoisomerase I (TOPO) binds to the oligoprobe and cleaves it at the 3’ end of the recognition sequence. The TOPO-activated FITC-labeled portion of the probe then religates to the blunt-ended DNA breaks with 5’OH in tissue section.
The assay uses an oligonucleotide probe and vaccinia DNA topoisomerase I. The assay utilizes the unique enzymatic properties of vaccinia DNA topoisomerase I, which can join two DNA molecules employing a mechanism different from those of ligases. This enzyme binds to duplex DNA having the CCCTT3’ recognition sequence, and creates a nick at its 3’ end (2). In normal conditions the enzyme then seals the nick, re-ligating the strand back to the acceptor DNA end with 5’OH.
In the assay, the topoisomerase CCCTT3’ recognition sequence is located in a hairpin oligonucleotide probe which has a 12-base-long single stranded region on its 3’ end. The enzyme recognition site is positioned at the end of the duplex-forming part of the probe and on the edge of an unhybridized 12 base overhang (Fig.2). When topoisomerase attaches to the probe, it cleaves the strand just after the recognition sequence. This cuts the 12-base long part of the oligonucleotide which then permanently separates, leaving vaccinia topoisomerase attached to the 3’ end of the blunt-ended hairpin. Now the oligonucleotide has a topoisomerase molecule strongly attached to its 3’ end. It can label DNase II type breaks because vaccinia topoisomerase I, which remains bound to the CCCTT motif, will religate the oligonucleotide to a double-strand DNA break possessing a complimentary 5’OH blunt-end. Therefore the breaks of DNase II type are detected specifically and directly.
The 12-bases overhang on the probe is required because the enzyme will not cut a shorter strand (19) and will therefore be unable to attach to the probe and activate its 3’end.
Although the topoisomerase-based assay can be used on its own, it can also be combined with in situ ligation. In this case, the T4 DNA ligase-based assay will label breaks with DNase I architecture, bearing 5’PO4 groups (Fig.1).
The combined detection, using both topoisomerase and ligase, is especially informative in the tissue section format because it can detect both phases of apoptotic cell disassembly: the self-driven cell disassembly and externally-controlled elimination of apoptotic cell corpses by phagocytizing cells.
When tested in tissue sections of dexamethazone-treated rat thymus, such a combined assay successfully detected both the primary DNase I-like cleavage in apoptotic thymocyte nuclei and the DNase II-like breaks in the cytoplasm of cortical macrophages ingesting apoptotic cells (16, 18).
The sensitivity of the assay depends on the method of signal registration and with the right microscope can potentially visualize very small numbers of DNase II-type breaks in individual cells (see Note 2).
In this chapter we present the complete protocol for topoisomerase-based detection applicable for fixed tissue sections.
2. MATERIALS
5–6µm-thick sections cut from paraformaldehyde-fixed, paraffin-embedded tissue blocks. Use slide brands which retain sections well, such as ProbeOn™ Plus charged and precleaned slides (Fisher Scientific, Pittsburgh, Pa) or similar product. Apoptotic tissue sections, such as dexamethazone treated rat thymus are recommended as controls (see Note 3
Xylene
80 and 96% Ethanol.
Oligoprobe 1 was synthesized by IDT (Integrated DNA Technologies, Inc., Coralville, IA). However it can be synthesized by many commercial oligonucleotide producers. PAGE or HPLC purification is recommended. Dilute with bidistilled water to 450 ng/µL stock concentration. Store at – 20°C protected from light. The probe is a labeled with a single fluorescein and detects blunt ended DNA breaks with 5’OH: 5’-AAG GGA CCT GCF GCA GGT CCC TTG ATA CGA TTC TA -3’F – FITC-dT
Vaccinia DNA topoisomerase l – 3000 U/µL (Millipore) (see Note 4).
50mM Tris-HCl, PH 7.4
Proteinase K (Roche Molecular Biochemicals, Indianapolis, IN) 20 mg/mL stock solution in distilled water. Store at - 20°C. In the reaction use 50µg/mL solution in PBS, prepared from the stock. Do not reuse (see Note 5).
Vectashield with DAPI (Vector Laboratories, Burlingame, CA)
100 ng/µL DNase I (Roche, Indianapolis, IN) in 50mM Tris-HCl, pH 7.4, 10mM MgCl2. DNase I (and DNase II) are needed for controls if verification of labeling specificity is required (see Note 6).
500 ng/µL DNase II in the buffer supplied with the enzyme (Sigma Chemicals, St. Louis, MO). Make the DNase II stock by diluting DNase II powder in water to the concentration of 1 mg/mL, aliquot in small volumes and keep the stock at −20° C. To run a reaction, dilute the stock solution 1:1 with the DNase II reaction buffer supplied with the enzyme.
Phosphate-buffered saline (1x PBS): dissolve 9g NaCl, 2.76 g NaH2PO4·H2O, 5.56 g Na2HPO4·7H2O in 800 mL of distilled water. Adjust to pH 7.4 with NaOH, and fill to 1L with distilled water.
Sodium bicarbonate buffer: 50mM NaHCO3, 15mM NaCl, pH 8.2
22×22mm or 22×40mm glass or plastic coverslips. Plastic coverslips are preferable as they are easier to remove from the section.
Fluorescent microscope with appropriate filters and objectives.
Video or photo camera for documentation.
3. METHOD
Labeling 5’OH blunt ended DNA breaks in tissue sections
Place the sections in a slide rack and dewax in xylene for 15 min, transfer to a fresh xylene bath for an additional 5 minutes.
Rehydrate by passing through graded ethanol concentrations: 96% Ethanol –2×5min; 80% Ethanol – 5min; water – 2×5 min.
Digest section with Proteinase K. Use 100µL of a 50µg/mL solution per section. Incubate 10 min at room temperature (23° C) in a humidified chamber (see Note 7).
Rinse in distilled water for 2×10 min.
While sections are rinsed in water, combine in a vial: 100 pmoles of Oligoprobe 1 and 100 pmoles (3.3 µg) of vaccinia topoisomerase I in solution of 50mM Tris-HCL, pH 7.4 (see Note 8). Incubate at room temperature for 15 min to allow for probe activation. Use 25µL of this reaction solution per section.
Aspirate water from sections and apply the reaction solution containing Oligoprobe 1 and vaccinia topoisomerase I.
Incubate for 15 min at room temperature (23° C) in a humidified chamber with a plastic coverslip, protected from light.
Remove coverslips by gently immersing the slides vertically in a coplin jar containing water at room temperature. Then wash section 3×10min in distilled water.
Rinse with sodium bicarbonate buffer (see Note 9).
Cover section with an antifading solution (Vectashield with DAPI), coverslip and analyze the signal using a fluorescent microscope. Double-strand DNA breaks with 5’OH will fluoresce green (see Note 10).
ACKNOWLEDGEMENT
This research was supported by grant R01 NS062842 from the National Institute of Neurological Disorders and Stroke, National Institutes of Health and by grants R21 NS064403 from the National Institute of Neurological Disorders and Stroke, National Institutes of Health through ARRA and R21 EB006301 National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
Footnotes
So far the only disease with suggested links to a disruption in apoptotic corpse disassembly is systemic lupus erythematosus (SLE). In SLE the apoptotic cells are not properly eliminated due to failures of the clearance system in the phase II of apoptotic DNA degradation. Cell corpses become the source of auto-antigens responsible for initiation and progression of the disease (20, 21).
The visualization of smaller numbers of breaks generated without apoptosis might require a biotin-labeled probe with enzymatic amplification of signal or a confocal microscope.
To make apoptotic thymus, inject Sprague-Dawley rats (150g) subcutaneously with 6 mg/kg dexamethasone (Sigma) dissolved in 30% dimethyl sulfoxide in water. Animals have to be sacrificed 24 h post injection. Fix the thymus tissue by incubating 18 hours in 4% paraformaldehyde. Then pass it through graded alcohols to 100% ethanol, place overnight in chloroform and embed in paraffin.
Highly concentrated vaccinia topoisomerase I, which works well with the described assay, can be obtained from Millipore, sold as a part of the ApopTag® ISOL Dual Fluorescence Apoptosis Detection Kit. The alternative commercial source of the highly concentrated preparation of this enzyme is Vivid Technologies (Houston, TX). Our initial enzyme preparation was obtained directly from Dr. Stewart Shuman (Sloan-Kettering Institute, New York) who originally described its purification (22). We have also received highly concentrated vaccinia topoisomerase I (3000 U/µL i.e. 0.2 µg /µL ~6 pmole/µL) from Epicentre Biotechnologies (Madison, WI) as a gift. The company’s regular preparation of 10 U/µL is too weak for the assay (1 pmole of the concentrated enzyme equals approximately 500 U).
Proteinase K is a very stable enzyme, when stored at concentrations higher than 1 mg/mL. However autolysis of the enzyme occurs in aqueous solutions at low concentrations (~ 10 µg/mL) (23).
DNase I and II treated sections can be used as positive and negative controls. Use sections of tissues with no preexisting DNA damage, such as normal bovine adrenal or rat heart. Not all normal tissues provide good controls. Normal rat brain sections, after treatment with DNase II, display high nonspecific background levels. Sections of such organs as normal thymus or small intestine contain cells with DNA cleavage.
The sections should be washed in water (2×10min), and treated with 100 ng/µL of DNase I (Roche) in 50mM Tris-HCL, pH 7.4, 10mM MgCl2 overnight at 37° C, or with 500 ng/µL DNase II in the buffer supplied with the enzyme (Sigma) for 30 min at 37° C. After washing in water (3×10min), and preblocking with 2% BSA (15 min, 23° C) the sections are ready for labeling.
Mock reactions without enzymes are also recommended as regular controls in order to assess nonspecific background staining. Sometimes additional controls might be needed to rule out possible contamination of vaccinia topoisomerase preparations with nucleases. For this the pretreatment of control sections with vaccinia topoisomerase I for 2 hours at 37° C is recommended followed by DNA breaks detection.
Proteinase K digestion time may need adjustment depending on the tissue type. Hard tissues might require longer digestion. Times of 10 min are usually used. Omitting the digestion step results in a weaker signal. On the other hand, overdigestion, results in signal disappearance and section disruption.
In the initial experiments we used 215 pmoles (7.1 µg) of the enzyme per every 25 µL of the reaction mix. However, the topoisomerase concentration can be significantly reduced without any loss of sensitivity. We later used four times less of the enzyme per section (1.76 µg in 25 µL of the reaction mix per section) with similar results. Reducing the amount of enzyme to 880 ng (in 25 µL of the reaction mix) resulted in a weaker signal and 8.8 ng of enzyme produced no signal.
Alkaline solution rinse enhances FITC fluorescence, which is pH sensitive and is significantly reduced below pH 7. This step is not needed if other non pH sensitive fluorophores are used in the probe.
Topoisomerase-based labeling and combined topoisomerase-ligase labeling can be performed using the ApopTag® ISOL Dual Fluorescence Kit (Millipore).
REFERENCES
- 1.Samejima K, Earnshaw WC. Trashing the genome: role of nucleases during apoptosis. Nature Reviews Molecular Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 2.Kawane K, et al. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nature Immunol. 2003;4:138–144. doi: 10.1038/ni881. [DOI] [PubMed] [Google Scholar]
- 3.Parrish JZ, Xue D. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma. 2006;115:89–97. doi: 10.1007/s00412-005-0038-0. [DOI] [PubMed] [Google Scholar]
- 4.Didenko VV, Hornsby PJ. Presence of double-strand breaks with single-base ’overhangs in cells undergoing apoptosis but not necrosis. J. Cell Biol. 1996;135:1369–1376. doi: 10.1083/jcb.135.5.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- 6.Staley K, Blaschke AJ, Chun J. Apoptotic DNA fragmentation is detected by a semi-quantitative ligation-mediated PCR of blunt DNA ends. Cell Death Differ. 1997;4:66–75. doi: 10.1038/sj.cdd.4400207. [DOI] [PubMed] [Google Scholar]
- 7.Weir AF. Deoxyribonuclease I and II. In: Burrell MM, editor. Enzymes of Molecular Biology. Totowa NJ: Humana; 1993. pp. 7–16. [Google Scholar]
- 8.Didenko VV, Ngo H, Baskin DS. In situ detection of double-strand DNA breaks with terminal 5’OH groups. In: Didenko VV, editor. In Situ Detection of DNA Damage: Methods and Protocols. Totowa NJ: Humana; 2002. pp. 143–151. [DOI] [PubMed] [Google Scholar]
- 9.Sikorska M, Walker PR. Endonuclease activities and apoptosis. In: Lockshin RA, Zakeri Z, Tilly JL, editors. When Cells Die. New York: Wiley-Liss; 1998. pp. 211–242. [Google Scholar]
- 10.Didenko VV. Detection of specific double-strand DNA breaks and apoptosis in situ using T4 DNA ligase. In: Didenko VV, editor. In Situ Detection of DNA Damage: Methods and Protocols. Totowa NJ: Humana; 2002. pp. 143–151. [DOI] [PubMed] [Google Scholar]
- 11.Chahory S, Padron L, Courtois Y, Torriglia A. The LEI/L-DNase II pathway is activated in light-induced retinal degeneration in rats. Neurosci. Lett. 2004;367:205–209. doi: 10.1016/j.neulet.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Krieser RJ, MacLea KS, Longnecker DS, Fields JL, Fiering S, Eastman A. Deoxyribonuclease IIalpha is required during the phagocytic phase of apoptosis and its loss causes perinatal lethality. Cell Death Differ. 2002;9:956–962. doi: 10.1038/sj.cdd.4401056. [DOI] [PubMed] [Google Scholar]
- 13.Loo DT. In Situ Detection of Apoptosis by the TUNEL Assay: An Overview of Techniques. In: Didenko VV, editor. In Vivo and In Situ Detection of DNA Damage: Methods and Protocols. Totowa NJ: Humana; 2009. pp. This volume. [Google Scholar]
- 14.Thiry M. In situ nick translation at the electron microscopic level. In: Didenko VV, editor. In Situ Detection of DNA Damage: Methods and Protocols. Totowa NJ: Humana; 2002. pp. 121–130. [DOI] [PubMed] [Google Scholar]
- 15.Hornsby PJ, Didenko VV. In situ oligo ligation (ISOL): A decade and a half of experience. In: Didenko VV, editor. In Vivo and In Situ Detection of DNA Damage: Methods and Protocols. Totowa NJ: Humana; 2009. pp. This volume. [Google Scholar]
- 16.Didenko VV, Minchew CL, Shuman S, Baskin DS. Semi-artificial fluorescent molecular machine for DNA damage detection. Nano Letters. 2004;4:2461–2466. doi: 10.1021/nl048357e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes B. Colour-coded tags show DNA damage. New Scientist. 2004;2477:23. [Google Scholar]
- 18.Didenko VV. Oscillating probe for dual detection of 5’PO4 and 5’OH DNA breaks in tissue sections. In: Didenko VV, editor. Fluorescent Energy Transfer Nucleic Acid Probes: Methods and Protocols. Totowa NJ: Humana; 2006. pp. 59–69. [DOI] [PubMed] [Google Scholar]
- 19.Shuman S. Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J. Biol. Chem. 1992;267:16755–16758. [PubMed] [Google Scholar]
- 20.Gabler C, Blank N, Winkler S, Kalden JR, Lorenz HM. Accumulation of histones in cell lysates precedes expression of apoptosis-related phagocytosis signals in human lymphoblasts. Ann. NY Acad. Sci. 2003;1010:221–224. doi: 10.1196/annals.1299.039. [DOI] [PubMed] [Google Scholar]
- 21.Cocca BA, Cline AM, Radic MZ. Blebs and apoptotic bodies are B cell autoantigens. J. Immunol. 2002;169:159–166. doi: 10.4049/jimmunol.169.1.159. [DOI] [PubMed] [Google Scholar]
- 22.Shuman S, Golder M, Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem. 1988;263:16401–16407. [PubMed] [Google Scholar]
- 23.Sweeney PJ, Walker JM. Proteinase K (EC 3.4.21.14) In: Burrell MM, editor. Enzymes of Molecular Biology. Totowa NJ: Humana; 1993. pp. 305–311. [DOI] [PubMed] [Google Scholar]