Abstract
The extracellular domain of p185c-neu can be viewed as a complex structure of four subdomains, two of which are cysteine-rich subdomains. We have investigated the contribution of these distinct p185c-neu extracellular subdomains to p185/epidermal growth factor receptor (EGFR) heteromer formation and EGF-induced heteromeric signaling. Our studies indicate that at least two separate p185 subdomains, a region spanning subdomains I and II and subdomain IV are involved in association of p185 with the EGFR. We also demonstrated that subdomain IV reduced the heteromeric signaling and transforming activities induced by EGF after associating with EGFR. When 126 aa were deleted from subdomain IV, this small subdomain IV-derived fragment could still lead to heterodimers with EGFR and suppress EGF-induced mitogen-activated protein kinase activation and subsequent transformation abilities. These data provide information about trans-inhibitory mechanisms of mutant p185 species and also indicate that both the entire and a part of subdomain IV may represent a therapeutic target for erbB-overexpressing tumors. Finally, these studies define a basic feature of receptor-receptor associations that are determined by cystine-knot containing subdomains.
Keywords: heterodimer, cysteine-rich domain
Epidermal growth factor receptor (EGFR), erbB-2 (the human homologue of rat p185c-neu), erbB-3, and the erbB-4 proteins are all members of the erbB receptor family of tyrosine kinases (1–9). Homodimerization or heterodimerization of these receptors plays an important role in receptor complex activation and signal transduction (7–15). Among these receptors, p185c-neu/erbB2 is the preferred heterodimerization partner with other erbB receptors (7, 11–14). In the assembly of p185c-neu/ EGFR heterodimers, ligands such as EGF facilitate the formation of the heteromeric structure, stimulate subsequent rapid phosphorylation of the ensemble, and ultimately cause activation of the p185 partners, leading in some cases to transformation (9, 16, 17). However, the mechanics of p185c-neu receptor activation that occurs in receptor dimerization with EGFR and the role of the discrete p185c-neu extracellular subdomains in these processes are not well understood.
p185c-neu has four extracellular subdomains (subdomains I–IV) that include two cysteine-rich domains (II, IV). The functional significance of any individual subdomain remains unknown (3, 8). Cysteine-rich domains generally are composed of cystine knots that are characterized by a distinct pattern of disulfide bonds (18, 19). In this regard, mutation of cysteine residues proximal to the membrane region in the p185c-neu subdomain IV promotes receptor dimerization and activation through the formation of disulfide bonds between the monomers (20, 21).
Recently, an alternative transcript product of c-erbB2 that consists of subdomains I and II followed by an additional 79 aa was found to be able to dimerize with wild-type p185erbB2 protein and was able to reduce signaling in a dominant negative manner (22). Our laboratory developed a panel of p185c-neu mutants to begin to clarify the role of individual p185c-neu subdomains in heterodimerization with EGFR. Previously we reported that p185c-neu lacking most of the cytoplasmic domain with (T691 stop) or without (N691 stop) single-point mutation (V664G) in transmembrane domain mediates a suppressive effect on the function of EGFR and that of p185c-neu in a trans-inhibitory manner (23–26). Analysis of the subdomain contributions to receptor formation is important for understanding the molecular mechanisms and general principles involved in receptor dimerization. Identification of sites that are necessary for dimerization may facilitate the development of therapeutic approaches against cancers characterized by the overexpression of either EGFR or erbB2 (27–30).
Materials and Methods
Construction of Expression Vectors.
A schematic representation of the expression vectors used in this study is shown in Fig. 1. pTNeu and pNeu contain full-length transforming p185neu or the cellular protooncogenic p185c-neu form lacking the single-point mutation (V664G) in the transmembrane region (3, 4, 16). pTex3–4, pTex4, pTex6CN, pNex1–3, pNex3–4, and pNex3 are truncated p185neu or p185c-neu expression vectors lacking most of the intracellular domain and portions of the extracellular subdomains. pTex encodes the full-length extracellular domain and the transmembrane domain of p185neu. All of these constructs were subcloned into pSectagB (Invitrogen). pMVEGFR is an expression vector of human EGFR containing vesticular stomatitis virus glycoprotein (VSVG) tag at C terminal, which was subcloned into pMV expression vector (Roche Molecular Biochemicals).
Figure 1.
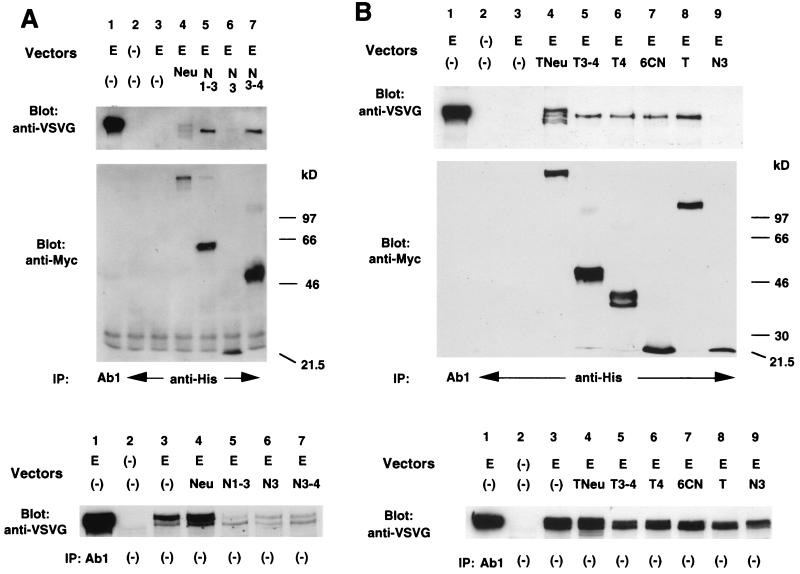
Schematic representation of the p185neu proteins and mutants used in this study. Numbers, amino acid positions from the first Met at N terminal of p185c-neu or EGFR. SP, signal peptide; I, subdomain I; II, subdomain II; III, subdomain III; IV, subdomain IV; TM, transmembrane domain; hatched squares, TM with single-point mutation (V664G) in transmembrane domain; TK, tyrosine kinase domain; Myc, Myc epitope; His, polyhistidine tag; VSVG, VSVG tag; broken line, deleted region.
Transfection.
Transfection was performed by using the DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) liposomal transfection reagent according to the manufacturer's protocol (Roche Molecular Biochemicals).
Cell Lines.
Cos7 and NR6 cells (31) were used in this study and were cultured in DMEM (Bio-Whittaker) supplemented with 10% FBS (HyClone), 100 units/ml penicillin, 50 μg/ml streptomyocin, and 2 mM l-glutamine and were maintained in a 5% CO2 humidified incubator at 37°C. pMVEGFR was transfected into NR6 cells followed by selection with 400 μg/ml of G418 to establish the NE99 cell line. NE99 is a stable NR6 cell line expressing EGFR. NE99 cells were next transfected with pTex, pTex4, or pTex6CN followed by the selection with 400 μg/ml of Zeocin (Invitrogen) to establish the NE/Tex, NE/Tex4, and NE/Tex6CN cell lines, respectively.
Heterodimerization Studies.
Cos7 cells were transfected with 5 μg of pMVEGFR and an equal amount of mutant p185 expression vectors. Forty eight hours later, medium was replaced with serum-free medium and cells were incubated for an additional 24 h. Cells were stimulated with 200 ng/ml of EGF (GIBCO/BRL) at 37°C for 10 min followed by crosslinker treatment of DTSSP (Pierce) at room temperature for 30 min as described (23). After stopping the crosslinking reaction with 10 mM Tris⋅HCl (pH 7.5), 0.9% NaCl, and 0.1 M glycine, cells were lysed in buffer containing 10 mM sodium phosphate (pH 7.4), 1% Triton X-100, 1% deoxycholic acid, 0.1% SDS, 150 mM NaCl, 1 μg/ml of aprotinin, 1 mM PMSF, 2 mM EDTA, 1 mM sodium orthovanadate, 10 mM NaF, 10 mM sodium pyrophosphate, and 10 mM iodoacetamide. Equal amounts of each cell lysate were immunoprecipitated with anti-His antibody (Invitrogen) or anti-EGFR antibody, Ab-1 (Oncogene Research Products, Cambridge, MA). Immunoprecipitated samples were subjected to SDS/PAGE analysis and transferred to nitrocellulose membranes. Immunoblotting was performed with anti-VSVG antibody (Roche Molecular Biochemicals) that recognizes the tagged EGFR. After stripping, the membranes were reprobed with anti-c-Myc antibody (Santa Cruz Biotechnology). Equal amounts of total cell lysate also were electrophoresed to determine the expression of exogenous EGFR as detected with anti-VSVG antibody.
Analysis of EGFR Phosphotyrosine Content.
Stable NR6 transfectants were starved for 24 h with serum-free medium and then were stimulated with EGF (50 ng/ml) at 37°C for 5 min. Cells then were lysed in lysis buffer as described above. Transiently transfected Cos7 cells were starved for 24 h and stimulated with EGF as described above, followed by treatment with the crosslinking reagent DTSSP at 4°C for 30 min. After quenching the reaction, cells were lysed with lysis buffer. Equal amounts of each cellular lysate were immunoprecipitated either with an anti-His antibody to detect heteromeric EGFR-p185 species in Cos7 cells or with anti-EGFR antibody R1 (Santa Cruz Biotechnology) to detect EGFR species in NR6 cell transfectants. Immunoprecipitated samples were separated on SDS/polyacrylamide gels and transferred to nitrocellulose membranes. After immunoblotting with either anti-Myc antibody or anti-VSVG, membranes were reblotted with antiphosphotyrosine antibody PY99 (Santa Cruz Biotechnology). The ratio of phosphorylated EGFR/total EGFR was approximated by laser scanning microdensitometry using National Institutes of Health image analysis software.
Mitogen-Activated Protein (MAP) Kinase Assay.
MAP kinase assays were performed as described with only slight modifications (32). In brief, Cos7 cells were transiently transfected with 3 μg of construct encoding each mutant p185neu species and an equal amount of pCDNA3-HA-ERK2 (a gift from Silvio Gutkind, National Institutes of Health, Bethesda, MD). Forty eight hours later, cells were starved for an additional 24 h, followed by stimulation with 50 ng/ml of EGF at 37°C for 5 min. Cells were lysed in buffer containing 10 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 2 mM EGTA, 2 mM DTT, 1 mM PMSF, 1 mM sodium orthovanadate, 10 μg/ml of aprotinin, and 1% Triton X-100. One thousand micrograms of each cell lysate was immunoprecipitated with 0.5 μg of anti-hemagglutinin antibody (Roche Molecular Biochemical). Immunoprecipitated samples were washed three times with lysis buffer and twice with 30 mM Hepes (pH 7.4), 10 mM MgCl2, 1 mM DTT. Immunoprecipitates were incubated with 2 μg of myelin basic protein (Sigma) in the 25 μl of reaction buffer containing 30 mM Hepes (pH 7.4), 10 mM MgCl2, 1 mM DTT, 10 μM ATP, and 1 μCi of γ32P ATP (NEN) at 30°C for 20 min. Reaction samples were electrophoresed in SDS/polyacrylamide gel, and phosphorylated myelin basic protein was visualized by autoradiography.
Anchorage-Independent Growth Assays.
Anchorage-independent growth was determined by analyzing colony formation of cells in soft agar as described (23) with only slight modifications. In brief, on day 0, 3,000 cells were suspended in 0.3% or 0.33% agarose in 5% FBS-DMEM and plated on the bottom agar (0.4%). Assays were performed in duplicate. From day 1, 0.3 ml of 5% FBS-DMEM with or without 10 ng/ml of EGF was added every 3 days. On day 21, cells were stained with p-indonitrotetrazolium violet (1 mg/ml) for 24 h, and visible colonies were counted.
Results
p185 Extracellular Subdomain Mutants Affect p185-EGFR-Heterodimer Formation.
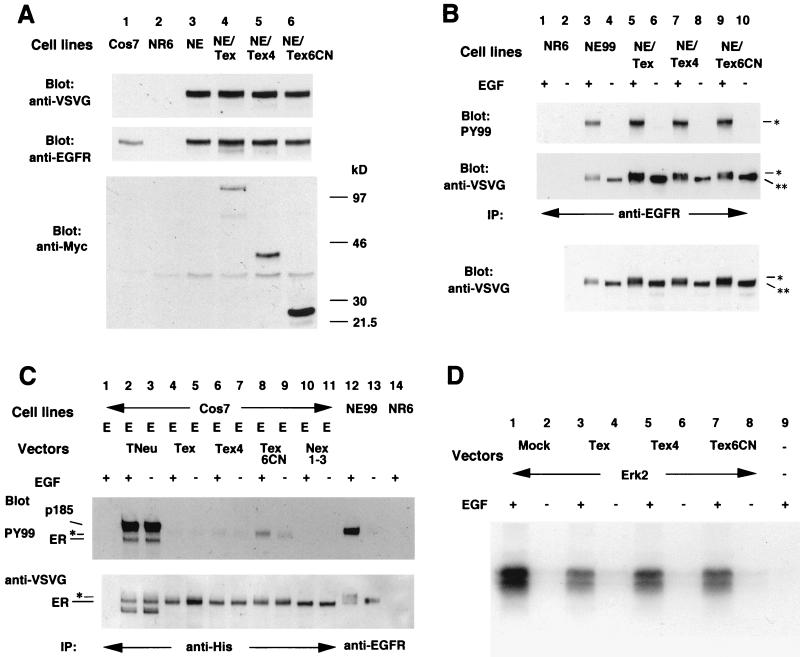
Cos7 cells were used for the heterodimer studies, and this cell line expresses some endogenous EGFR (see Fig. 3A). EGFR constructs containing a VSVG tag at the carboxyl terminal were used to distinguish the endogenous from the exogenous EGFR species. A variety of p185 mutants (see Fig. 1) were transfected along with the EGFR constructs, and dimerization behavior was analyzed. As shown in Fig. 2A, pNex3 (lane 6) showed reduced dimerization with EGFR compared with pNex3–4 or pNex1–3. We also evaluated p185neu and mutant p185neu species that possessed a point mutation in the transmembrane domain. pTex, pTex3–4, and pTex4 all were able to heterodimerize with EGFR (Fig. 2B, lanes 5, 6, and 8). These data indicate that certain extracellular subdomains of p185 play an important role in dimerization. At least two areas including subdomain IV and subdomains I and II affected heterodimerization. Furthermore, part of the subdomain IV containing only a small 46-aa region extending from the cell membrane (pTex6CN) was still able to dimerize with EGFR.
Figure 3.
Analysis of the effect of each mutant p185neu form on EGF-mediated signaling. (A) Analysis of the expression of EGFR and each mutant p185neu form in each cell line. The antibodies used were anti-VSVG (Top) followed by the second immunoblot with anti-EGFR, 1005 (Santa Cruz Biotechnology, Middle). Another membrane was immunoblotted with anti-Myc to determine the expression of each mutant p185neu form (Bottom). (B) Phosphorylation of EGFR in each NE99 clone with (+) or without (−) EGF stimulation. Membrane was immunoblotted with anti-VSVG (Middle), then reblotted with antiphosphotyrosine antibody, PY99 (Top). The same total cell lysates were transferred to the nitrocellulose membrane and blotted with anti-VSVG (Bottom). IP, immunoprecipitation; **, EGFR species with an approximate molecular mass of 170 kDa; *, the slower-migrating EGFR species. (C) Phosphorylation of heteromeric EGFR in transiently transfected Cos7 cells with (+) or without (−) EGF stimulation followed by the crosslinker treatment. Membrane was immunoblotted with anti-VSVG (Bottom), then reblotted with antiphosphotyrosine antibody, PY99 (Middle). E, pMVEGFR; ER, EGFR species with an approximate molecular mass of 170 kDa; *, slower-migrating EGFR species. (D) MAP kinase assay. Transiently transfected Cos7 cells with (+) or without (−) EGF stimulation were used for MAP kinase assay as described in Materials and Methods. Phosphorylated myelin basic protein was visualized by autoradiography. Mock, pSectagC as an empty vector; Erk2, pCDNA3-HA-ERK2.
Figure 2.
Analysis of heterodimer formation between EGFR and each mutant p185c-neu protein. Cos7 cells transiently transfected with the indicated vectors were stimulated with EGF followed by the crosslinker treatment as described in Materials and Methods. Cell lysates were immunopretipitated (IP) with the antibodies as indicated. Immunoblottings (Blot) were performed with the antibodies as indicated. E, pMVEGFR; Neu, pNeu; N, pNex; N3–4, pNexD3–4; N1–3, pNex1–3; N3, pNex3; Tex, pTex; T3–4, pTexD3–4; T4, pTexD4; 6CN, pTex6CN. Membranes indicated (Top) were reblotted with anti-VSVG as indicated (Middle), and the same total cell lysates was subjected to the nitrocellulose membrane to determine the exogenous expression of EGFR (Bottom).
When we evaluated all of the mutants with a transmembrane mutation (Fig. 2B), pTex, which has the entire extracellular domain of p185neu, was found to heterodimerize the most efficiently.
p185 Extracellular Subdomain Mutants Effect on EGF-Induced Phosphorylation of EGFR.
We next analyzed whether subdomain IV or a part of this subdomain could suppress EGF-induced signaling after heterodimerization with EGFR. We transfected pMVEGFR into NR6 cells that express no detectable endogenous EGFR to establish the NE99 cell line. NE99 cells then were transfected with either pTex, pTex4, or pTex6CN to establish the NE/Tex, NE/Tex4, and NE/Tex6CN stably transfected cell lines, respectively. We analyzed the expression of both exogenous EGFR and mutant p185neu proteins in each cell line. As shown in Fig. 3A Top and Middle, under nonstimulated conditions, NE99, NE/Tex, NE/Tex4, and NE/Tex6CN expressed almost equal levels of EGFR. The EGFR expression was even higher than that seen in Cos7. On the other hand, the expression of the mutant p185neu species in NE/Tex6CN was somewhat higher than in NE/Tex or NE/Tex4 (Fig. 3A Bottom, which detects p185neu forms that are myc-tagged).
The phosphotyrosine content of EGFR in the NR6 clones was analyzed next. Upon EGF stimulation, EGFR in each cell line became phosphorylated (Fig. 3B). We contrived a ratio of phosphorylated to total EGFR to gauge relative kinase activity. The ratio of phosphorylated/total EGFR in NE99 was the highest of all and was the lowest in NE99/Tex (39% of the ratio in NE99), whereas those in NE99/Tex4 or in Ne99/Tex6CN were intermediate (57% and 63%, respectively). These data suggest that mutant p185neu forms suppressed EGF-induced signaling by affecting the extent of tyrosine phosphorylation of the EGFR. Furthermore, the EGFR species in NE99, NE/Tex, NE/Tex4, and NE/Tex6CN cell lines displayed a molecular weight change after EGF stimulation. These changes were observed in both immunoprecipitated samples and after electrophoresis of the total cell lysates (Fig. 3B Middle and Bottom). Upon detailed review of the gels, we noticed that the molecular weight range of the EGFR species in NE/Tex, NE/Tex4, and NE/Tex6CN was consistently greater when compared with that seen in NE99. The slower migrating EGFR species in the cells with mutant p185neu (Fig. 3B Top and Middle, marked *) seemed to be reactive with phosphotyrosine antibody, whereas the faster migrating EGFR species in each cell line (marked **) was less phosphorylated after stimulation with EGF. These data indicate the possibility that EGFR-mutant p185neu complexes contain a poorly phosphorylated species of EGFR, which might account for the slightly lower molecular weight forms.
To investigate this pattern further, we used transiently transfected Cos7 cells to analyze the phosphotyrosine content of EGFR that heterodimerized with mutant p185 forms. As shown in Fig. 3C, each major EGFR species that underwent dimerization with truncated p185 showed the same molecular weight as that of nonstimulated EGFR and less than that of the slow-migrating EGFR species in NE99. This latter form was highly reactive with antiphosphotyrosine antibody after EGF stimulation. The major EGFR species dimerizing with truncated p185 did not change their molecular weight nor acquire additional phosphorylation after stimulation with EGF. In addition to the major EGFR species, other slow-migrating species can be detected as very faint bands just above the major band (Fig. 3C Bottom, lanes 4–9, marked *). These slow-migrating species were phosphorylated under nonstimulated conditions. Among these species, only EGFR dimerizing with Tex6CN showed an increase of phosphorylation after EGF stimulation (Fig. 3C Top, lane 8).
Collectively, these data suggest that subdomains I, II, and IV suppress EGFR function in a trans-inhibitory manner, whereas a smaller fragment of subdomain IV suppresses EGFR function less effectively.
Suppression of EGF-Induced MAP Kinase Activation by Subdomain IV of Neu.
To evaluate whether pTex4 can suppress downstream signals emanating from the activated EGFR, we used the MAP kinase assay as a screen. These studies used transiently transfected Cos7 cells that express endogenous EGFR (Fig. 3A). We transfected pCDNA3-HA-ERK2 with the mutant p185neu expression vector or empty vector, pSectagC (Invitrogen). As shown in Fig. 3D, each transiently transfected Cos7 cell line showed activated ERK2 activity after EGF stimulation. Therefore, the mutant p185 forms could not completely suppress ERK2 activation triggered by this level of EGF stimulation. However, the degree of ERK2 activation was elevated the most in Cos7 cells with ERK2 alone (Fig. 3D, lanes 1 and 2), and the least in cells with ERK2 and pTex (Fig. 3D, lanes 3 and 4, 47% of the result obtained by ERK2 alone), and only intermediate in cells with ERK2 and either pTex4 (62%) or pTex6CN (56%). These data are consistent with our assessment of the phosphotyrosine content of the EGFR that reflects the overall tyrosine kinase activity of EGFR.
Effect of p185 Extracellular Subdomain IV on the Growth of the Cells Transformed by EGFR.
We evaluated the suppressive effect of various kinds of mutant p185neu forms, including the smallest mutant species in anchorage-independent growth assays. As shown in Table 1, NR6 cells were transformed and formed foci in response to EGF stimulation by the transfection of EGFR (NE99). NE/Tex, NE/Tex4, and NE/Tex6CN also retained colony-forming abilities albeit at a somewhat reduced level in 0.3% agar. We assessed colony assays with a higher percentage of agar (0.33%), and only NE99 cells showed more than 25 colonies in response to EGF stimulation, whereas others showed fewer than three colonies. Therefore, subdomain IV of p185neu or a fragment of this domain suppressed not only EGF-induced signaling but also EGF-induced transforming capabilities of EGFR-expressing cells. We consider these data to reflect the potent affects of phenotypic modulation that mutant species of p185neu are able to exert.
Table 1.
Summary of studies of the transforming efficiency of mutant p185neu forms coexpressed with the EGFR
| Cell lines | 0.3% agar
|
0.33% agar
|
||
|---|---|---|---|---|
| EGF (+) | (−) | (+) | (−) | |
| NE99 | 67 | 7 | 31 | 9.5 |
| NE/Tex | 25.5 | 2.5 | 1.5 | 1.5 |
| NE/Tex4 | 29.5 | 5.5 | 0 | 2 |
| NE/Tex6CN | 7.5 | 1 | 0.5 | 0 |
| NR6 | 1 | 1 | 1.5 | 1.5 |
Anchorage-independent growth was determined as described in Materials and Methods. The number of average colonies obtained by each clone is indicated.
Discussion
Downstream signaling emanating from erbB receptor complexes has been investigated (10, 14, 33), but the role of distinct extracellular subdomains of p185c-neu in creating signaling receptor ensembles with EGFR has not been fully defined. Previous efforts from our laboratory suggested that the extracellular domain is involved in homodimer and heterodimer formation (23–25), even in the context of naturally occurring truncated EGFR forms (34). We focused most closely on the role of subdomain IV of p185neu receptors because mutagenesis studies of this cysteine-rich domain of EGFR and p185c-neu have implicated this region in the formation and stabilization of dimerization events (20, 21, 35).
The function of the p185neu subdomain IV and a portion of the subdomain IV (containing six cysteine residues) in heterodimerization and EGF-induced signaling was assessed, and we found that both subdomain IV and the fragment derived from it can bind EGFR. Although complexes of the larger extracellular domain fragment of p185neu are effective for suppression of EGFR phosphorylation, a subportion of subdomain IV (Tex6CN) was still able to suppress EGFR activation. This species also suppressed EGF-induced MAP kinase activity to some extent. In anchorage-independent growth assays, pTex, pTex4, and pTex6CN all suppressed the transforming ability of EGFR. Among all transfectants containing mutant p185neu species, NE/Tex6CN had the least transforming ability although its binding affinity to EGFR or suppressive effect against EGF-induced signaling is certainly less than that of another p185 form or the entire extracellular domain of p185neu. One reason to account for these properties might be the high expression level of the mutant p185neu form in the NE/Tex6CN cell lines. These results suggest the possibility that a part of the p185c-neu subdomain IV might be useful for the design of therapeutic peptidomimetics against tumors overexpressing EGFR and perhaps other members of the erbB family (18).
Extracellular domains lacking subdomain IV also heterodimerized with EGFR. This finding suggests that additional distinct areas in the extracellular domain of p185c-neu may be involved in heterodimerization with EGFR. Doherty et al. (22) found a form of the extracellular domain consisting of subdomain I and II followed by an additional 79-aa sequence was an autoinhibitor of p185erbB2 protein. We showed that subdomain III with transmembrane domain of p185c-neu (Nex3) had reduced affinity against heterodimerization, whereas subdomains I–III (Nex1–3) dimerize efficiently with EGFR. Moreover, the increase of phosphotyrosine content of this heterodimer was not observed upon EGF stimulation, suggesting that subdomains I and II stabilized the heteromer, and this association leads to inhibition of EGF-induced signaling. However, the role of subdomain III of p185c-neu remains unclear because Nex3 may have lost its inherent function due to conformational changes induced by the deletion of subdomain IV.
Investigation of the phosphotyrosine content of the EGFR heterodimerized with our mutant p185neu species also provided information about erbB heteromeric complexes. The majority of the EGFR heterodimerized with entire extracellular domain of p185neu (Tex), subdomain IV (Tex4), or subportion of subdomain IV (Tex6CN) displayed the same molecular weight as that of nonstimulated EGFR in NE99. These species did not show any phosphorylation nor increase of molecular weight upon EGF stimulation. We have argued that heterodimeric complexes comprised of two kinase-active subunits (p185 and EGFR) are probably activated predominantly in trans (11). p185c-neu/erbB2 is not simply a substrate for EGFR but a transactivator as well because EGFR dimerizing with p185c-neu lacking most of the cytoplasmic domain (N691 stop) is inactivated even upon EGF stimulation (23).
On the other hand, the slow-migrating species of heteromeric EGFR has a basal level phosphotyrosine content. In addition, the species of the EGFR that heterodimerized with a subportion of p185neu-subdomain IV (EGFR-Tex6CN) showed an increase of phosphotyrosine content upon EGF stimulation. As Tex6CN does not have a tyrosine kinase domain, the increase of phosphorylation may be due to the existence of another undefined tyrosine kinase. Thus the slow-migrating EGFR species may be involved in even more receptor complex assemblies and are not merely simple monomeric forms.
In conclusion, p185 extracellular subdomains I, II and IV are involved in association of the p185 species with the EGFRs. Subdomain IV and a fragment of subdomain IV reduced the EGF-induced heteromeric signaling and transforming activities. In addition, these data provide additional information about trans-inhibitory mechanisms of erbB receptor complexes and suggest that subdomain IV, or even a subportion of subdomain IV, may possess therapeutic potential as a target for erbB-driven transformation. We have further delineated the role of distinct p185c-neu subdomains in p185c-neu/EGFR heteromeric association and activation. Several peptidomimetics based on a putative structural model of subdomain IV have been prepared, and their suppressive effects against erbB-mediated transformation will be evaluated.
Acknowledgments
This work was supported by grants from the National Cancer Institute, the American Cancer Society, the U.S. Army, and Abramson Family Cancer Center to M.I.G. This work also was supported by grants from the Veterans Administration Merit Review Program and The Brain Tumor Society to D.M.O.
Abbreviations
- EGF
epidermal growth factor
- EGFR
EGF receptor
- VSVG
vesticular stomatitis virus glycoprotein
- MAP
mitogen-activated protein
References
- 1.Ullrich A, Coussens L, Hayflick J S, Dull T J, Gray A, Tam A W, Lee J, Yarden Y, Libermann T A, Schlessinger J, et al. Nature (London) 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto T, Ikawa S, Akiyama T, Semba K, Nomura N, Miyajima N, Saito T, Toyoshima K. Nature (London) 1986;319:230–234. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- 3.Bargmann C I, Hung M C, Weinberg R A. Nature (London) 1986;319:226–230. doi: 10.1038/319226a0. [DOI] [PubMed] [Google Scholar]
- 4.Hung M-C, Schechter A L, Chevray P-Y M, Stern D F, Weinberg R A. Proc Natl Acad Sci USA. 1986;83:261–264. doi: 10.1073/pnas.83.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus M H, Issing W, Miki T, Popescu N C, Aaronson S A. Proc Natl Acad Sci USA. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plowman G D, Whitney G S, Neubauer M G, Green J M, McDonald V L, Todaro G J, Shoyab M. Proc Natl Acad Sci USA. 1990;87:4905–4909. doi: 10.1073/pnas.87.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plowman G D, Culouscou J-M, Whitney G S, Green J M, Carlton G W, Foy L, Neubauer M G, Shoyab M. Proc Natl Acad Sci USA. 1993;90:1746–1750. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ullrich A, Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 9.Wada T, Qian X, Greene M I. Cell. 1990;61:1339–1347. doi: 10.1016/0092-8674(90)90697-d. [DOI] [PubMed] [Google Scholar]
- 10.Carraway K L, III, Cantley L C. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 11.Heldin C-H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 12.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin B J, Sela M, Yarden Y. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 13.Graus-Porta D, Beerli R R, Daly J M, Hynes N E. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olayioye M A, Neve R M, Lane H A, Hynes N E. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson K M, Darling P J, Mohan M J, Macatee T L, Lemmon M A. EMBO J. 2000;19:4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. Nature (London) 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 17.Kokai Y, Myers J N, Wada T, Brown V I, LeVea C M, Davis J G, Dobashi K, Greene M I. Cell. 1988;58:287–292. doi: 10.1016/0092-8674(89)90843-x. [DOI] [PubMed] [Google Scholar]
- 18.Murali R, Greene M I. Immunol Res. 1998;17:163–169. doi: 10.1007/BF02786441. [DOI] [PubMed] [Google Scholar]
- 19.Ward C W, Hoyne P A, Flegg R H. Proteins. 1995;22:141–153. doi: 10.1002/prot.340220207. [DOI] [PubMed] [Google Scholar]
- 20.Siegel P M, Muller W J. Proc Natl Acad Sci USA. 1996;93:8878–8883. doi: 10.1073/pnas.93.17.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel P M, Ryan E D, Cardiff R D, Muller W J. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty J K, Bond C, Jardim A, Adelman J P, Clinton G M. Proc Natl Acad Sci USA. 1999;96:10869–10874. doi: 10.1073/pnas.96.19.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X, LeVea C M, Freeman J K, Dougall W C, Greene M I. Proc Natl Acad Sci USA. 1994;91:1500–1504. doi: 10.1073/pnas.91.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian X, Dougall W C, Hellman M E, Greene M I. Oncogene. 1994;9:1507–1514. [PubMed] [Google Scholar]
- 25.Qian X, O'Rourke D M, Zhao H, Greene M I. Oncogene. 1996;13:2149–2157. [PubMed] [Google Scholar]
- 26.O'Rourke D M, Qian X, Zhang H, Davis J G, Nute E, Meinkoth J, Greene M I. Proc Natl Acad Sci USA. 1997;94:3250–3255. doi: 10.1073/pnas.94.7.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrich A, Schlessinger J. Nature (London) 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 28.Slamon D J, Clark G M, Wong S G, Levin W J, Ullrich A, McGuire W L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 29.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, Press M F. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 30.Schlegel J, Merdes A, Stumm G, Albert F K, Forsting M, Hynes N, Kiessling M. Int J Cancer. 1994;56:72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- 31.Pruss R M, Herschman H R. Proc Natl Acad Sci USA. 1977;74:3918–3921. doi: 10.1073/pnas.74.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian X, O'Rourke D M, Fei Z, Zhang H, Kao C, Greene M I. J Biol Chem. 1999;274:574–583. doi: 10.1074/jbc.274.2.574. [DOI] [PubMed] [Google Scholar]
- 33.Hunter T. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 34.O'Rourke D M, Nute E J, Davis J G, Wu C, Lee A, Murali R, Zhang H, Qian X, Kao C, Greene M I. Oncogene. 1998;16:1197–1207. doi: 10.1038/sj.onc.1201635. [DOI] [PubMed] [Google Scholar]
- 35.Saxon M L, Lee D C. J Biol Chem. 1999;274:28356–28362. doi: 10.1074/jbc.274.40.28356. [DOI] [PubMed] [Google Scholar]