Abstract
Here, we describe a Drosophila melanogaster transposon insertion mutant with tolerance to V. cholerae infection and markedly decreased transcription of diptericin as well as other genes regulated by the IMD innate immunity signaling pathway. We present genetic evidence that this insertion affects a locus previously implicated in pupal eclosion. This genetic locus, which we have named mustard (mtd), contains a LysM domain, often involved in carbohydrate recognition, and a TLDc domain of unknown function. Over twenty Mtd isoforms containing one or both of these conserved domains are predicted. We establish that the mutant phenotype represents a gain of function and can be replicated by increased expression of a short, nuclearly localized Mtd isoform comprised almost entirely of the TLDc domain. We show that this Mtd isoform does not block Relish cleavage or translocation into the nucleus. Lastly, we present evidence suggesting that the eclosion defect previously attributed to the Mtd locus may be the result of the unopposed action of the NF-κB homolog, Relish. Mtd homologs have been implicated in resistance to oxidative stress. However, this is the first evidence that Mtd or its homologs alter the output of an innate immunity signaling cascade from within the nucleus.
Keywords: Drosophila melanogaster, innate immunity, IMD pathway, Relish, TLDc domain, NF-kB inhibition, Vibrio cholerae
Introduction
The epithelia of most animals are continually exposed to microorganisms. By necessity, these animals have evolved signaling pathways that enable symbiotic interactions with beneficial microbes and activate an innate immune defense against invading pathogens.
The common fruit fly Drosophila melanogaster harbors two distinct innate immune signaling pathways, namely the immune deficiency pathway (Imd), which responds primarily to Gram-negative bacteria, and the Toll pathway, which responds primarily to Gram-positive bacteria and fungi (1, 2). Both these pathways regulate nuclear translocation of NF-κB homologs, which activate the transcription of many genes including those that encode small cationic antimicrobial peptides (AMPs).
Three NF-κB homologs are encoded in the Drosophila genome. Two of these, Dorsal and Dorsal-related immunity factor (Dif), respond to signaling through the Toll pathway (3). The third NF-κB homolog Relish, comprised of an N-terminal Rel homology domain (RHD) and a C-terminal ankyrin repeat domain, responds to signaling through the IMD pathway, which is initiated by receptor protein recognition of meso-diaminopimelic acid-type peptidoglycan. Through a series of intermediates, this leads to activation of the caspase 8 homolog Dredd and the IΚΚ complex consisting of the catalytic subunit IKΚβ, encoded by ird5, and the regulatory subunit IKΚγ, encoded by Kenny (key). Phosphorylation of Relish by the IKK complex at residues 528 and 529 is required for activation of Relish targets. Dredd is required for cleavage of the Relish C-terminal inhibitory ankyrin repeat domain, allowing nuclear translocation of a 68 kDa N-terminal RHD-containing fragment and activation of an immune response against Gram-negative and some Gram-positive bacterial pathogens (4–7).
Many studies have demonstrated that, in addition to generation of reactive oxygen species by the host (8, 9), IMD pathway signaling plays a role in the Drosophila innate immune response to both commensal and pathogenic intestinal bacteria (10–12). To protect the host against its own immune response, signaling through this pathway is tightly regulated (12–18).
Vibrio cholerae, a Gram-negative, halophilic bacterium, is responsible for the severe diarrheal disease cholera (19). Furthermore, it has been associated with both marine and terrestrial arthropods in the environment (20–24), and arthropods have been proposed as both reservoirs and vectors of V. cholerae. We developed Drosophila melanogaster as a model in which to study the interaction of V. cholerae with the arthropod intestine (25) and previously reported that IMD pathway mutants have increased tolerance to oral V. cholerae infection (26).
As part of a genetic screen for host susceptibility factors, we identified a mutant with increased tolerance to oral V. cholerae infection and a transcriptional profile similar to that of an IMD pathway mutant. This mutant, which we have named Mustard (Mtd), carries a P-element insertion in a complex genetic locus encoding conserved LysM and TLDc domains. LysM domains are often involved in carbohydrate recognition (27). While the TLDc domain has been associated with resistance to oxidative stress, its mechanism of action is unknown (28). The Mtd locus is predicted to give rise to 21 transcripts containing one or both of these conserved domains (29). We show that the phenotype of the Mtd mutant represents a gain of function and can be reproduced by expression of a nuclearly-localized isoform containing only the TLDc domain. TLDc domains are widespread in eukaryotes. Here we present the first evidence for modulation of the IMD innate immune signaling pathway by a TLDc domain-containing protein.
Materials and Methods
Bacterial strains, fly stocks, and growth media
MO10, a V. cholerae O139 clinical isolate, was used in all Drosophila infections (30). Either w1118 or yw stocks obtained from the Bloomington Drosophila stock center at Indiana University were used as controls. The details of other fly lines used in these experiments are available upon request. Other than the decreased rates of eclosion observed for insertion mutants with global defects in Mtd transcription, no developmental defects were noted for the fly lines utilized in these experiments. Drosophila melanogaster strains were reared at 24°C on standard Drosophila medium, and bacterial strains were propagated in Luria–Bertani (LB) broth supplemented with streptomycin (100 µg/ml) at 27 °C.
Transposon localization
Inverse PCR was used to identify the P-element insertion site of the mtdEY04695 mutant as follows. Genomic DNA was prepared from 25 adult flies using a QIAamp DNA Mini Kit (QIAGEN) and then digested with Msp I. The resulting DNA fragments were self-ligated overnight at 16°C using T4 DNA ligase (Invitrogen) to create a mixture of circular fragments. DNA flanking the 5’ end of the transposon was amplified from this mixture using the outward facing primer pair Pwht1 (GTAACGCTAATCACTCCGAACAGGTCACA) and Plac1 (CACCCAAGGCTCTGCTCCCACAAT). DNA flanking the 3’ end of the transposon was amplified using the primer pair Pry4 (CAATCATATCGCTGTCTCACTCA) and Pry1 (CCTTAGCATGTCCGTGGGGTTTGAAT). PCR products were sequenced and aligned with the Drosophila genome sequence available from the Berkeley Drosophila Genome Project (BDGP).
Transgenic fly construction
The mtd-RC transcript was reverse-transcribed from Drosophila RNA prepared as described below using a primer specific to the 3’ end of the RC transcript and the SuperScript III First-Strand synthesis System (Invitrogen). The resulting cDNA was amplified with a 5’ primer including an Eco R1 site and a 3’ primer including an Xho l site. This PCR product was inserted into pCR2.1-TOPO using a TOPO TA cloning kit (Invitrogen) and then excised and ligated into pUAST using the Eco R1 and Xho l restriction sites. A pUAST vector encoding HA-tagged Mtd-RC was created similarly except that an HA tag was added in two steps by PCR amplification with primers encoding the tag. We first confirmed the presence of mtd-RH by reverse-transcribing, amplifying, and sequencing this transcript. Then, mtd-RH and mtd-RH-HA transcripts were amplified from the pUAST plasmids containing either mtd-RC or mtd-RC-HA, respectively, using a forward primer including a 5’ Eco Rl site and a reverse primer including a 3’ Xho l site. After digestion with Eco Rl and Xho l, these products were ligated into pUAST. All inserts were confirmed by sequence analysis. Plasmids were sent to Rainbow Transgenics for injection into w1118 embryos. Homozygous transgenic fly stocks were generated from these offspring.
Oral and systemic bacterial infections
Flies were infected orally as previously described (25). Briefly, three fly vials were prepared with a cellulose acetate plug containing 2 mls of an overnight culture of V. cholerae diluted in a 1:10 ratio in LB broth. 30 male flies were divided evenly between these three vials. Numbers of viable flies in each vial were recorded at least once each day. Survival curves were constructed, and log-rank analysis was used to determine statistical significance. Reproducibility of all survival data was confirmed in at least two independent experiments.
Septic injury was used to produce systemic infection for experiments as noted. In this model, 30 flies were anesthetized with CO2 and pricked in the dorsal thorax with a fine needle (Fine Science Tools) dipped in a solution containing either fresh LB broth or LB broth harboring an overnight culture of V. cholerae. After septic injury, flies were returned to food vials, where they remained until the specified time.
Measurements of bacterial load
Quantification of bacterial load within infected flies was assayed as previously described (26). After 24 hr of V. cholerae ingestion, 10 flies were collected and homogenized in 200 µl of PBS. Dilutions of the suspension were plated on LB agar supplemented with streptomycin. Colony-forming units per fly were enumerated. No colonies were cultured from flies fed LB broth alone, suggesting that all bacteria retrieved from LB agar plates supplemented with streptomycin were V. cholerae. Statistical significance was calculated using a Mann-Whitney U test.
qRT-PCR
Flies were homogenized in TRIzol reagent (Invitrogen). Total RNA was extracted once, treated with DNase I, and then extracted a second time. 1 µg of purified RNA was used as a template for synthesis of cDNA using a QuantiTect reverse transcription system (Qiagen). The resulting cDNA was used for quantitative PCR with iTaq SYBR green supermix with carboxy-X-rhodamine (ROX; Bio-Rad) and 2 pmol of the relevant primers in a 20-µl reaction volume. The sequence of primers used for qRT-PCR is available on request. The experiments were conducted with a StepOnePlus PCR system (Applied Biosystems). The transcript level of each gene was quantified by comparison with a standard curve and normalized using the level of the reference gene rp49. Ten to fifteen flies were used per biological replicate, and three biological replicates were performed for each experiment. Error bars represent the standard error of the mean, and statistical significance was calculated using a student’s t-test. Each experiment was performed at least twice with the exception of those in Figure S3, which were performed once.
In control yw flies, we found that, after 24 hours of V. cholerae ingestion, dipt expression increased approximately 35-fold and remained at this level over several days. Therefore, in oral infections, dipt transcription was measured after 24 hours. Six hours after septic injury, we found that dipt transcription had increased approximately 230-fold. It continued to increase over the next three hours and then was maintained at 300 times baseline (data not shown). Therefore, for septic injury, dipt expression was measured after 6 hours.
Western analysis
Protein extracts for Western analysis were prepared as follows. 10 adult flies were homogenized in a 200 µl volume of NP-40 cell lysis buffer (Invitrogen) supplemented with 1 mM PMSF and a protease inhibitor tablet (Roche). Extracts were spun at 14,000 RPM for 10 minutes at 4 °C to pellet debris. 60 µl of the resulting supernatant was then combined with 15 µl of Lane Marker Reducing Sample Buffer (Pierce), boiled for 5 minutes, and spun at 14,000 RPM for 1 minute. Proteins in this mixture were separated on an 8% Precise protein gel (Pierce). Full length Relish and the products of its proteolysis were visualized with the following two primary antibodies: (i) a mouse IgG monoclonal antibody to the C-terminus of Relish (Developmental Studies Hybridoma bank) and (ii) a polyclonal rabbit IgG antibody raised against a peptide fragment within the N-terminal RHD domain (Pacific Immunology Corporation). Each primary antibody was used in a 1:1,000 dilution. Anti-mouse or anti-rabbit IgG secondary antibody conjugated to horse radish peroxidase (Amersham) was used as appropriate in a 1:5,000 dilution. The HRP-conjugated secondary antibodies were visualized by chemiluminescence with the ECL Plus Western Blotting system (Amersham Biosciences).
Immunofluorescence
Fly tissues were dissected in PBS, transferred immediately to 4% formaldehyde in phosphate buffered saline (PBS), and incubated for 30 minutes at room temperature. Tissues were then rinsed three times for 30 minutes each in PBS supplemented with 0.1% Triton and 1% BSA and incubated overnight with one of the following primary antibodies: mouse anti-HA (1:100, Santa Cruz Biotech), mouse anti-Flag (1:500, Sigma), or rabbit anti-HA (1:100, Santa Cruz Biotech). After rinsing, a second overnight incubation was performed with one of the following secondary antibodies: Alexa 488 goat anti-mouse IgG (1:100, Invitrogen), Dylight 549 goat anti-mouse IgG (1:100, Jackson ImmunoResearch), or Alexa 488 goat anti-rabbit IgG (1:200, Invitrogen). The tissues were rinsed again with addition of DAPI (1 µg/ml) to the last wash. Tissues were mounted in Vectashield and examined using either an UltraView Vox spinning disc confocal microscope (Perkin Elmer) or an LSM700 confocal microscope.
Microarray analysis
RNA was prepared as described above except that 100 flies were used. This RNA was supplied to the Molecular Genetics Core Facility at Children’s Hospital Boston where it was labeled and hybridized to a GeneChip Drosophila Genome 2.0 Array (Affymetrix) according to the manufacturer's instruction. Data analysis was carried out using GeneSpring software (Silicon Genetics, Redwood City, CA). Statistical significance was determined using a one-way ANOVA with a threshold p-value of 0.05. Microarray data have been deposited in the GEO database (Accession number: GSE35439; http://www.ncbi.nlm.nih.gov/geo/).
Results
Identification of a transposon-insertion mutant with increased tolerance to oral V. cholerae infection
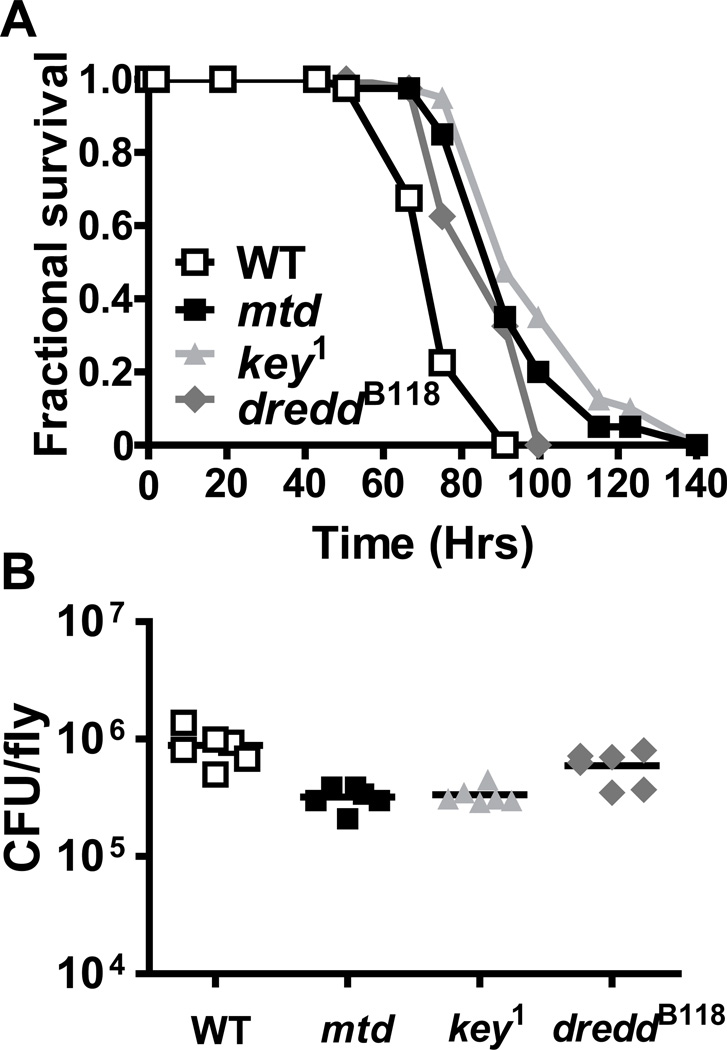
In a forward genetic screen of homozygous viable Drosophila transposon-insertion mutants, we identified a line, P{EPgy2}l(3)82FdEY04695, that, like IMD pathway mutants, showed prolonged survival in the face of oral V. cholerae infection (Figure 1A) (26). We named this mutant Mustard (Mtd). The bacterial load of the mtdEY04695 mutant was similar to that of IMD pathway mutants and slightly less than that of control flies (Figure 1B). Because the mtdEY04695 mutant was comparably susceptible to colonization by V. cholerae but less susceptible to lethal infection than control flies, we describe it as tolerant to V. cholerae infection (31).
Figure 1. The mtdEY04695 mutant phenocopies IMD pathway mutants.
(A) Fractional survival during oral V. cholerae infection of the control strain yw (WT), the transposon insertion line mtdEY04695, and the IMD pathway mutants, dreddB118 and key1. Log-rank analysis demonstrated a statistically significant difference between survival of control flies and mtdEY04695 mutants (p < 0.001), key1 mutants (p < 0.001), and dreddB118 mutants (p < 0.05). (B) Bacterial burden of yw, mtdEY04695, dreddB118 and key1 flies after 48 hours of infection. There was a statistically significant difference in bacterial burden between control flies and mtdEY04695 (p=0.0022) or key1 (p=0.0022) mutant flies. However, the difference in bacterial burden between control flies and dreddB118 mutant flies was not statistically significant (p=0.0931).
Although Relish cleavage proceeds normally, diptericin transcription is decreased in the mtdEY04695 mutant
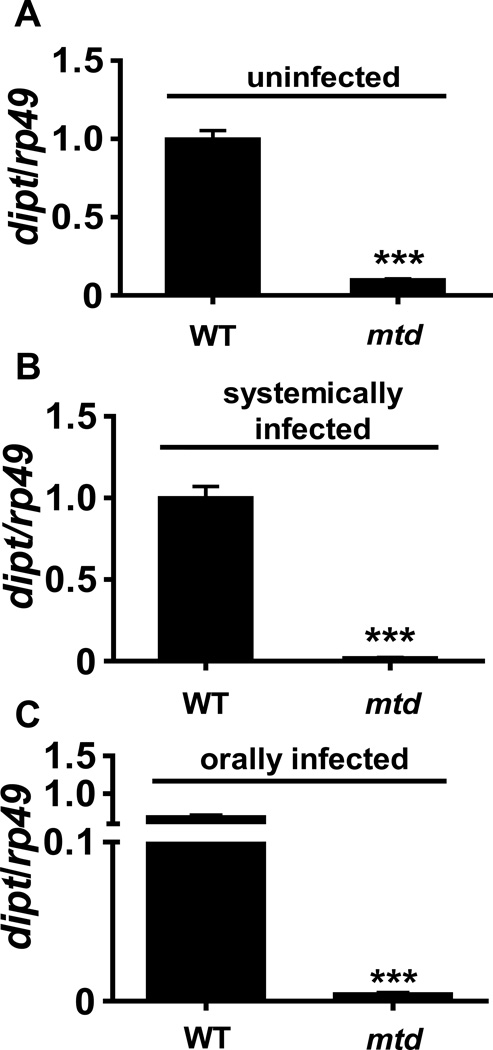
We questioned whether IMD pathway signaling might be blocked in the mtdEY04695 mutant. The IMD pathway directly regulates expression of the antimicrobial peptide diptericin (32). Therefore, to evaluate signaling through the IMD pathway, we compared dipt transcription in the mtdEY04695 mutant to that in control flies in the presence and absence of oral or systemic infection with V. cholerae. As shown in Figure 2, dipt levels were dramatically reduced in uninfected, orally infected, and systemically infected mtdEY04695 mutant flies.
Figure 2. dipt transcription is decreased in the mtdEY04695 mutant.
qRT-PCR analysis of dipt levels in (A) uninfected, (B) systemically, and (C) orally infected yw (WT) and mtdEY04695 mutant flies. *** indicates p < 0.001
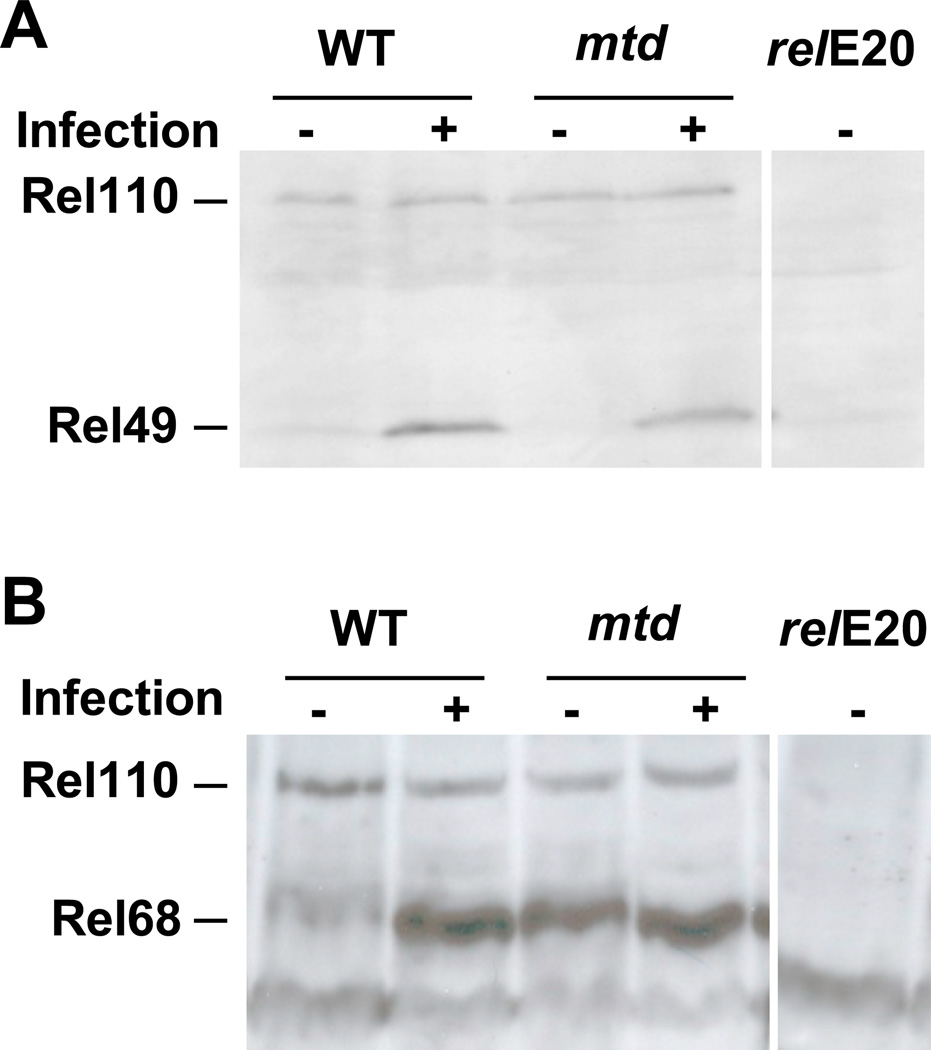
Because dipt transcription was decreased in the mtdEY04695 mutant, we hypothesized that IMD pathway signaling might be blocked prior to Relish cleavage. To examine this possibility, we performed Western analysis on protein extracts derived from control flies and mtdEY04695 mutant flies before and after systemic infection with V. cholerae. As shown in Figure 3, Relish and its cleavage products were equally abundant in extracts from wild-type and mtdEY04695 mutant flies after infection. As a control, neither full length Relish nor its cleavage products were present in a relE20 mutant. This suggests that, in the mtdEY04695 mutant, signaling through the IMD pathway proceeds past the point of Relish cleavage.
Figure 3. Proteolytic cleavage of Relish is similar in control flies and the mtdEY04695 mutant.
Western analysis of Relish cleavage fragments in uninfected or infected w1118 (WT), mtdEY04695, or relE20 flies by probing with anti-Rel49 (A) and anti-Rel68 antibodies (B).
In multiple experiments, Rel68 but not Rel49 appeared to be increased in the uninfected mtdEY04695 mutant as compared with control flies. This may suggest that the Rel68 peptide is less susceptible to proteolytic degradation in the mtdEY04695 mutant.
Similarities and differences in gene regulation in an IMD pathway mutant and the mtdEY04695 mutant
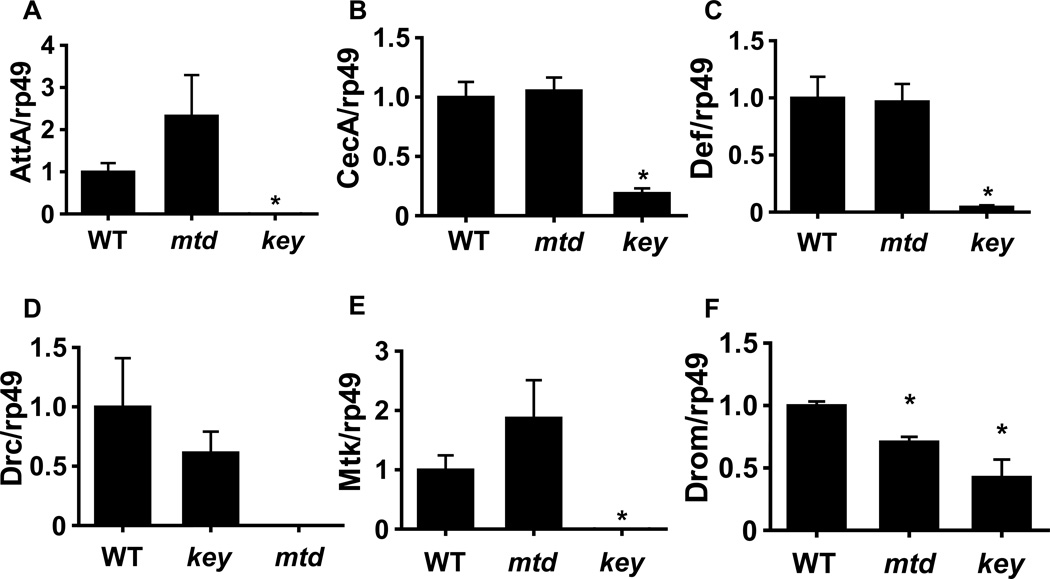
Because cleavage of Relish proceeds normally in the mtdEY04695 mutant, we questioned whether transcription of other AMP’s was repressed in this mutant. Therefore, we measured the transcript levels of a number of AMPs in flies orally infected with V. cholerae (Figure 4). Interestingly, although attacin, cecropin, defensin, drosocin, and metchnikowin transcripts were decreased in the IMD pathway mutant, levels of these transcripts in the mtdEY04695 mutant were similar to those in control flies. Transcription of drosomycin, which is regulated by the Toll pathway, was slightly but significantly decreased in both mutants.
Figure 4. Transcript abundance of genes encoding antimicrobial peptides in control flies, an IMD pathway, and a mtdEY04695 mutant orally infected with V. cholerae.
qRT-PCR analysis of (A) attacin (Att), (B) cecropin A (CecA), (C) defensin (Def), (D) drososin (Drc), (E) metchnikowin (Mtk), and (F) drosomycin (Drom) transcription in control Drosophila melanogaster (WT), a mtdEY04695 mutant (mtd), and an IMD pathway mutant (key). * indicates p < 0.05.
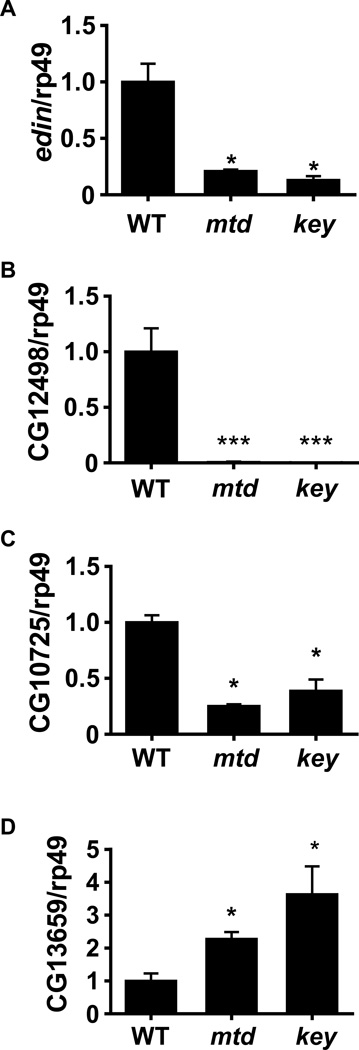
To determine whether there might be other genes whose transcription was similarly altered in an IMD pathway mutant and the mtdEY04695 mutant, we compared the transcriptomes of orally infected key1 and mtdEY04695 mutant flies with those of w1118 control flies by microarray analysis. 398 genes were differentially regulated in the mtdEY04695 mutant and 434 genes were differentially regulated in the key1 mutant as compared with control flies. Of these, 40% were similarly regulated in mtdEY04695 and key1 mutant flies (Table S1). For a small group of the most highly regulated genes, we confirmed the microarray results by qRT-PCR. As shown in Figure 5, transcript levels of Edin, CG12498, and CG10725 were similarly decreased in the two mutants, while levels of CG13659 were increased.
Figure 5. Additional genes that are similarly regulated in an IMD pathway mutant and a mtdEY04695 mutant as compared with control flies orally infected with V. cholerae.
qRT-PCR analysis of (A) edin, (B) CG12498, (C) CG10725, (D) CG13659. * indicates p < 0.05; *** indicates p < 0.001.
The effect of the transposon insertion on the mtd locus is complex
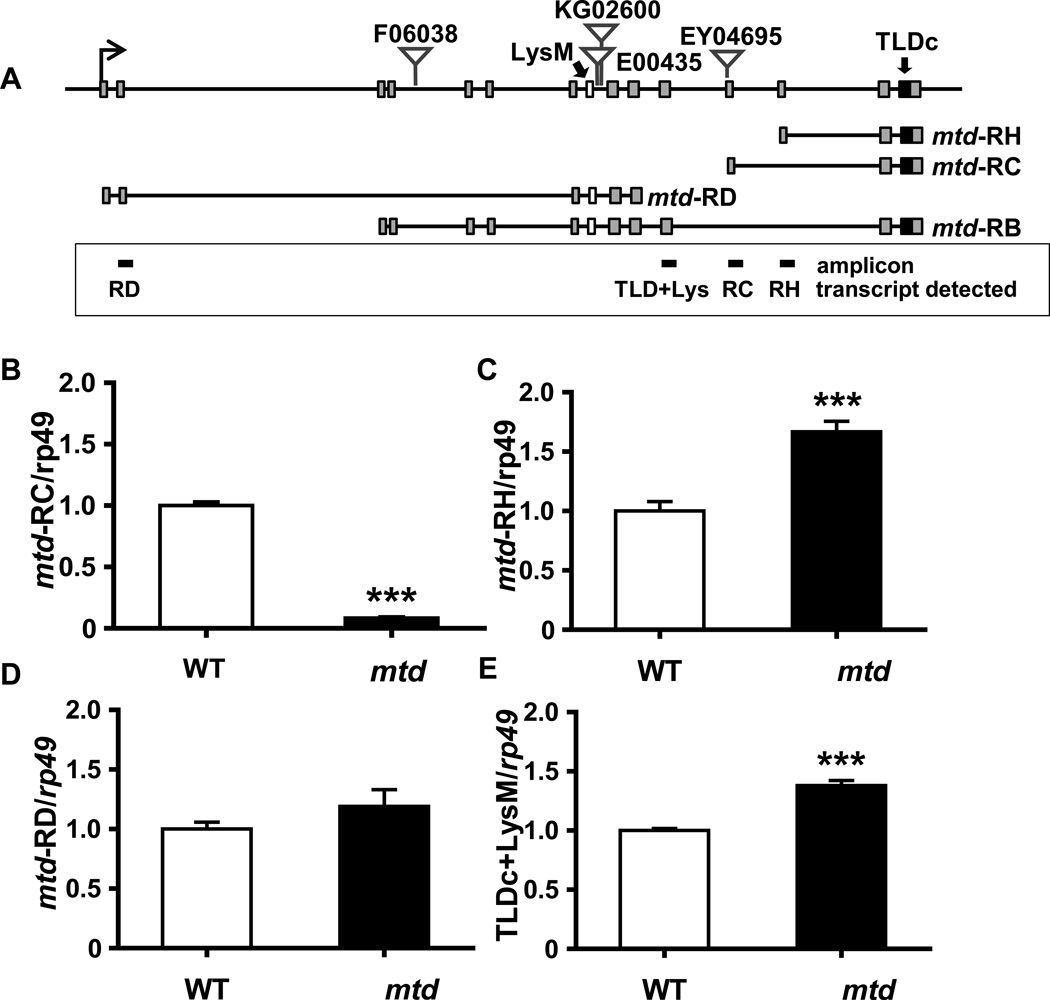
The mtdEY04695 mutant harbors a P-element insertion within the mtd locus, which is predicted to encode twenty unique isoforms as a result of alternative splice sites and alternative promoters (29). Mtd isoforms include one or both of the conserved LysM and TLDc domains encoded within the Mtd locus (Figure 6A). The LysM domain is present in prokaryotic and eukaryotic proteins that bind carbohydrates such as peptidoglycan and chitin (27, 33–35), while the TLDc domain has no known function but has been associated with resistance to oxidative stress (28, 36–41). Eleven of the predicted mtd transcripts include both LysM and TLDc domains (LysM/TLDc), nine of the predicted shorter transcripts include only the TLDc domain (TLDc-only), and one predicted transcript, RD, includes only the LysM domain (LysM-only). Transposon mapping by inverse PCR and subsequent sequence analysis indicated that the P-element was located 200 bp upstream of the start codon of a transcript encoding the TLDc-only isoform, mtd-RC (Figure 6A).
Figure 6. Impact of the mtdEY04695 insertion on transcription of representative Mtd transcripts.
(A) Diagram of the TLDc-only Mtd transcripts mtd-RH and mtd-RC, the LysM-only transcript mtd-RD, and a representative LysM/TLDc transcript mtd-RB encoded in the mtd locus. Small rectangles represent exons. The exons containing the conserved LysM and TLDc domains are labeled and filled in white and black, respectively. Triangles indicate the sites of transposon insertions studied here. The box below contains the locations of the amplicons generated in qRT-PCR experiments shown in B–E and the corresponding transcript detected. (B–E) Relative transcript levels of mtd-RC, mtd-RH, mtd-RD and all long transcripts (TLDc+LysM) in the mtdEY04695 mutant as compared with yw flies (WT). *** indicates p < 0.001.
Given the complexity of the mtd locus, our first goal was to identify mtd transcripts whose abundance was altered in the mtdEY04695 mutant. We hypothesized that the insertion would decrease levels of mtd-RC and might also alter the abundance of other mtd transcripts. To test this, we identified amplicons (shown in Figure 6A) that allowed us to uniquely measure the abundance of the TLDc-only transcripts, mtd-RC and mtd-RH, and the LysM-only transcript mtd-RD. In addition, we measured the overall abundance of LysM/TLDc transcripts using primers just downstream of the LysM domain. We compared transcript levels in control flies and the mtdEY04695 mutant. While transcription of mtd-RC in the mutant was 12-fold less than that in control flies, levels of mtd-RH and all long transcripts were increased by 66.8% and 37.9%, respectively (Figure 6B, C, and E). Mtd-RD transcription was unchanged in the mtdEY04695 mutant (Figure 6D). Based on these data, we concluded that the mtdEY04695 mutant phenotype could result either from decreased levels of mtd-RC or from increased levels of mtd-RH and the longer LysM/TLDc transcripts.
Because the mtdEY04695 mutant was more tolerant to oral infection with V. cholerae, we were particularly interested in mtd transcripts that were expressed in the intestine. We found high levels of mtd-RH, -RC, and full length transcripts in the intestine. In contrast, expression of mtd-RD in the intestine was negligible (Figure S1). Furthermore, oral infection did not alter the levels of any of the measured transcripts either in whole flies or specifically in the intestine (Figure S1).
Precise excision suggests that the transposon insertion is responsible for the mtdEY04695 mutant phenotype
We first used sequence analysis to establish that the extremely low expression of dipt in the mtdEY04695 mutant was not the result of a mutation in the promoter or coding sequence of the dipt gene (data not shown). We then confirmed that the phenotype of the mtdEY04695 mutant was, in fact, due to the P-element insertion by crossing the mtdEY04695 mutant with flies carrying the P-element transposase. PCR amplification of the insertion site was used to screen for precise excision lines. This was confirmed by sequence analysis. Precise excision lines were then backcrossed to the parental strain to generate flies with a comparable genetic background. Two independent precise excision lines (EP13 and EP18) were studied in detail.
Transcription of both mtd-RC and mtd-RH in the EP13 and EP18 precise excision lines was significantly different from that of the mtdEY04695 mutant and closer to that of control flies (Figure S2 A and B). dipt transcription in precise excision lines was also closer to that of control flies (Figure S 2C). We compared the susceptibility of precise excision lines to oral V. cholerae infection with that of control flies. The susceptibility to infection of both the EP13 and EP18 precise excision lines was significantly greater than that of the mtdEY04695 mutant. However, only the susceptibility of the EP18 line approached that of control flies (Figures S2D and E). Taken together, these results strongly suggest that the P-element insertion in the mtd locus is responsible for the phenotype of the mtdEY04695 mutant.
The mtdEY04695 mutant phenotype is not due to a decrease in the mtd-RC transcript
We hypothesized that the mtdEY04695 mutant phenotype could be due either to a decrease in transcription of mtd-RC or to increased levels of another transcript. To determine whether decreased mtd levels could be responsible for the mtdEY04695 mutant phenotype, we generated a hemizygous mutant by crossing the mtdEY04695 fly line with a deficiency strain lacking the mtd gene region (Df(3R)3-4, Df). As expected, transcription of mtd-RC in the resulting hemizygous fly (mtdEY04695/Df) was decreased (Figure S3 A). However, dipt transcription in this hemizygote was similar to that of the heterozygote control (Figure S3 B). Furthermore, susceptibility to V. cholerae infection was comparable to that of control flies (Figure S3 C). This suggested to us that a decrease in the level of the mtd-RC transcript was not the basis of the mtdEY04695 mutant phenotype.
To further support this conclusion, we examined a series of mutants from the Harvard Exelixis stock collection carrying insertions in the mtd locus (Figure 6A). In these mutants, levels of the mtd-RC and LysM/TLDc transcripts were reduced, while mtd-RH transcription was not altered (Figure S3 D–F). In contrast to the mtdEY04695 mutant, these mutants were all more susceptible to infection (Figure S3 G). Taken together, these results suggest that the mtdEY04695 phenotype is not the result of a decrease in mtd-RC transcription.
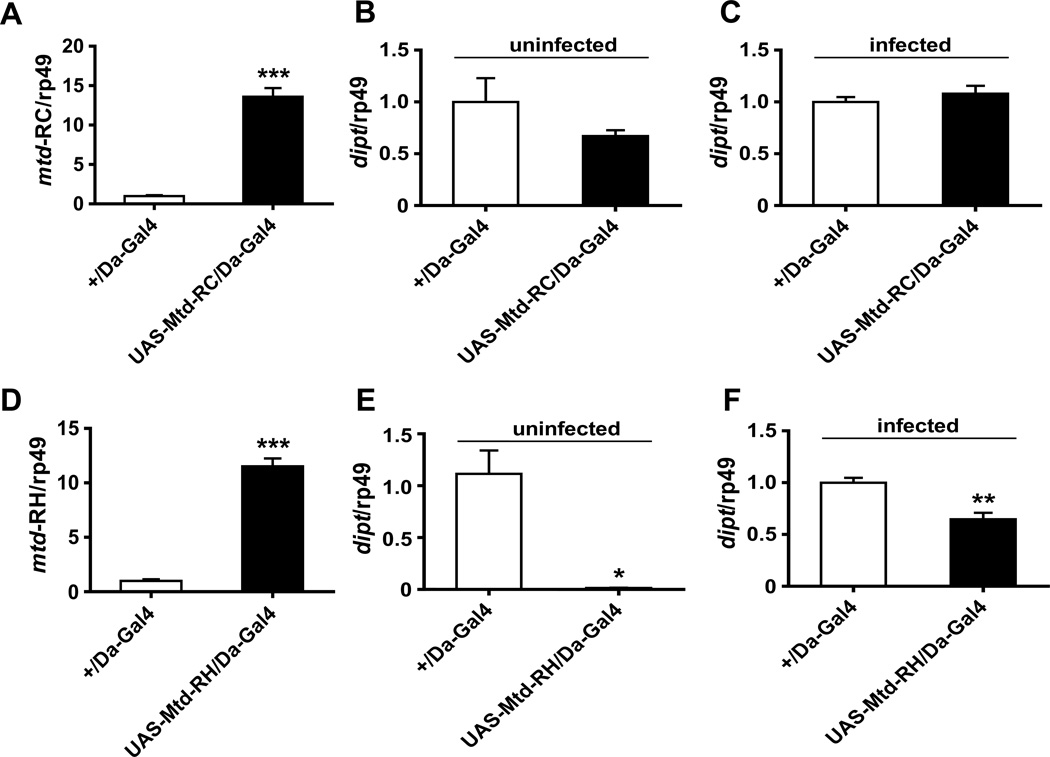
Overexpression of mtd-RH but not mtd-RC phenocopies the mtdEY04695 mutant
Next, we examined the possibility that the mtdEY04695 phenotype might result from an increase in the level of mtd-RH. We confirmed the presence of RC and RH transcripts and generated transgenic flies encoding a wild-type mtd-RH allele downstream of a UAS element controlled by Gal4. As a control, we generated transgenic flies carrying a similar mtd-RC construct. Mtd isoform C (Mtd-PC) differs from isoform H (Mtd-PH) only by the presence of 70 additional amino acids at its N-terminus.
When induced by the ubiquitous driver Daughterless (Da-Gal4), transcription of mtd-RC and mtd-RH increased more than 10 fold in comparison with the Gal4 control (Figures 7A and D). Overexpression of mtd-RC did not alter dipt transcription either in uninfected or systemically infected flies (Figures 7B and C). In contrast, overexpression of mtd-RH significantly repressed dipt transcription in uninfected and systemically infected flies (Figures 7E and F).
Figure 7. Overexpression of mtd-RH but not mtd-RC blocks activation of dipt transcription.
(A) qRT-PCR measurement of mtd-RC transcript levels in a strain with ubiquitous overexpression of mtd-RC (UAS-Mtd-RC/Da-Gal4) and a driver-only control (+/Da-Gal4). (B) qRT-PCR measurement of dipt transcript levels in a strain with ubiquitous overexpression of mtd-RC (UAS-Mtd-RC/Da-Gal4) and a driver-only control (+/Da-Gal4). (C) Same experiment as (B) performed with flies infected with V. cholerae by septic injury. (D) qRT-PCR measurement of mtd-RH transcript levels in a strain with ubiquitous overexpression of mtd-RH (UAS-Mtd-RH/Da-Gal4) and a driver-only control (+/Da-Gal4). (E) qRT-PCR measurement of dipt transcript levels in a strain with ubiquitous overexpression of mtd-RH (UAS-Mtd-RH/Da-Gal4) and a driver-only control (+/Da-Gal4). (F) Same experiment as (D) performed with flies infected with V. cholerae by septic injury. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001.
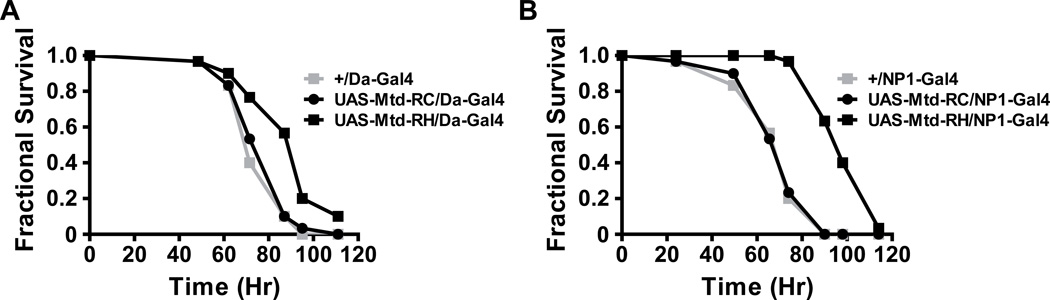
We also tested the susceptibility of these flies to V. cholerae infection. Overexpression of mtd-RH but not mtd-RC both ubiquitously and in the midgut prolonged survival (Figure 8). These results support the conclusion that the phenotype of the mtdEY04695 mutant is due, at least in part, to an increase in the level of the mtd-RH transcript.
Figure 8. Overexpression of mtd-RH but not mtd-RC increases tolerance to oral V. cholerae infection.
Fractional survival of flies with (A) ubiquitous expression of mtd-RC (UAS-Mtd-RC/Da-Gal4) or mtd-RH (UAS-Mtd-RH/Da-Gal4) compared with driver-only control flies (+/Da-Gal4) and (B) midgut-only expression of mtd-RC (UAS-Mtd-RC/NP1-Gal4) or mtd-RH (UAS-Mtd-RH/NP1-Gal4) compared with driver-only control flies (+/NP1-Gal4). Log-rank analysis demonstrated a statistically significant difference between survival of control flies and flies expressing mtd-RH either ubiquitously (p<0.0001) or in the midgut alone (p < 0.0001) but not between survival of control flies and flies expressing mtd-RC ubiquitously (p>0.05) or in the midgut alone (p > 0.05).
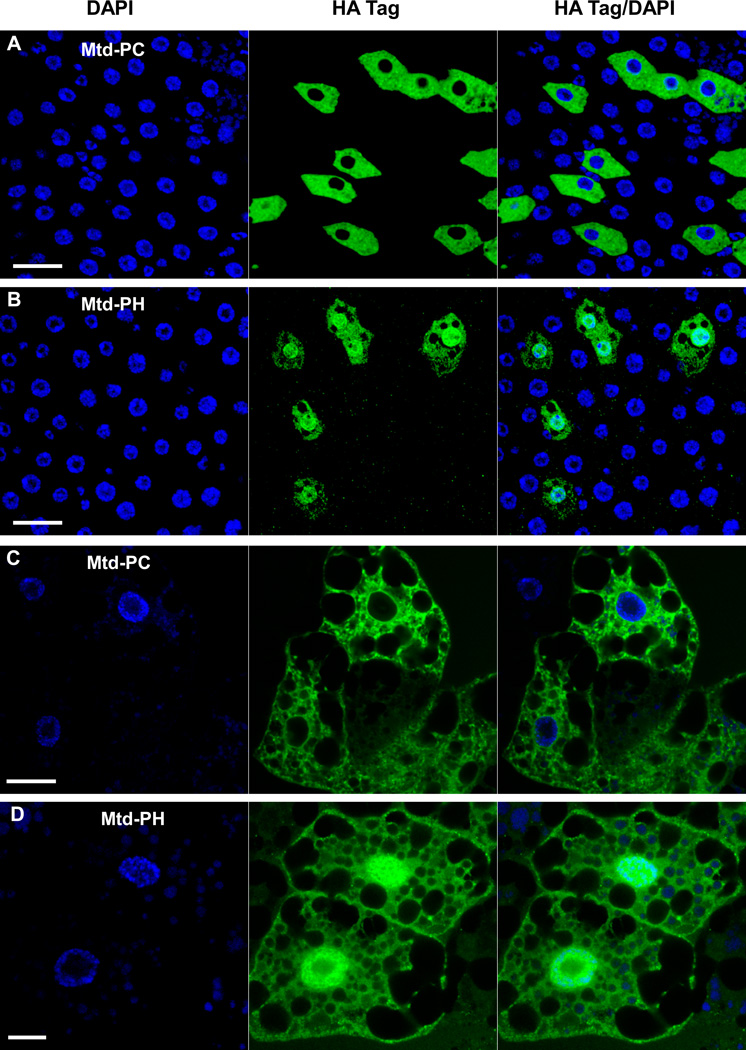
Mtd-PC and Mtd-PH exhibit distinct subcellular distributions
Mtd-PC and Mtd-PH differ only in 70 amino acids at their N-termini; yet overexpression of these two proteins leads to very different effects on IMD pathway signaling and resistance to oral infection. Because Rel68 relocates to the nucleus, we hypothesized that the different functions of these two transcripts could be the result of distinct subcellular distributions. To examine this hypothesis, we generated flies carrying wild-type mtd-RC or mtd-RH alleles with a C-terminal Human influenza hemagglutinin (HA) tag added downstream of a Gal4-UAS promoter. We used the Da-Gal4 driver to express these proteins in the midgut. Interestingly, although Da-Gal4 is considered a ubiquitous driver, the transgenes were not equally expressed in all midgut epithelial cells, leading to a mosaic pattern (Figure 9). A similar pattern was noted when Da-Gal4 was used to drive a UAS-GFP transgene, demonstrating that this mosaic pattern of expression is not specific to Mtd (data not shown).
Figure 9. Mtd-PH is nuclearly localized, while Mtd-PC is excluded from the nucleus.
Immunofluorescence of Mtd-PC-HA (A) and Mtd-PH-HA (B) in the adult midgut. Immunofluorescence of Mtd-PC-HA (C) and Mtd-PH-HA (D) in the larval fat body. In all cases, expression was driven by Da-Gal4. The tagged protein is visualized with Alexa 488 (green), and nuclear DNA is stained with DAPI (blue). The mosaic pattern of expression is a characteristic of the Da-Gal4 driver. Scale bar = 20 µm.
Mtd-PC and Mtd-PH were visualized by immunofluorescence both in the larval fat body and in the adult midgut. While isoform C was observed exclusively in the cytoplasm of larval fat body and adult midgut epithelial cells (Figure 9A and C), isoform H was also present in the nucleus (Figure 9B and D). Thus, despite the high similarity of Mtd-PC and Mtd-PH, these proteins have distinct distributions within the cell. This is likely the basis for their distinct effects on targets of the IMD pathway.
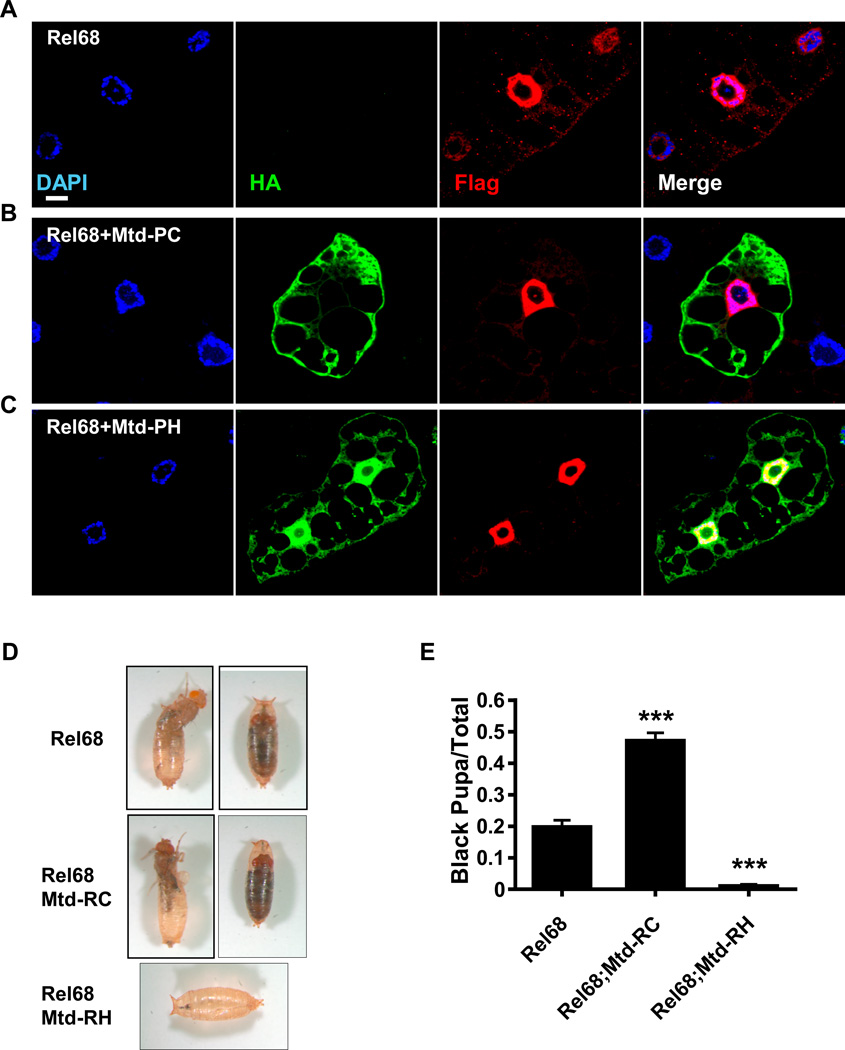
mtd-RH rescues the eclosion defect that results from rel68 overexpression
In order to determine whether Mtd-PC or Mtd-PH altered the subcellular distribution of Rel68, we co-expressed tagged alleles of mtd-RH and rel68 and visualized the subcellular locations of these two proteins in larval fat body cells by immunofluorescence. As shown in Figure 10 A–C, neither Mtd-PC nor Mtd-PH blocked nuclear localization of Rel68. However, while rearing Rel68 overexpression flies, we noticed that, although these flies developed normally, many of them were unable to eclose (Figure 10 D–E). Interestingly, co-overexpression of mtd-RH and rel68 rescued this eclosion defect. Co-expression of mtd-RC with rel68 did not mitigate the eclosion defect and, in fact, appeared to exacerbate it. These data support a genetic interaction of Mtd-PH but not Mtd-PC with the NF-κB homolog Relish.
Figure 10. Overexpression of mtd-RH does not prevent nuclear localization of Rel68 in the larval fat body but does rescue the eclosion-lethal phenotype resulting from overexpression of rel68.
Immunofluorescence of larval fat bodies overexpressing (A) rel68-FLAG (Rel68), (B) rel68-FLAG and mtd-RC-HA (Rel68+Mtd-RC), or (C) rel68-FLAG and mtd-RH-HA (Rel68 + Mtd-RH) driven by Da-Gal4. Nuclei are stained with DAPI. (D) Photographs of the eclosion phenotypes in flies expressing rel68, rel68 and mtd-RC together (Rel68+Mtd-RC), or rel68 and mtd-RH together (Rel68+Mtd-RH) driven by Da-Gal4. (E) Quantification of the fraction of abnormal pupa in flies overexpressing rel68 (Rel68), rel68 and mtd-RC (Rel68+Mtd-RC), or rel68 and mtd-RH (Rel68+Mtd-RH) driven by Da-Gal4. *** indicates p < 0.001 as compared to rel68 overexpression alone.
Discussion
In a screen for factors that alter Drosophila susceptibility to oral V. cholerae infection, we identified a infection-tolerant transposon-insertion mutant in the complex l(3)82Fd locus, which we have re-named mustard (mtd). Subsequent experiments suggested that the phenotype of this mutant was the result of increased expression of a short Mtd transcript containing only a TLDc domain. The mtd locus was previously described as an ecdysone-activated, late puff gene whose mutation led to the inability of adult flies to eclose (42, 43). It encodes at least 20 unique isoforms including a LysM-only isoform, several TLDc-only isoforms, and several with both domains. Here we report a novel function for a short, nuclearly-localized Mtd isoform in tailoring of the IMD pathway-activated innate immune response.
The Mtd homolog Oxr1 has been shown to protect cells against reactive oxygen species (ROS), which are produced during intestinal infection as well as in neurodegenerative disease, by increasing the transcription of genes that scavenge ROS such as catalase and glutathione peroxidase (9, 41, 44). Furthermore, investigators have shown that C753 of the mouse Mtd homolog itself can be oxidized by H2O2 (41). This residue (C167) is conserved in Mtd-PH. We previously showed that breakdown of the Drosophila midgut epithelium occurs during V. cholerae infection of wild-type Drosophila and presented evidence that increased cell turnover within the intestines of IMD pathway mutants might enhance tolerance to infection (26). Therefore, we initially hypothesized that breakdown of the fly intestinal epithelium during V. cholerae infection was the result of ROS production by dual oxidase (Duox) and that Mtd increased host tolerance by protecting the intestinal epithelium against oxidative stress. However, RNAi knockdown of the duox transcript, overexpression of the immune regulated catalase, and oral administration of glutathione did not prolong survival of orally infected control flies (data not shown), suggesting that ROS itself is not an important factor in fly mortality and that Mtd does not increase host tolerance by directly or indirectly scavenging ROS generated by the host immune response. However, it is possible that oxidation of Mtd by host-derived ROS could modulate its function.
We also considered that the low level of diptericin transcription in the mtd mutant reflected a less invasive infection leading to decreased activation of the IMD pathway. However, we observed that, in oral infection, the abundance of other IMD-regulated AMP transcripts was similar to that of control flies, suggesting that the IMD pathway is similarly activated in control and mtdEY04695 mutant flies. Alternative mechanisms for the observed effect of Mtd-PH on transcription of genes in the Relish regulon include (i) interaction with Rel68 at specific promoters, (ii) interaction with DNA at a subset of Rel68-regulated promoters, (iii) interaction with another protein that modulates transcription of genes in the Relish regulon, or (iv) direct modulation of transcription of a protein that affects Relish function by binding to DNA. These hypotheses are currently under investigation.
The loci encoding the two human homologs of Mtd, Oxr1 and NCOA7, are also predicted to give rise to multiple transcripts and isoforms (28, 38, 40, 45). Interestingly, while there is considerable variability in the central regions of the fly and human homologs, the LysM and TLDc domains of these proteins are more than 50% identical. Furthermore, the TLDc domains of these proteins are much more similar to each other than they are to other TLDc domain-containing proteins in their respective genomes, suggesting that they may function similarly in flies and humans. Therefore, this work provides a rationale for the systematic investigation of the short isoforms of Oxr1 and NCOA7. Such studies may yield additional nuclearly-localized isoforms that have new and important functions in regulation of gene transcription and modulation of the innate immune response in mammals.
Supplementary Material
Acknowledgements
We would like to acknowledge Drs. Neil Silverman, Norbert Perrimon, Lorri Marek, Jon Kagan, Alexandra Purdy, and Michael Volkert for helpful discussions during the course of this work. We would also like to thank Drs. Neil Silverman, Norbert Perrimon, Svenja Stoven, Won Jae Lee, and the Exelixis Collection at Harvard Medical School for generously providing us with fly stocks and Dr. Qing Ji for her valuable contributions to program development and data processing. Microscopy was performed at the Intellectual and Developmental Disabilities Research Center Imaging Core, Children’s Hospital Boston with expertise provided by Lihong Bu and Anthony Hill. The Molecular Genetics Core Facility at Children’s Hospital carried out microarray hybridizations.
Footnotes
This work was supported by NIH AI071147 to PIW. The Intellectual and Developmental Disabilities Research Center Imaging Core, Children’s Hospital Boston is supported by NIH-P30-HD-18655. The Children's Hospital, Boston Molecular Genetics Core Facility is supported by NIH-P50-NS40828 and NIH-P30-HD18655.
References
- 1.Aggrawal K, Silverman N. Peptidoglycan recognition in Drosophila. Biochem Soc Trans. 2007;35:1496–1500. doi: 10.1042/BST0351496. [DOI] [PubMed] [Google Scholar]
- 2.Kim T, Kim YJ. Overview of innate immunity in Drosophila. J Biochem Mol Biol. 2005;38:121–127. doi: 10.5483/bmbrep.2005.38.2.121. [DOI] [PubMed] [Google Scholar]
- 3.Ashok Y. Drosophila toll pathway: the new model. Sci Signal. 2009;2 doi: 10.1126/scisignal.252jc1. jc1. [DOI] [PubMed] [Google Scholar]
- 4.Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, Oh BH, Kim J, Lee WJ. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in the Drosophila gut. Nat Immunol. 2009;10:949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 9.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 10.Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, Lee DG, Kim J, Lee D, Lee WJ. An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. Embo J. 2006;25:3693–3701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 13.Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005 doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, Lemaitre B, Gstaiger M, Meier P, Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal K, Rus F, Vriesema-Magnuson C, Erturk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Kleino A, Myllymaki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Ramet M. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- 18.Ryu JH, Nam KB, Oh CT, Nam HJ, Kim SH, Yoon JH, Seong JK, Yoo MA, Jang IH, Brey PT, Lee WJ. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol Cell Biol. 2004;24:172–185. doi: 10.1128/MCB.24.1.172-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 20.Shukla BN, Singh DV, Sanyal SC. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol. and Medical Microbiol. 1995;12:113–120. doi: 10.1111/j.1574-695X.1995.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 21.Colwell RR, Huq A, Islam MS, Aziz KM, Yunus M, Khan NH, Mahmud A, Sack RB, Nair GB, Chakraborty J, Sack DA, Russek-Cohen E. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci U S A. 2003;100:1051–1055. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotedar R. Vector potential of houseflies (Musca domestica) in the transmission of Vibrio cholerae in India. Acta Trop. 2001;78:31–34. doi: 10.1016/s0001-706x(00)00162-5. [DOI] [PubMed] [Google Scholar]
- 23.Halpern M, Broza YB, Mittler S, Arakawa E, Broza M. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb Ecol. 2004;47:341–349. doi: 10.1007/s00248-003-2007-6. [DOI] [PubMed] [Google Scholar]
- 24.Broza M, Gancz H, Halpern M, Kashi Y. Adult non-biting midges: possible windborne carriers of Vibrio cholerae non-O1 non-O139. Environ Microbiol. 2005;7:576–585. doi: 10.1111/j.1462-2920.2005.00745.x. [DOI] [PubMed] [Google Scholar]
- 25.Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A, Jackson FR, Watnick PI. Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 2005;1:e8. doi: 10.1371/journal.ppat.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkey CD, Blow N, Watnick PI. Genetic analysis of Drosophila melanogaster susceptibility to intestinal Vibrio cholerae infection. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 28.Durand M, Kolpak A, Farrell T, Elliott NA, Shao W, Brown M, Volkert MR. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol. 2007;8:13. doi: 10.1186/1471-2121-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McQuilton P, St Pierre SE, Thurmond J. FlyBase 101 - the basics of navigating FlyBase. Nucleic Acids Res. 2011;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldor MK, Colwell R, Mekalanos JJ. The Vibrio cholerae O139 serogroup antigen includes an O-polysaccharide capsule and lipopolysaccharide virulence determinant. Proc Natl Acad Sci USA. 1994;91:11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayres JS, Schneider DS. Tolerance of Infections. Annu Rev Immunol. 2011 doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 32.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149:2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 34.de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XC, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics. 2008;180:1671–1678. doi: 10.1534/genetics.108.089748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shkolnik K, Ben-Dor S, Galiani D, Hourvitz A, Dekel N. Molecular characterization and bioinformatics analysis of Ncoa7B, a novel ovulation-associated and reproduction system-specific Ncoa7 isoform. Reproduction. 2008;135:321–333. doi: 10.1530/REP-07-0402. [DOI] [PubMed] [Google Scholar]
- 38.Elliott NA, Volkert MR. Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol Cell Biol. 2004;24:3180–3187. doi: 10.1128/MCB.24.8.3180-3187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer H, Zhang XU, O'Brien KP, Kylsten P, Engvall E. C7, a novel nucleolar protein, is the mouse homologue of the Drosophila late puff product L82 and an isoform of human OXR1. Biochem Biophys Res Commun. 2001;281:795–803. doi: 10.1006/bbrc.2001.4345. [DOI] [PubMed] [Google Scholar]
- 40.Volkert MR, Elliott NA, Housman DE. Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc Natl Acad Sci U S A. 2000;97:14530–14535. doi: 10.1073/pnas.260495897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver PL, Finelli MJ, Edwards B, Bitoun E, Butts DL, Becker EB, Cheeseman MT, Davies B, Davies KE. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS Genet. 2011;7:e1002338. doi: 10.1371/journal.pgen.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thummel CS. Ecdysone-regulated puff genes 2000. Insect Biochem Mol Biol. 2002;32:113–120. doi: 10.1016/s0965-1748(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 43.Stowers RS, Russell S, Garza D. The 82F late puff contains the L82 gene, an essential member of a novel gene family. Dev Biol. 1999;213:116–130. doi: 10.1006/dbio.1999.9358. [DOI] [PubMed] [Google Scholar]
- 44.Jaramillo-Gutierrez G, Molina-Cruz A, Kumar S, Barillas-Mury C. The Anopheles gambiae oxidation resistance 1 (OXR1) gene regulates expression of enzymes that detoxify reactive oxygen species. PLoS One. 2010;5:e11168. doi: 10.1371/journal.pone.0011168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol Cell Biol. 2002;22:3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.