Abstract
Since TGF-β/Smad signaling inhibits chondrocyte maturation, endogenous negative regulators of TGF-β signaling are likely also important regulators of the chondrocyte differentiation process. One such negative regulator, Ski, is an oncoprotein that is known to inhibit TGF-β/Smad3 signaling via its interaction with phospho-Smad3 and recruitment of histone deacetylases (HDACs) to the DNA binding complex. Based on this, we hypothesized that Ski inhibits TGF-β signaling and accelerates maturation in chondrocytes via recruitment of HDACs to transcriptional complexes containing Smads. We tested this hypothesis in chick upper sternal chondrocytes (USCs), where gain and loss of Ski expression experiments were performed. Over-expression of Ski not only reversed the inhibitory effect of TGF-β on the expression of hypertrophic marker genes such as type × collagen (colX) and osteocalcin, it induced these genes basally as well. Conversely, knockdown of Ski by RNA interference led to a reduction of colX and osteocalcin expression under basal conditions. Furthermore, Ski blocked TGF-β induction of cyclinD1 and caused a basal up-regulation of Runx2, consistent with the observed acceleration of hypertrophy. Regarding mechanism, not only does Ski associate with phospho-Smad2 and 3, but its association with phospho-Smad3 is required for recruitment of HDAC4 and 5. Implicating this recruitment of HDACs in the phenotypic effects of Ski in chondrocytes, the HDAC inhibitor SAHA reversed the up-regulation of colX and osteocalcin in Ski over-expressing cells. These results suggest that inhibition of TGF-β signaling by Ski, which involves its association with phospho-Smad3 and recruitment of HDAC4 and 5, leads to accelerated chondrocyte differentiation.
Keywords: Ski, Chondrocyte, TGF-β, Smad3, Histone deacetylase, Endochondral ossification
Transforming growing factor-β (TGF-β) family members are multifunctional growth modulators that control cellular proliferation, differentiation, matrix synthesis and apoptosis [Shi and Massague, 2003]. It is known that TGF-β signaling, which potently inhibits chondrocyte maturation, is required for maintenance of articular cartilage and regulation of the hypertrophic program during endochondral ossification [Ballock et al., 1993; Ferguson et al., 2000; Serra et al., 1997; Yang et al., 2001]. Thus, endogenous mechanisms that inhibit TGF-β signaling in chondrocytes are important in the control of the maturational program in chondrocytes.
Members of the TGF-β/BMP superfamily induce signaling via binding to a receptor complex and the initiation of transcription factor phosphorylation by the activated receptors. TGF-β/BMP receptors, which function as serine/threonine kinases, are composed of trimeric heterodimers of type I and type II receptor subtypes [Massague et al., 2000; Massague and Chen, 2000; Massague et al., 1997]. Ligand binding to the type II receptor results in phosphorylation and activation of the type I receptor, which subsequently recruits other signaling molecules including receptor-associated Smads (2 and 3 for TGF-β; 1,5, and 8 for the BMPs) and the co-factor Smad4 [Heldin et al., 1997; Ishisaki et al., 1999; Massague et al., 2000; Massague and Chen, 2000]. The receptor-associated Smads are phosphorylated following receptor activation, form heteromeric complexes with the co-factor Smad4 and translocate to the nucleus where they influence gene transcription [Massague et al., 1997].
Regarding TGF-β signaling, there are a number of endogenous negative regulators that are likely important for modulating TGF-β effects on chondrocyte differentiation. One such mediator is the oncoprotein Ski, which antagonizes TGF-β signaling by recruiting various transcriptional co-repressors, including the nuclear hormone receptor co-repressor N-CoR [Shinagawa et al., 2001], mSin3A [Khan et al., 2001], HIPK2 [Harada et al., 2003] and methyl-CpG-binding protein MeCP2 [Kokura et al., 2001] to Smad-containing complexes. Ski also represses TGF-β signaling via recruitment of the histone deacetylases (HDACs) to Smad-containing transcriptional complexes [Liu et al., 2001; Luo, 2004; Nomura et al., 1999]. It should also be noted that Ski can negatively regulate signaling on the BMP axis via interaction with BMP R-Smads [Luo, 2003; Wang et al., 2000; Wu et al., 2002]. However, regarding specificity, Ski may have a more dominant effect on the TGF-β pathway due to its strong interaction with Smad3 and 4 but only a weak interaction with Smad1 and 5 [Akiyoshi et al., 1999; Mizuide et al., 2003; Takeda et al., 2004].
Several studies suggest that Ski may have an important role to play in musculoskeletal development and growth. For example, over-expression of Ski induces oncogenic transformation of chick and quail embryo fibroblasts as well as skeletal muscle differentiation of quail embryo cells [Colmenares and Stavnezer, 1989; Stavnezer et al., 1986]. Ski over-expression in the skeletal muscle of transgenic mice results in increased hypertrophy, an effect occurring well after terminal muscle cell differentiation [Leferovich et al., 1995; Sutrave et al., 1990]. Furthermore, among other defects, Ski knockout mice have a cleft palate, craniofacial malformations and polydactyly [Colmenares et al., 2002]. Despite these clear skeletal phenotypes in various genetic models, the function of Ski in bone and cartilage is unknown.
In this report, the functional role of Ski in the skeleton is partially evaluated by examining how its negative regulation of TGF-β signaling affects chondrocyte differentiation. Based on what is known about its signaling effects and function in various tissues, we hypothesize that Ski inhibits TGF-β signaling in chondrocytes, leading to accelerated differentiation/maturation. Results presented here suggest that Ski inhibits TGF-β signaling in chondrocytes through an association with phosphorylated Smads 2 and 3 (pSmad2 and 3) enabling recruitment of HDAC4 and 5 to the transcriptional complex. Furthermore, this action of Ski induces hypertrophic progression of the cells, evidenced by enhanced expression of type × collagen(colX) and osteocalcin in Ski over-expressing cells. This effect on differentiation may be due to the inhibition of cyclinD1 expression and induction of Runx2, suggesting that Ski activity can directly regulate the key molecular modulators of chondrocyte maturation. These results provide initial insights into how Ski is an important regulator of TGF-β signaling and chondrocyte maturation and suggest how it may play a role in normal skeletal development and growth processes as well as in pathologic states such as osteoarthritis.
MATERIALS & METHODS
Cell model
Cephalic (upper) sternal chondrocytes (USCs) were isolated from day 15 chick embryos (Gallus Gallus Domesticus) via a previously described method [Leboy et al., 1989]. Briefly, sternae were dissected from the embryos and connective tissue was removed with the aid of a dissecting microscope. The cephalic region (upper third of the sternum) was recovered, minced, and digested for 4 hours at 37°C in F12 medium containing 3.6 mg/mL collagenase and 0.1% trypsin. Dispersed cells were spun to a pellet, resuspended in DMEM containing 10% NuSerum IV and cultured in a 5% CO2/37°C incubator for 5–7 days. After this primary culturing period, floating cells were collected, re-plated and grown in fresh sternal chondrocyte medium. USCs were plated in either 12-well plates at a density of 2×105 cells per well or in 60mm dishes at a density of 2×106 cells per dish.
RNA harvest and real time RT-PCR
Total cellular RNA was extracted from USCs plated in 60mm dishes using Trizol reagent (Invitrogen) according to the manufacturer's instructions. For the reverse transcription reaction, 1 μg aliquots of total RNA were reverse transcribed into cDNA using the reverse-iT™ kit (AB gene).Real time RT-PCR was performed using the Rotor Gene real-time DNA amplification system (Corbett Research) and the fluorescent dye SYBR green was used to monitor cDNA synthesis (AB Gene). Table 1 lists sense and antisense primer sequences for the various genes of interest. PCR conditions included a 95°C denaturation step for 15 minutes followed by 50 cycles of 94°C for 20 sec, 50°C for 30 sec, and 72°C for 20 sec. The data were analyzed and quantified with the RotorGene analysis software. The expression level of each gene of interest was normalized to either the GAPDH or β-actin mRNA pools.
Table 1.
Forward and reverse primer sequences used for real time RTPCR.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Ski | 5'-GAGGTTGCAGCACATTCTGA-3' | 5'-GCCACTCTGAGGAACTCCAG-3' |
| Type × collagen | 5'-ACATGCATTTACAAATATCGTT-3' | 5'-AAAATAGTAGACGTTACCTTG-3' |
| Runx2 | 5'-ACTTTGACAATAACTGTCCT-3' | 5'-GACACCTACTCTCATACTGG-3' |
| Osteocalcin | 5'-CTG CTC ACA TTC AGC CTC TG-3' | 5'-CTCAGCTCACACACCTCTCG-3' |
| Cyclin D1 | 5'-CCTCTCCTATCAATGCCTCAC-3' | 5'-GGTCTGCTTCGTCCTCTAC-3' |
| GAPDH | 5'-TATGATGATATCAAGAGGGTAGT-3' | 5'-TGTATCCAAACTCATTGTCATAC-3' |
Luciferase assay
Twenty four hours after plating in 12 well plates, USCs were transiently transfected with various DNAs of interest using Superfect reagent (Invitrogen). To assess TGF-β signaling, the transfection reactions included the P3TP-luc reporter along with the SV40 renilla plasmid to control for transfection efficiency. Three hours after transfection, the medium was changed and the cells were treated with 5ng/mL TGF-β or vehicle for 48hours. Cells were then harvested and assayed for luciferase activity using the Dual Luciferase Reporter Assay Kit from Promega as described in the manufacturer's protocol.
Retrovirus-mediated Ski RNA interference
293P cells were cultured in DMEM medium containing 5% fetal bovine serum and 100 units/mL penicillin/streptomycin. Cells were transiently transfected with pRetro-shSki(Cellogenetics) using FuGENE 6 reagent (Roche Diagnostics). Forty eight hours after transfection, viral supernatants were collected and filtered with a 0.45 μm filter. USCs were infected with fresh viral supernatant mixed in a 1:1 ratio with plating medium supplemented with 6 μg/mL Polybrene. Phenotypic assessments were generally performed 48 hours post-infection.
Immunoprecipitation and Western blotting
USCs cultured in 60mm dishes were transiently transfected with either an expression plasmid containing flag-tagged human Ski (cmv-Ski) or empty vector using SuperFect reagent. Forty eight hours after transfection, the transfected cells were treated with 5 ng/mL TGF-β for 30 minutes. Cellular proteins were extracted with lysis buffer (20 mMTris-HCl, pH 7.5, 150 mMNaCl, 1% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium fluoride, and 1 mM sodium orthovanadate containing protease inhibitors) and centrifuged at 14,000 rpm for 10 minutes at 4°C. Lysates were pre-cleared for 1 hour with immobilized protein-G Sepharose beads (Amersham Pharmacia Biotech) at 4°C and then were incubated with the IP antibody of interest (Sigma) overnight at 4°C. Immunocomplexes were collected by centrifugation following a two hour incubation with 50 μl of freshly immobilized protein-G Sepharose beads to facilitate immunoprecipitation of Ski-associated proteins. To detect proteins present in the immunoprecipitate, SDS-PAGE and Western blot analyses were performed with a 1:1000 dilution of primary antibody followed by a 1:2000 dilution of horseradish peroxidase-conjugated anti-rabbit secondary antibody (Bio-Rad Laboratories). Immunoprecipitated proteins were visualized by ECL reagent (Amersham Pharmacia Biotech).
RESULTS
Ski is expressed in chick cartilage
To determine the comparative Ski protein expression pattern in chick tissues, we performed Western blot analysis of chicken lung, brain, heart, liver and sternal cartilage (Figure 1). As previously reported, lung and brain tissues possess high levels of Ski protein [Lyons et al., 1994], with the antibody detecting a 95kD band consistent with the molecular weight of Ski (Lanes 1 and 2). This was in contrast to liver which was Ski-negative (Lane 4). Cartilage showed robust expression of Ski (Lane 5), suggesting that it has a functionally important role in this tissue. It should be noted that the apparent molecular weight of the detected proteins in heart and cartilage was different from that seen in lung or brain. This may be due to tissue-specific differences in the level of Ski glycosylation as has been suggested previously with respect to the production of recombinant protein [Nagase et al., 1990]. These results provide a basis for further assessment of Ski expression in cartilage and a study of its possible regulatory role in this tissue.
Figure 1. Ski is expressed in cartilage.
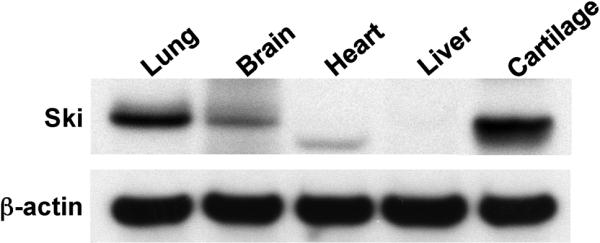
Chick tissues including lung, brain, heart, liver and sternal cartilage were harvested and proteins were extracted. Fifty μg quantities of solubilized protein were loaded and run on polyacrylamide gels, transferred to nylon membranes and Western analyses were performed to detect Ski, with detection of β-actin confirming equal loading of the gel. The analysis was performed 3 times, with the representative blot shown.
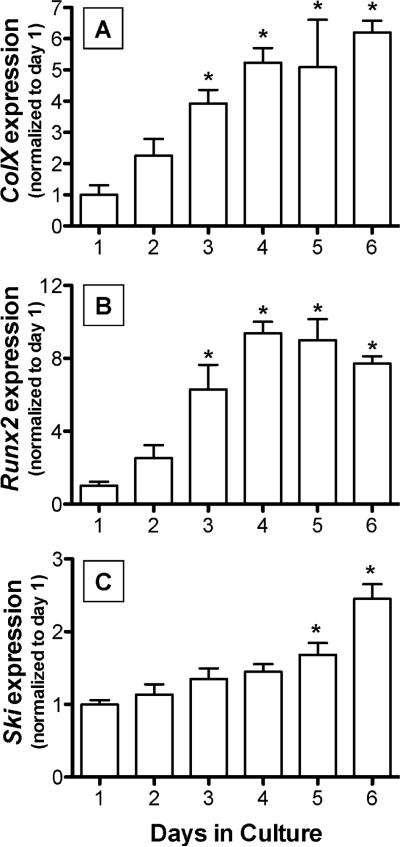
To determine if there is a relationship between chondrocyte hypertrophy and Ski expression, Ski mRNA levels were measured in chick USCs at various time points after seeding of the cells. Consistent with previous studies [Dong et al., 2006; Grimsrud et al., 2001], these cells displayed maturational progression over a 6 day period in culture based on enhanced expression of colX (Figure 2A) and Runx2 (Figure 2B). Comparatively, Ski was up-regulated progressively with time as the cells matured, with a 2-fold increase observed by day 6 (Figure 2C). These results demonstrate that Ski expression is enhanced during the later stages of chondrocyte hypertrophy.
Figure 2. Ski mRNA expression is increased in hypertrophic USCs.
Chick USCs were isolated and cultured in medium containing 50 μg/mL ascorbic acid for between 1 and 6 days. mRNA was harvested and qPCR was performed to detect the hypertrophy-associated genes colX(A) and Runx2(B) as well as Ski (C). Statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from the day 1 value for each gene assessed.
Ski regulates chondrocyte maturation
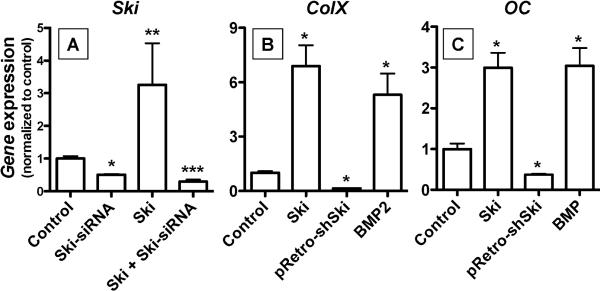
To determine if differential regulation of the Ski gene has functional relevance in the control of chondrocyte maturation, we established methods to perform Ski loss and gain of function experiments. A pRetro-shSki retrovirus was designed to interfere with both human and chicken transcripts, and its effect on Ski mRNA expression was assessed. Twenty four hours after plating, USCs were transfected with an expression plasmid containing the human Ski cDNA (cmv-Ski) or the empty vector control. After an additional 24 hours, cells were infected with pRetro-shSki or a nonsense control virus. Total mRNA was harvested 36 hours later and real time RT-PCR was performed. Confirming successful Ski over-expression, cmv-Ski-transfected cells showed a greater than 3-fold increase in the Ski pool compared to the empty vector-transfected control (Figure 3A). UCSs infected with pRetroshSki showed a greater than 50% reduction in basal Ski expression and a 90% reduction of Ski levels in cells transfected with the human Ski construct (Figure 3A). These results establish cmv-Ski and the pRetro-shSki as useful reagents in Ski gain and loss of function experiments respectively.
Figure 3. Gain and loss of Ski expression affects USC differentiation.
Chick USCs were isolated and cultured in medium containing 50 μg/mL ascorbic acid. To confirm that experimental strategies for gain and loss of Ski expression were effective, USCs were infected with pRetro-shSkiretrovirus, transfected with cmv-Ski (Ski), treated with both reagents, or infected with nonsense shRNA virus (control). Twenty four hours post infection/transfection, mRNA was harvested and qPCR was performed to detect Ski (A). Using these Ski gain and loss of expression strategies, USCs were subsequently infected with pRetro-shSkiretrovirus, transfected with cmv-Ski or treated for 50 ng/mL BMP-2 (positive control). Control cells were infected with a nonsense shRNA retrovirus. Twenty four hours post-treatment, mRNA was harvested and qPCR was performed to quantify colX (B) and osteocalcin (C) expression levels. For all three panels, statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from control value for each gene assessed.
To assess the impact of Ski on maturational progression in USCs, we utilized the above reagents to either over-express Ski or knock down its expression in monolayer cultures. Twenty four hours after initial plating, USCs were either left untreated (negative control), transfected with cmv-Ski, infected with pRetro-shSki, or treated with 50 ng/mL BMP-2 (positive control). Forty eight hours after these treatments, mRNA was harvested and the expression level of colX and osteocalcin was assessed via real time RT-PCR. Compared to the untreated control, over-expression of Ski led to a better than 6-fold increase in colX, an effect that was as strong as that seen with BMP-2 treatment (Figure 3B). Conversely, cells infected with pRetroshSki showed a better than 70% reduction in colX expression relative to control (Figure 3B). Comparatively, Ski over-expression and BMP-2 treatment induced osteocalcin approximately 3-fold,while pRetro-shSki infection reduced osteocalcin expression greater than 50% compared to the untreated control (Figure 3C). It should be noted that transfection of USCs with the empty vector or infection with the nonsense shRNA did not affect colX or osteocalcin expression levels relative to the untreated negative control or the BMP-2 positive control (data not shown). Overall, these results suggest that Ski is a likely participant in the complex regulation of chondrocyte maturation by various signaling pathways.
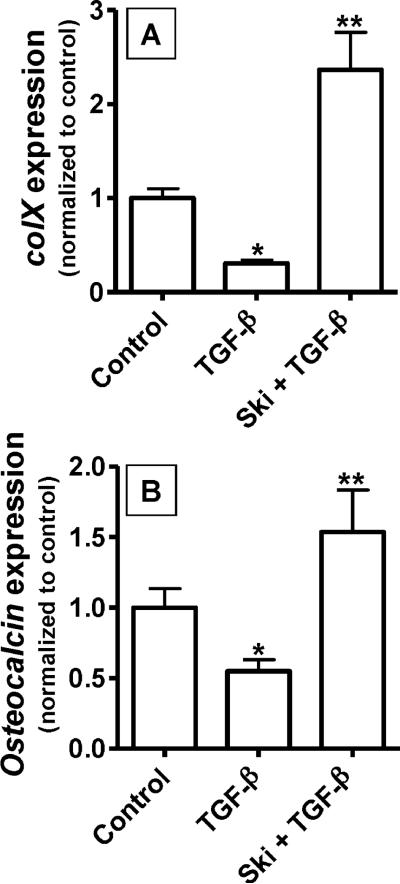
Since Ski is known to be a potent inhibitor of TGF-β/Smad signaling, we assessed the effect of Ski over-expression on inhibition of chondrocyte hypertrophy by TGF-β. USCs were plated in monolayer and transfected with either cmv-Ski or the empty vector negative control 24 hours post-seeding. Twenty four hours after transfection, cells were treated with 5 ng/mL TGF-β or vehicle and total mRNA was harvested 48 hours later. Real time RT-PCR was employed to assess colX and osteocalcin mRNA levels. Interestingly, the inhibition of colX and osteocalcin expression by TGF-β was completely reversed in cells over-expressing Ski (Figure 4A and 4B). These results support the hypothesis that acceleration of chondrocyte maturation by Ski is at least partially due to an inhibition of the influence of TGF-β.
Figure 4. Ski reverses the inhibitory effect of TGF-β on colX and osteocalcin.
As before, chick USCs were isolated and cultured in medium containing 50 μg/mL ascorbic acid. Twenty four hours after plating, cells were transfected with cmv-Ski (Ski) or an empty vector (Control). Twenty four hours post-transfection, cultures were treated with or without 5 ng/mL TGF-β. After an additional 24 hours in culture, mRNA was harvested and qPCR was performed to quantify colX (A) and osteocalcin (B) expression levels. For both panels, statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from the control value for each gene assessed and double asterisks (**) denoting significance from the TGF-β-treated group.
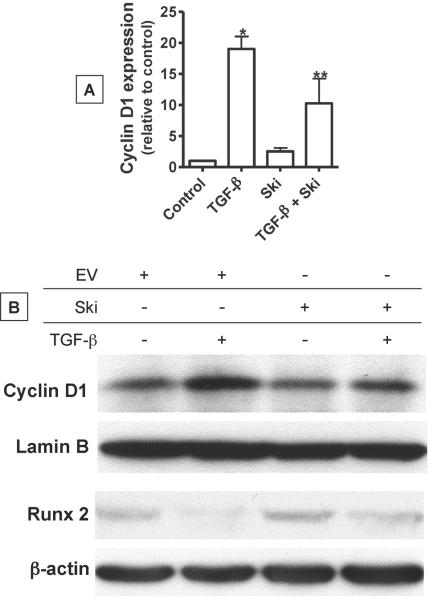
One possible mechanism underlying the inhibitory effect of TGF-β on chondrocyte maturation is its direct induction of cyclin D1 and concomitant activation of cell cycle progression [Beier et al., 2001; Beier et al., 1999; Li et al., 2006a]. Since the cyclin D1 gene is a direct target of TGF-βsignaling, we assessed the impact of Ski over-expression on its mRNA and protein levels in UCSs. Twenty four hours after USCs were transfected with cmv-Ski or empty vector, cultures were exposed to a 48 hr treatment with 5 ng/mL TGF-β or vehicle. Total mRNA was harvested and real time RT-PCR was performed to assess cyclin D1 transcript levels. As expected, TGF-β up-regulated cyclin D1 expression levels by nearly 20-fold (Figure 5A). While Ski over-expression did not significantly affect cyclin D1 expression, its induction by TGF-β was blunted by 50% (Figure 5A). This effect was also observed at the protein level, where Western blot analysis revealed that induction of the cyclin D1 pool by TGF-β was significantly blunted in USCs over-expressing Ski (Figure 5B). Lamin B was detected to confirm equal loading. Not only do these findings demonstrate the influence of Ski on the expression of a key TGF-β target gene, but they also suggest a possible mechanism behind how Ski contributes to the control of the hypertrophic process.
Figure 5. Ski stimulates Runx2 expression and reverses the effect of TGF-β on cyclin D1 and Runx2 expression.
Chick USCs were isolated and cultured in medium containing 50 μg/mL ascorbic acid for 24 hours. Following this, cells were transfected with cmv-Ski (Ski) or an empty vector (Control). Twenty four hours post-transfection, cultures were treated with or without 5 ng/mL TGF-β for 24 additional hours. mRNA and total cellular protein were harvested and qPCR and Western blotting was performed to quantify cyclin D1 mRNA levels (A) and both cyclin D1 (Lamin B load control) and Runx2 (β-actin load control) protein levels (B). In panel (A), statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from the control value and double asterisks (**) denoting significance from the TGF-β-treated group. Blots shown in panel (B) are representative of experiments that were repeated 3 times.
It has been previously shown that in addition to driving cell cycle progression, cyclin D1 likely affects differentiation of bone and cartilage cells by inducing degradation of Runx2 [Shen et al., 2006a], a transcription factor required for terminal hypertrophy and mineralization of the matrix [Komori, 2002]. Thus, we further assessed the influence of TGF-β and Ski on the Runx2 protein levels in USCs. As in the cyclin D1 experiment described above, cells were transfected with cmv-Ski or empty vector and treated with either TGF-β or vehicle for 48 hours. Harvested proteins were used in a Western blot analysis of Runx2 expression. As would be predicted from the published data indicating that cyclin D1 induces Runx2 degradation, we observed a significant reduction of Runx2 levels in USCs treated with 5 ng/mL TGF-β (Figure 5B, lanes 1 and 2). Not only did over-expression of Ski slightly enhance Runx2 protein pools (Lane 3), the TGF-β associated reduction in Runx2 was partially reversed in USCs over-expressing Ski (Lane 4). Equal loading in this experiment was confirmed via blotting for β-actin. These results provide further mechanistic insight into how Ski over-expression could accelerate maturation in chondrocytes, leading to the up-regulation of marker genes such as colX and osteocalcin as shown in Figure 3.
Ski inhibits TGF-β signaling
Since Ski was found to strongly suppress the influence of TGF-β on USC maturation, we performed a mechanistic analysis of the interaction between Ski and the TGF-β/Smad signaling pathway. To begin, we performed a TGF-β signaling reporter assay to assess inhibition by Ski in chondrocytes. USCs were co-transfected with the P3TP-luc reporter, SV40 renilla (efficiency control) and cmv-Ski or the empty vector negative control. After treatment with or without 5 ng/mL TGF-β for 48 hours, cell extracts were harvested for assessment of luciferase expression. As expected, TGF-β induced a robust (6-fold) signaling response compared to control-treated cultures (Figure 6A). Consistent with the purported role of Ski, cultures over-expressing Ski showed reduced luciferase expression both under basal conditions (50% decrease) and following TGF-β treatment (85% decrease). These results demonstrate that Ski inhibits TGF-β signaling in chondrocytes.
Figure 6. Ski inhibits TGF-β signaling and co-immunoprecipitates with pSmad2, pSmad3 and Smad4.
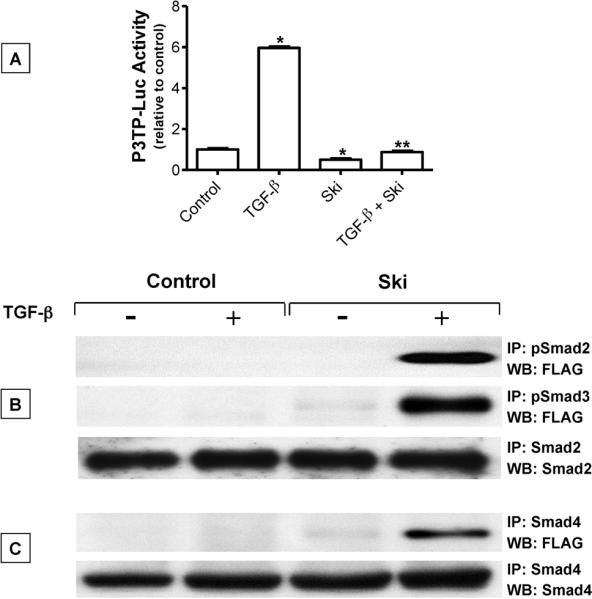
(A) Following an initial 24 hr culture period with medium containing 50 μg/mL ascorbic acid, USCs were co-transfected with P3TP-luc and SV40-renilla (transfection efficiency control) and either cmv-Ski (Ski) or an empty vector plasmid (Control). Twenty four hours later, cells were treated with or without 5 ng/mL TGF-β for 36 hours. Cell extracts were then collected and luciferase activity was measured using the Promega dual luciferase detection kit as directed by the manufacturer. Statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from the control group and double asterisks (**) denoting significance from the TGF-β-treated group. (B) Following an initial 24 hr culture period in medium containing 50 μg/mL ascorbic acid, USCs were transfected with cmv-Ski (Ski) or empty vector (Control). Twenty four hours later, cells were treated with or without 5 ng/mL TGF-β for 4 hours and then total cellular proteins were harvested. Immunoprecipitations were performed using either pSmad2, pSmad3, Smad2 (B) or Smad4 (C) antibody. Immunoprecipitated proteins were run out on polyacrylamide gels, transferred to nylon membranes and Western blotting was performed to detect the flag epitope (at the N-terminus of the Ski cDNA). To confirm equal loading of protein at the immunoprecipitation step, Smad2 or Smad4 immunoprecipitations were followed by detection of Smad2 (B) or Smad4 (C) respectively by Western. Blots shown in panels (B) and (C) are representative of experiments that were repeated 3 times.
To determine if these signaling effects of Ski involve its interaction with various TGF-β/Smads, we performed immunoprecipitation and Western blotting. Cells transfected with cmv-Ski (flag-tagged) or empty vector were treated with or without 5 ng/mL TGF-β for 30 minutes. Harvested proteins were immunoprecipitated with either anti-pSmad2 or anti-pSmad3, run on polyacrylamide gels, transferred to nylon membranes and western blotting was performed with an anti-flag monoclonal antibody. To confirm equal protein loading onto the resin at the immunoprecipitation step, IPs were performed with anti-Smad2 followed by western blotting with anti-Smad2. While no immune complexes were detected in un-transfected cells (negative control), flag-tagged Ski protein was detected in TGF-β treated cultures following immunoprecipitation with anti-pSmad2 and anti-pSmad3 antibodies (Figure 6B). Since nuclear localization of pSmad2 and 3 is dependent on association with Smad4, we also determined if Ski was present in complexes containing Smad4. Cell extracts from cultures treated as described above were examined by immunoprecipitation with anti-Smad4 antibody to detect flag-tagged Ski. Consistent with the pSmad2 and pSmad3 immunoprecipitates, flag-tagged Ski also associated with Smad4. As expected, interactions between Smad4 and Ski were only detected with the anti-flag antibody in cells transfected with cmv-Ski (Figure 6C). More importantly, the Smad4-Ski association was only detected following TGF-β treatment of cells, suggesting that Ski only associates with a Smad4-containing complex when TGF-β signaling is active. Overall, these results suggest that the inhibitory effect of Ski on TGF-β/Smad signaling may be due to its interaction with transcriptional complexes containing pSmad2 or pSmad3.
Association of HDAC4 and 5 with pSmad3 is Ski-dependent
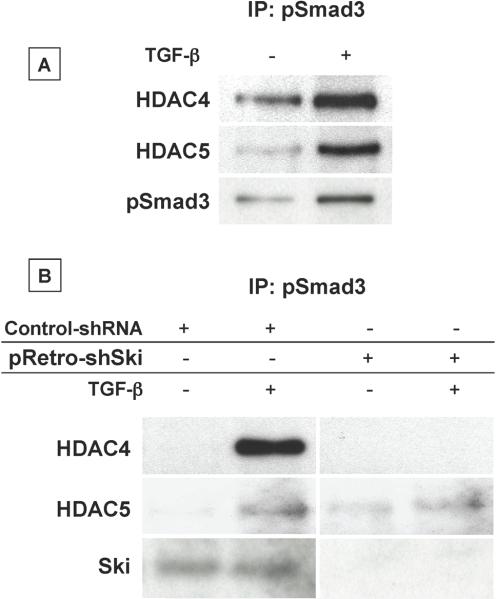
Since the inhibitory influence of Ski on TGF-β signaling is known to be via binding of HDAC family members to Smad-containing transcriptional complexes [Liu et al., 2001; Luo, 2003; Nomura et al., 1999], we assessed the association of HDAC4 and 5 with pSmad3. We focused on pSmad3 in particular because of the well established role it plays in TGF-β regulation of chondrocyte maturation [Li et al., 2006b; Yang et al., 2001]. First, we examined if stimulation of Smad3 phosphorylation leads to the recruitment of HDAC4 and 5. USCs were treated with or without 5 ng/mL TGF-β for 30 minutes and protein extracts were immunoprecipitated with anti-pSmad3 antibody. Immunoprecipitated proteins were analyzed by western blotting using antibodies against HDAC4, HDAC5 and pSmad3 (control). An association between pSmad3 and HDAC4 and 5 was found under basal conditions, and this association was significantly enhanced in cells treated with TGF-β (Figure 7A). These results not only confirm that pSmad3 and HDAC4 and 5 interact following stimulation of TGF-β signaling, but also provide rationale for assessing the dependence of this Smad/HDAC association on Ski.
Figure 7. Ski is required for the association between pSmad3 and HDAC4 and HDAC5.
(A) Following an initial 24 hr culture period with medium containing 50 μg/mL ascorbic acid, USCs were treated with or without 5 ng/mL TGF-β for 4 hours. Total cellular proteins were harvested and immunoprecipitations were performed using pSmad3 antibody. Immunoprecipitated proteins were run out on polyacrylamide gels, transferred to nylon membranes and Western blotting was performed to detect HDAC4 and HDAC5. (B) Following initial culturing in medium containing 50 μg/mL ascorbic acid, USCs were infected with pRetro-shSki or a nonsense shRNA virus. Twenty four hours later, cells were treated with or without 5 ng/mL TGF-βfor 4 hours. Total cellular protein was harvested and immunoprecipitations were performed as described in Experimental Procedures using pSmad3 antibody. Immunoprecipitated proteins were run out on polyacrylamide gels, transferred to nylon membranes and Western blotting was performed to detect HDAC4 and HDAC5. Blots shown in panels (A) and (B) are representative of results obtained from experiments that were repeated 3 times.
To assess if Ski is required for pSmad3 to interact with HDAC4 and 5, we performed immunoprecipitation/Western blotting to assess complex formation in USCs with reduced Ski expression. Twenty four hours after plating, USCs were infected with either pRetro-shSki or a nonsense shRNA (control). Forty eight hours post-infection, cells were treated with 5 ng/mL TGF-β for 30 minutes followed by harvest of cellular proteins. Immunoprecipitations were performed with the anti-pSmad3 antibody followed by detection of immune complexes via Western blotting with antibodies against HDAC4, HDAC5 and Ski (to confirm knockdown in pRetro-shSki infected cultures). While there were clear associations between pSmad3 and both HDAC4 and 5 in TGF-β-treated cells infected with the control shRNA retrovirus, the pSmad3 and HDAC4 interaction was completely ablated with Ski knockdown (Figure 7B). A less robust reduction in the pSmad3-HDAC5 association was also detected in pRetro-shSki-infected USCs. These results establish that Ski is involved in the recruitment of HDAC4 and 5 to pSmad3-containing transcriptional complexes. This provides an insight into a possible mechanism underlying the acceleration of chondrocyte hypertrophy seen in cells over-expressing Ski.
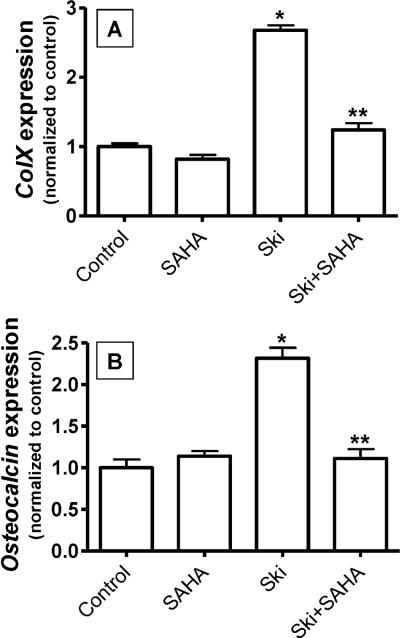
If the influence of Ski on chondrocyte maturation is dependent on repression of TGF-β signaling via its recruitment of HDAC4 and 5, blockade of HDAC activity should reverse the effects of Ski. To evaluate this, freshly isolated USCs were pre-treated with or without the broadly effective HDAC inhibitor suberoylanilidehydroxamic acid (SAHA) for 24 hours and then were transfected with cmv-Ski or empty vector. After an additional 24 hours of culture, mRNA was harvested, reverse transcribed and real time RT-PCR was performed to detect colX and OC. Consistent with results shown in Figure 3B and 3C, over-expression of Ski significantly enhanced colX and OC expression in USCs (Figure 8). While SAHA did not affect the basal expression of these genes, it did reverse the up-regulation of colX and OC induced by Ski over-expression (Figure 8). This finding further implicates that recruitment of HDACs as the mechanism underlying the effects of Ski on chondrocyte phenotype.
Figure 8. Induction of colX and osteocalcin expression by Ski is blocked by HDAC inhibition.
Chick USCs were isolated and cultured in medium containing 50 μg/mL ascorbic acid. Twenty four hours after plating, cells were pre-treated with the HDAC inhibitor suberoylanilidehydroxamic acid (SAHA, 5μM). Following this, cells were transfected with cmv-Ski (Ski) or empty vector (Control) and maintained in culture for an additional 24 hours in the presence of SAHA. Messenger RNA was then harvested and qPCR was performed to quantify colX (A) and osteocalcin (B) expression levels. For both panels, statistically significant differences were identified via ANOVA (N=3, p<0.05) with asterisks (*) denoting significance from the control value for each gene assessed and double asterisks (**) denoting significance from the Ski-transfected group.
DISCUSSION
The control of endochondral ossification is exerted by a number of key growth factors and their respective signaling pathways. In particular, the TGF-β/BMP superfamily of factors and their downstream signaling responses are known to be critical in the orchestration of chondrocyte hypertrophy. Specifically, Smad3 signaling initiated by TGF-β is a potent inhibitory signal that leads to deceleration of chondrocyte maturation in vitro [Ferguson et al., 2000; Ionescu et al., 2003; Pateder et al., 2001] and delayed endochondral ossification in vivo [Serra et al., 1997]. Consistent with this, loss of TGF-β/Smad3 signaling leads to accelerated chondrocyte hypertrophy and the development of osteoarthritis in the mouse [Yang et al., 2001]. Conversely, BMP/Smad signaling strongly accelerates chondrocyte hypertrophy in a number of cell [Denker et al., 1999; Grimsrud et al., 1999; Grimsrud et al., 2001; Leboy et al., 2001] and animal models [Li et al., 2006b; Zhang et al., 2003]. Given the integral role of these pathways in controlling chondrocyte differentiation, understanding their negative regulation is an important step toward fully characterizing the process of hypertrophy in development and growth as well as in pathologic circumstances such as arthritis.
There are a number of well characterized endogenous negative regulators of the TGF-β and BMP signaling pathways. For example, Smad7 can bind to the TGF-β receptor complex and block Smad2/3 phosphorylation [Nakao et al., 1997] while Smad6 binds to Smads 1, 5 and 8 to block their phosphorylation by the active BMP receptor [Ishida et al., 2000; Nakayama et al., 1998]. Comparatively, Smurf1 and Smurf2 are E3 ubiquitin ligases which target a number of proteins in both pathways for proteasomal degradation. Smurf1 ubiquitinates and degrades Runx2 and Smad1 [Shen et al., 2006a; Zhu et al., 1999], while Smurf2 targets include Smad2, phospho-Smad3 and the TGF-β type I receptor (Smad7-dependent) and Smad1 [Kavsak et al., 2000; Lin et al., 2000; Wu et al., 2008; Zhang et al., 2001]. Additional negative regulatory influence is exerted by noggin and chordin, secreted factors that bind and sequester BMPs in the matrix, thereby abrogating interaction with the receptor and blocking induction of signaling [Canalis et al., 2003]. As an extension to these mechanistic studies, work has been published identifying the potential involvement of several of these negative regulatory mechanisms in normal tissue homeostasis and in pathological states [Fontaine et al., 2005; Horiki et al., 2004; Semonin et al., 2001].
In addition to the negative regulatory mechanisms mentioned above, the oncoprotein Ski has been identified as a potent inhibitor of signaling that involves Smads. Ski was initially identified as the transforming protein of the avian Sloan-Kettering retrovirus that induces oncogenesis in chick embryonic fibroblasts [Li et al., 1986]. Ski orthologs have been identified in a number of species including chicken, mouse and human, with the human ortholog being a nuclear protein of 728 amino acids [Liu et al., 2001]. Functionally, Ski interacts with Smad proteins on the TGF-β and BMP signaling pathways including Smads 3/4 and Smads 1/4, 5/4 respectively [Liu et al., 2001], leading to transcriptional repression of TGF-β- and BMP-responsive promoters [Sun et al., 1999; Wang et al., 2000]. Significant effort has been spent to elucidate the binding configuration of Ski to Smad complexes, revealing that a primary candidate mechanism underlying the inhibitory effect of Ski on signaling is via interference with the interaction between Smad4 and R-Smads [Sun et al., 1999]. Additionally, Ski binds various co-repressors including N-CoR, mSin3A and HIPK2, leading to the recruitment of HDAC proteins to transcriptional complexes [Harada et al., 2003; Khan et al., 2001; Kokura et al., 2001; Shinagawa et al., 2001]. By binding to Smads, Ski can thus recruit this repressor complex to Smad-responsive elements in key target promoters. These mechanisms likely function cooperatively to inhibit TGF-β and BMP signaling [Liu et al., 2001].
Given the ability of Ski to inhibit TGF-β and BMP signaling, the nature of its potential role in the regulation of chondrocyte differentiation becomes an important question. Along this line, a number of the other negative regulators of these signaling pathways have been studied, including Smad7, Smad6, Smurf1, Smurf2 and noggin. For example, inhibition of the TGF-β pathway by Smad7 leads to reduced chondrocyte proliferation and proteoglycan synthesis in chondrocyte cultures [Scharstuhl et al., 2003]. Inhibition of BMP signaling with either Smad6 or noggin leads to deceleration of the chondrocyte hypertrophic program in vitro [Chen et al., 2004; Li et al., 2003]. While transgenic mice utilizing the type 11 collagen promoter to drive expression of either Smad6 or Smurf1 in cartilage show no obvious phenotype, crossing of these lines leads to reduced BMP signaling and significantly delayed chondrocyte hypertrophy in the growth plate [Horiki et al., 2004]. Finally, our group has shown that blockade of TGF-β signaling in cartilage via over-expression of Smurf2 under control of the type II collagen promoter leads to accelerated chondrocyte hypertrophy in sternal chondrocytes isolated from transgenic mice [Wu et al., 2008]. These mice also show inappropriate chondrocyte hypertrophy coupled with development of an osteoarthritis-like disease of their joints [Wu et al., 2008]. These reports establish the important role of negative regulatory mechanisms of the TGF-β/BMP axis in normal chondrocyte physiology.
As mentioned, constitutive deletion of the Ski gene causes a number of skeletal defects including altered formation or deletion of craniofacial bones, polydactyly and fusion of some vertebrae [Colmenares et al., 2002]. Therefore, we aimed to determine how negative regulation of TGF-β/BMP signaling by Ski affects skeletal tissues. As a first step toward understanding the functional relevance of Ski in these tissues, we employed the chick upper sternal chondrocyte model to evaluate the role of Ski during chondrocyte maturation. Not only did we identify robust endogenous expression of Ski in chick cartilage that is upregulated in association with maturation, we demonstrate that gain of Ski expression leads to enhanced hypertropic progression, evidenced by the upregulation of colX and OC. We also observed abrogation of the inhibition of these genes by TGF-β in cells over-expressing Ski. Complementing this, knock-down of Ski via RNA interference leads to reduced colX and OC expression. These results suggest that the dominant effect of Ski in USCs is via inhibition of the TGF-β pathway.
It is known that cyclin D1 is a direct target of TGF-β signaling in chondrocytes [Beier et al., 2001; Beier et al., 1999; Li et al., 2006b]. Its activity as a driver of proliferation as well as an inducer of Runx2 proteasomal degradation [Shen et al., 2006b] may lead to its ability to delay chondrocyte maturation overall. Based on this, we hypothesized that Ski may drive hypertrophy by inhibiting the TGF-β induction of cyclin D1 leading to a concomitant enhancement of the Runx2 pool. Supporting this hypothesis, over-expression of Ski indeed led to a significant inhibition of the TGF-β induction of cyclin D1 (mRNA and protein) and a net increase in Runx2 protein levels. These data provide a potential mechanism underlying the induction of hypertrophy by up-regulation of Ski.
While we observed potent inhibition of TGF-β signaling on the P3TP-luc reporter in USCs over-expressing Ski, it should be noted that Ski was also found to inhibit BMP induction of signaling as measured by the 12XSBE-OC promoter/luciferase reporter (data not shown). However, since the dominant phenotypic effect of Ski in chondrocytes suggests its function may primarily be due to its inhibition of the TGF-β pathway, we focused on the interaction of Ski with signaling proteins downstream of the TGF-β receptor. First, IP/Western blot experiments identified a strong interaction between Ski and pSmad2 or pSmad3. Given that Smad4 dimerizes with these phosphorylated Smads, we also saw a strong interaction between Ski and Smad4 following TGF-β treatment of the cultures. Furthermore, we confirmed a strong participation of HDAC4 and HDAC5 in these complexes that was lost when Ski levels were ablated via RNA interference. These results are consistent with previously published work which proposes that Ski interacts with pSmad complexes and is required for recruitment of HDAC proteins to pSmad-containing transcriptional complexes [Harada et al., 2003; Khan et al., 2001; Kokura et al., 2001; Shinagawa et al., 2001]. Two previous studies have identified association of HDACs with pSmad3 on the osteocalcin promoter in an osteoblast cell model and on the Nkx3.2 promoter in chondrocytes treated with TGF-β [Kang et al., 2005; Kim and Lassar, 2003]. Based on our findings, it is possible that this type of interaction is dependent on the involvement of Ski in the complex.
In conclusion, we establish that reduction of TGF-β signaling leads to acceleration of chondrocyte differentiation. We demonstrate that over-expression of Ski causes increased expression of hypertrophic marker genes, possibly by its direct inhibition of TGF-β induction of cyclin D1 leading to increased Runx2 levels. The mechanism of the inhibition of the TGF-β pathway by Ski is likely facilitated by its interaction with pSmads 2 and 3 and its recruitment of HDAC4 and 5 to the complex. Overall, our results support the hypothesis that Ski is an important modulator of the regulation of chondrocyte differentiation by TGF-β. As chondrocytes progress through endochondral ossification, the enhanced expression of Ski could serve to reduce the inhibitory influence of TGF-β permitting hypertrophic differentiation to proceed. This would have strong relevance during normal development and growth processes in the skeleton, during repair of skeletal injury such as in fracture healing, and in pathological processes that involve inappropriate recapitulation of endochondral ossification such as in osteoarthritis. In conclusion, results reported here establish that Ski is an important endogenous negative regulator of TGF-β pathway in chondrocytes whose overall function contributes significantly to the regulation of the hypertrophic program.
ACKNOWLEDGEMENTS
The authors wish to acknowledge excellent technical support from Erica Dussmann and Michael Thullen for the generation of sternal chondrocytes. This work was supported by an Arthritis Investigator Award from the Arthritis Foundation (MJZ), R01 AR045700 (RNR),P50 AR054041 (RNR), R01 AR055915 (DC), R01 AR057022 (MJH), 2T32 AR053459 (ERS, trainee) and P30 AR061307.
Grants: Arthritis Foundation, 5AI-Zus-A-5, NIH/NIAMS R01 AR045700, NIH/NIAMS P50 AR054041, NIH/NIAMS R01 AR055915, NIH/NIAMS R01 AR057022, NIH/NIAMS T32 AR053459 and NIH/NIAMS P30 AR061307.
ABBREVIATIONS
- (TGF-β)
Transforming growing factor-β
- (HDACs)
histone deacetylases
- (pSmad2 and 3)
phosphorylated Smads 2 and 3
- (colX)
type × collagen
- (USCs)
Cephalic (upper) sternal chondrocytes
- (SAHA)
suberoylanilidehydroxamic acid
REFERENCES
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. Journal of Biological Chemistry. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- Ballock RT, Heydemann A, Wakefield LM, Flanders KC, Roberts AB, Sporn MB. TGF-beta1 prevents hypertrophy of epiphyseal chondrocytes: Regulation of gene expression for cartilage matrix proteins and metalloproteases. Developmental Biology. 1993;158:414–429. doi: 10.1006/dbio.1993.1200. [DOI] [PubMed] [Google Scholar]
- Beier F, Ali Z, Mok D, Taylor AC, Leask T, Albanese C, Pestell RG, LuValle P. TGFbeta and PTHrP control chondrocyte proliferation by activating cyclin D1 expression. Molecular Biology of the Cell. 2001;12:3852–3863. doi: 10.1091/mbc.12.12.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc.Natl.Acad.Sci.U.S.A. 1999;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr.Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Colmenares C, Heilstedt HA, Shaffer LG, Schwartz S, Berk M, Murray JC, Stavnezer E. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski−/− mice. Nat.Genet. 2002;30:106–109. doi: 10.1038/ng770. [DOI] [PubMed] [Google Scholar]
- Colmenares C, Stavnezer E. The ski oncogene induces muscle differentiation in quail embryo cells. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- Denker AE, Haas AR, Nicoll SB, Tuan RS. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation. 1999;64:67–76. doi: 10.1046/j.1432-0436.1999.6420067.x. [DOI] [PubMed] [Google Scholar]
- Dong YF, Soung dY, Schwarz EM, O'Keefe RJ, Drissi H. Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J.Cell Physiol. 2006;208:77–86. doi: 10.1002/jcp.20656. [DOI] [PubMed] [Google Scholar]
- Ferguson CM, Schwarz EM, Reynolds PR, Puzas JE, Rosier RN, O'Keefe RJ. Smad2 and 3 mediate TGF-á1-induced inhibition of chondrocyte maturation. Endocrinology. 2000;141:4728–4735. doi: 10.1210/endo.141.12.7848. [DOI] [PubMed] [Google Scholar]
- Fontaine K, Semonin O, Legarde JP, Lenoir G, Lucotte G. A new mutation of the noggin gene in a French Fibrodysplasia ossificans progressiva (FOP) family. Genet.Couns. 2005;16:149–154. [PubMed] [Google Scholar]
- Grimsrud CD, Romano PR, D'Souza M, Puzas JE, Reynolds PR, Rosier RN, O'Keefe RJ. BMP-6 is an autocrine stimulator of chondrocyte differentiation. Journal of Bone and Mineral Research. 1999;14:475–482. doi: 10.1359/jbmr.1999.14.4.475. [DOI] [PubMed] [Google Scholar]
- Grimsrud CD, Romano PR, D'Souza M, Puzas JE, Schwarz EM, Reynolds PR, Rosier RN, O'Keefe RJ. BMP signaling stimulates chondrocyte maturation and the expression of indian hedgehog. Journal of Orthopaedic Research. 2001;19:18–25. doi: 10.1016/S0736-0266(00)00017-6. [DOI] [PubMed] [Google Scholar]
- Harada J, Kokura K, Kanei-Ishii C, Nomura T, Khan MM, Kim Y, Ishii S. Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation. Journal of Biological Chemistry. 2003;278:38998–39005. doi: 10.1074/jbc.M307112200. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, Ten Dijke P. TGF-á signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Horiki M, Imamura T, Okamoto M, Hayashi M, Murai J, Myoui A, Ochi T, Miyazono K, Yoshikawa H, Tsumaki N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. Journal of Cell Biology. 2004;165:433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu AM, Schwarz EM, Zuscik MJ, Drissi H, Puzas JE, Rosier RN, O'Keefe RJ. ATF-2 cooperates with Smad3 to mediate TGF-beta effects on chondrocyte maturation. Experimental Cell Research. 2003;288:198–207. doi: 10.1016/s0014-4827(03)00181-2. [DOI] [PubMed] [Google Scholar]
- Ishida W, Hamamoto T, Kusanagi K, Yagi K, Kawabata M, Takehara K, Sampath TK, Kato M, Miyazono K. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. Journal of Biological Chemistry. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- Ishisaki A, Yamato K, Hashimoto S, Nakao A, Tamaki K, Nonaka K, Ten Dijke P, Sugino H, Nishihara T. Differential inhibition of Smad6 and Smad7 on bone morphogenetic protein- and activin-mediated growth arrest and apoptosis in B cells. Journal of Biological Chemistry. 1999;274:13637–13642. doi: 10.1074/jbc.274.19.13637. [DOI] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO Journal. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF-á receptor for degradation. Molecular Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Khan MM, Nomura T, Kim H, Kaul SC, Wadhwa R, Shinagawa T, Ichikawa-Iwata E, Zhong S, Pandolfi PP, Ishii S. Role of PML and PML-RARalpha in Mad-mediated transcriptional repression. Molecular Cell. 2001;7:1233–1243. doi: 10.1016/s1097-2765(01)00257-x. [DOI] [PubMed] [Google Scholar]
- Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol.Cell Biol. 2003;23:8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. Journal of Biological Chemistry. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- Komori T. Runx2, a multifunctional transcription factor in skeletal development. J.Cell Biochem. 2002;87:1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- Leboy P, Grasso-Knight G, D'Angelo M, Volk SW, Lian JV, Drissi H, Stein GS, Adams SL. Smad-Runx interactions during chondrocyte maturation. J.Bone Joint Surg.Am. 2001;83-A(Suppl 1):S15–S22. [PubMed] [Google Scholar]
- Leboy PS, Vaias L, Uschmann B, Golub E, Adams SL, Pacifici M. Ascorbic acid induces alkaline phosphatase, type × collagen, and calcium deposition in cultured chick chondrocytes. Journal of Biological Chemistry. 1989;264:17281–17286. [PubMed] [Google Scholar]
- Leferovich JM, Lana DP, Sutrave P, Hughes SH, Kelly AM. Regulation of c-ski transgene expression in developing and mature mice. Journal of Neuroscience. 1995;15:596–603. doi: 10.1523/JNEUROSCI.15-01-00596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TF, Chen D, Wu Q, Chen M, Sheu TJ, Schwarz EM, Drissi H, Zuscik M, O'Keefe RJ. Transforming growth factor-beta stimulates cyclin D1 expression through activation of beta-catenin signaling in chondrocytes. Journal of Biological Chemistry. 2006a;281:21296–21304. doi: 10.1074/jbc.M600514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TF, Darowish M, Zuscik MJ, Chen D, Schwarz EM, Rosier RN, Drissi H, O'Keefe RJ. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. Journal of Bone and Mineral Research. 2006b;21:4–16. doi: 10.1359/JBMR.050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ionescu AM, Schwarz EM, Zhang X, Drissi H, Puzas JE, Rosier RN, Zuscik MJ, O'Keefe RJ. Smad6 is induced by BMP-2 and modulates chondrocyte differentiation. Journal of Orthopaedic Research. 2003;21:908–913. doi: 10.1016/S0736-0266(03)00008-1. [DOI] [PubMed] [Google Scholar]
- Li Y, Turck CM, Teumer JK, Stavnezer E. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J.Virol. 1986;57:1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteosome-dependent degradation of Smad2 in transforming growth factor-beta signaling. Journal of Biological Chemistry. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Weinberg RA, lodish HF. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Luo K. Negative regulation of BMP signaling by the ski oncoprotein. J.Bone Joint Surg.Am. 2003;85-A(Suppl 3):39–43. doi: 10.2106/00004623-200300003-00008. [DOI] [PubMed] [Google Scholar]
- Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Current Opinion in Genetics and Development. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Herr MJ, Horrigan SK, Namciu S, Shardy D, Stavnezer E. Protooncogene c-ski is expressed in both proliferating and postmitotic neuronal populations. Dev.Dyn. 1994;201:354–365. doi: 10.1002/aja.1002010407. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGF-beta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen Y. Controlling TGF-beta signaling. Genes and Development. 2000;14:627–640. [PubMed] [Google Scholar]
- Massague J, Hata A, Liu F. TGF-beta signaling through the Smad pathway. Trends in Cell Biology. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- Mizuide M, Hara T, Furuya T, Takeda M, Kusanagi K, Inada Y, Mori M, Imamura T, Miyazawa K, Miyazono K. Two short segments of Smad3 are important for specific interaction of Smad3 with c-Ski and SnoN. Journal of Biological Chemistry. 2003;278:531–536. doi: 10.1074/jbc.C200596200. [DOI] [PubMed] [Google Scholar]
- Nagase T, Mizuguchi G, Nomura N, Ishizaki R, Ueno Y, Ishii S. Requirement of protein co-factor for the DNA-binding function of the human ski proto-oncogene product. Nucleic Acids Res. 1990;18:337–343. doi: 10.1093/nar/18.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, Ten Dijke P. Identification of Smad7, a TGF-beta-inducible antagonist of TGF-beta signaling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Gardner H, Berg LK, Christian JL. Smad6 functions as an intracellular antagonist of some TGF-beta family members during Xenopus embryogenesis. Genes Cells. 1998;3:387–394. doi: 10.1046/j.1365-2443.1998.00196.x. [DOI] [PubMed] [Google Scholar]
- Nomura T, Khan MM, Kaul SC, Dong HD, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes and Development. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateder DB, Ferguson CM, Ionescu AM, Schwarz EM, Rosier RN, Puzas JE, O'Keefe RJ. PTHrP expression in in chick sternal chondrocytes is regulated by TGF-á through Smad-mediated signaling. Journal of Cellular Physiology. 2001;188:343–351. doi: 10.1002/jcp.1118. [DOI] [PubMed] [Google Scholar]
- Scharstuhl A, Diepens R, Lensen J, Vitters E, van Beuningen H, van der KP, van den BW. Adenoviral overexpression of Smad-7 and Smad-6 differentially regulates TGF-beta-mediated chondrocyte proliferation and proteoglycan synthesis. Osteoarthritis.Cartilage. 2003;11:773–782. doi: 10.1016/s1063-4584(03)00165-1. [DOI] [PubMed] [Google Scholar]
- Semonin O, Fontaine K, Daviaud C, Ayuso C, Lucotte G. Identification of three novel mutations of the noggin gene in patients with fibrodysplasia ossificans progressiva. Am.J.Med.Genet. 2001;102:314–317. doi: 10.1002/ajmg.1504. [DOI] [PubMed] [Google Scholar]
- Serra R, Johnson M, Filvaroff EH, LaBorde J, Sheehan DM, Derynck R, Moses HL. Expression of truncated. kinase-defective TGF-beta type II receptor in mouse skeletal muscle promotes terminal chondrocyte differentiation and osteoarthritis. Journal of Cell Biology. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O'Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. Journal of Biological Chemistry. 2006a;281:3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Wang X, Drissi H, Liu F, O'Keefe RJ, Chen D. Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. Journal of Biological Chemistry. 2006b;281:16347–16353. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shinagawa T, Nomura T, Colmenares C, Ohira M, Nakagawara A, Ishii S. Increased susceptibility to tumorigenesis of ski-deficient heterozygous mice. Oncogene. 2001;20:8100–8108. doi: 10.1038/sj.onc.1204987. [DOI] [PubMed] [Google Scholar]
- Stavnezer E, Barkas AE, Brennan LA, Brodeur D, Li Y. Transforming Sloan-Kettering viruses generated from the cloned v-ski oncogene by in vitro and in vivo recombinations. J.Virol. 1986;57:1073–1083. doi: 10.1128/jvi.57.3.1073-1083.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu X, Eaton EN, Lane WS, lodish HF, Weinberg RA. Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling. Molecular Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- Sutrave P, Kelly AM, Hughes SH. ski can cause selective growth of skeletal muscle in transgenic mice. Genes and Development. 1990;4:1462–1472. doi: 10.1101/gad.4.9.1462. [DOI] [PubMed] [Google Scholar]
- Takeda M, Mizuide M, Oka M, Watabe T, Inoue H, Suzuki H, Fujita T, Imamura T, Miyazono K, Miyazawa K. Interaction with Smad4 is indispensable for suppression of BMP signaling by c-Ski. Molecular Biology of the Cell. 2004;15:963–972. doi: 10.1091/mbc.E03-07-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mariani FV, Harland RM, Luo K. Ski represses bone morphogenic protein signaling in Xenopus and mammalian cells. Proc.Natl.Acad.Sci.U.S.A. 2000;97:14394–14399. doi: 10.1073/pnas.97.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell. 2002;111:357–367. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- Wu Q, Kim KO, Sampson ER, Chen D, Awad H, O'Brien T, Puzas JE, Drissi H, Schwarz EM, O'Keefe RJ, Zuscik MJ, Rosier RN. Induction of an osteoarthritis-like phenotype and degradation of phosphorylated Smad3 by Smurf2 in transgenic mice. Arthritis and Rheumatism. 2008;58:3132–3144. doi: 10.1002/art.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. Journal of Cell Biology. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Schwarz EM, Rosier RN, Zuscik MJ, Puzas JE, O'Keefe RJ. ALK2 functions as a BMP type I receptor and induces Indian hedgehog in chondrocytes during skeletal development. Journal of Bone and Mineral Research. 2003;18:1593–1604. doi: 10.1359/jbmr.2003.18.9.1593. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehlin DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proceedings of the National Academy of Sciences USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A Smad ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]