ABSTRACT
The increasing antibiotic resistance of pathogenic Escherichia coli species and the absence of a pan-protective vaccine pose major health concerns. We recently identified, by subtractive reverse vaccinology, nine Escherichia coli antigens that protect mice from sepsis. In this study, we characterized one of them, ECOK1_0290, named FdeC (factor adherence E. coli) for its ability to mediate E. coli adhesion to mammalian cells and extracellular matrix. This adhesive propensity was consistent with the X-ray structure of one of the FdeC domains that shows a striking structural homology to Yersinia pseudotuberculosis invasin and enteropathogenic E. coli intimin. Confocal imaging analysis revealed that expression of FdeC on the bacterial surface is triggered by interaction of E. coli with host cells. This phenotype was also observed in bladder tissue sections derived from mice infected with an extraintestinal strain. Indeed, we observed that FdeC contributes to colonization of the bladder and kidney, with the wild-type strain outcompeting the fdeC mutant in cochallenge experiments. Finally, intranasal mucosal immunization with recombinant FdeC significantly reduced kidney colonization in mice challenged transurethrally with uropathogenic E. coli, supporting a role for FdeC in urinary tract infections.
IMPORTANCE
Pathogenic Escherichia coli strains are involved in a diverse spectrum of diseases, including intestinal and extraintestinal infections (urinary tract infections and sepsis). The absence of a broadly protective vaccine against all these E. coli strains is a major problem for modern society due to high costs to health care systems. Here, we describe the structural and functional properties of a recently reported protective antigen, named FdeC, and elucidated its putative role during extraintestinal pathogenic E. coli infection by using both in vitro and in vivo infection models. The conservation of FdeC among strains of different E. coli pathotypes highlights its potential as a component of a broadly protective vaccine against extraintestinal and intestinal E. coli infections.
Introduction
Escherichia coli strains are versatile microorganisms that constantly acquire and lose virulence attributes, leading to the emergence of successful new genetic combinations that can confer an increased ability to colonize new niches and to cause a broad spectrum of intestinal and extraintestinal diseases (1). Strains with successful combinations of virulence factors and that cause similar diseases have become pathotypes. Intestinal E. coli pathotypes appear to be unable to persist in the human intestine and cause diarrheal diseases only when ingested in sufficient quantities by a naive host. On the other hand, extraintestinal pathogenic E. coli (ExPEC), while not inducing enteric disease, can asymptomatically colonize the human intestinal tract as the predominant species in ~20% of healthy individuals (2, 3). Extraintestinal infections resulting from these strains, however, include neonatal meningitis, urinary tract infections (UTIs), diverse intra-abdominal infections, pneumonia, intravascular-device infections, osteomyelitis, soft tissue infections, bacteremia, and sepsis (4). In particular, UTIs, which can be either asymptomatic or symptomatic, are characterized by a wide spectrum of manifestations ranging from mild dysuria to bacteremia, sepsis, or even death (5). Uncomplicated UTI is confined to the bladder, while severely complicated UTIs include pyelonephritis and urosepsis. Recurrent UTIs occur as result of reinfection by bacteria from outside the urinary tract or from persistent bacteria (6). Virulence factors most commonly associated with uropathogenic E. coli (UPEC) include adhesive fimbriae, iron acquisition systems, and toxins such as hemolysin and cytotoxic necrotizing factor (7). After bacterial attachment, UPEC may invade epithelial cells and form small clusters of intracellular bacteria, termed intracellular bacterial communities (IBCs) (8). Bacteria may persist in these protective niches, creating a chronic quiescent reservoir in the bladder.
UPEC strains contribute significantly to the burden of ExPEC-associated diseases, being the causative agent in 70% to 95% of community-acquired UTIs and 50% of all cases of nosocomial infections (9). Due to the emergence of an increasing number of antibiotic-resistant strains, the development of an efficacious ExPEC vaccine would have both a significant impact on public health and great economic benefit. Recently, we determined the genome sequence of an ExPEC K1 strain, IHE3034 (ST95), isolated from a case of neonatal meningitis, and compared it to the available genome sequences of other ExPEC strains and a few nonpathogenic E. coli strains (10). By a subtractive reverse vaccinology approach, nine antigens were identified and demonstrated to be protective in a mouse model of sepsis. Their conservation in other E. coli pathotypes indicated their potential as candidates for a universal E. coli vaccine (10). In this report, we describe the structural and functional properties of one of these protective antigens, ECOK1_0290, renamed FdeC (for factor adherence E. coli). In this analysis, we elucidated its putative role during ExPEC pathogenesis by using both in vitro and in vivo infection models. Our findings, corroborated by epidemiological data on antigen conservation in other pathotypes, strongly support the importance of FdeC in E. coli colonization of host tissues and the relevance of the protein as a vaccine target.
RESULTS
Genomic characterization, distribution, and conservation of the fdeC gene.
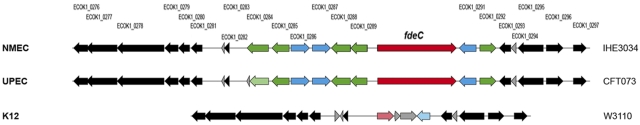
FdeC, previously annotated in the ExPEC strain IHE3034 as bacterial immunoglobulin-like domain (group 1) protein (ECOK1_0290) (10), is a 1,416-amino-acid (aa) protein that has low sequence similarity with Yersinia pseudotuberculosis invasin (11) and enteropathogenic E. coli (EPEC) intimin (12) (conserved up to 35% over selected regions). In addition, FdeC shares 95% identity with EaeH, a putative adhesin identified by subtractive hybridization from the genome sequence of the enterotoxigenic E. coli (ETEC) strain H10407 (13). A closer examination of the region encompassing the fdeC gene showed that in ExPEC strains (Fig. 1) and other E. coli pathotypes (see Fig. S1 in the supplemental material) the gene resides in a locus containing up to three putative regulatory genes and five putative reductases and hydrolases. In the nonpathogenic K-12 strains, the fdeC gene is disrupted and is present as a shorter pseudogene, corresponding only to the N-terminal region of the full-length protein (Fig. 1). To assess the distribution of the fdeC gene among different E. coli isolates, we evaluated the prevalence of the gene in 128 sequenced E. coli genomes available in public databases and performed PCR amplification of the gene in a collection of 143 E. coli isolates, including distinct pathotypes of human and animal origin. The fdeC gene was found to be highly distributed among all strains with an overall presence of approximately 99% in ExPEC isolates and 92 to 100% in intestinal E. coli pathotypes (Table 1). When present, the gene is highly conserved with >91% identity at the amino acid sequence level among these strains.
FIG 1 .
Comparative genome analysis of the fdeC cluster. Graphical representation of the fdeC cluster, containing the fdeC gene (red), putative transcriptional regulators (blue), putative reductases and hydrolases (green), insertion sequences, and variable genes (gray). Regions highly conserved among all the strains are considered core regions (black). Truncated genes are represented in light colors. Linear visualization of the fdeC cluster was generated by a noncommercial software program for research purposes, GECO (36). NMEC, neonatal meningitis-associated E. coli.
TABLE 1 .
fdeC gene distribution analysis
| Pathotypea | No. positive | Total no. | Prevalence, % |
|---|---|---|---|
| ExPEC | 116 | 117 | 99.1 |
| ETEC | 13 | 14 | 92.9 |
| EHEC | 34 | 38 | 89.5 |
| EPEC | 5 | 5 | 100.0 |
| AIEC | 3 | 3 | 100.0 |
| EAEC | 3 | 3 | 100.0 |
| STEC | 6 | 6 | 100.0 |
| Other | 53 | 55 | 96.4 |
| Commensal/fecal | 19 | 23 | 82.6 |
| Nonpathogenic | 2 | 7 | 28.6 |
The fdeC gene presence was evaluated in 128 sequenced genomes (based on >90% identity) and in 143 clinical isolates by PCR amplification. ExPEC, avian pathogenic E. coli; UPEC, uropathogenic E. coli; NMEC, neonatal meningitis-associated E. coli; SEPEC, septicemic E. coli; ETEC, enterotoxigenic E. coli; EHEC, enterohemorrhagic E. coli; EPEC, enteropathogenic E. coli; AIEC, adherent-invasive E. coli; EAEC, enteroaggregative E. coli; STEC, Shiga toxin-producing E. coli; other, unknown pathotypes; commensal/fecal, E. coli isolated from healthy individuals; nonpathogenic, E. coli lab strains.
Sequence organization and crystal structure of FdeC.
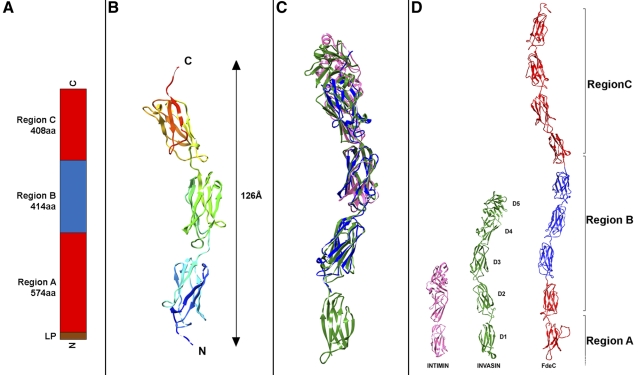
Sequence analysis revealed that FdeC is predicted to contain a number of bacterial Ig-like domains. The N-terminal region is identified as a domain of unknown function (DUF3442) and is likely to form a β-barrel multidomain structure. Considering this prediction and the inability to purify soluble full-length recombinant protein, FdeC was divided into three regions: region A, predicted as the transmembrane domain, and regions B and C, predicted to contain the extracellular Ig-like domains (Fig. 2A). While no expression was obtained for region A alone, we successfully purified soluble recombinant polypeptides for the following fragments: AB (FdeCAB), B (FdeCB), and C (FdeCC). Fragment C, however, was unstable and rapidly degraded postpurification.
FIG 2 .
FdeC regions and crystal structure. (A) Schematic of the regional organization of FdeC. (B) Ribbon diagram of FdeCB (amino acids 678 to 991); N and C termini are indicated. (C) Superimposition of FdeCB, in blue, with intimin (PDB_ID 1F00), in purple, and invasin (PDB_ID 1CMV), in green. (D) Structural comparison between the crystal structures of the surface-exposed domains of intimin and invasin with a model of the potential surface-exposed region of FdeC. Modeled regions are colored in red, while the crystal structure of FdeCB is colored in blue.
Among these fragments, we solved the crystal structure of region B (residues 597 to 1008), previously reported to be responsible for the antigenic properties of the protein (10). The FdeCB crystal structure was solved at 1.9-Å resolution. Electron density could be identified only for residues 678 to 991 of the structure, and crystal packing showed no space available to contain the first 81 residues of the construct, leading us to conclude that these residues were lost during crystallization via a proteolytic or degradation event. The structure of residues 678 to 991 revealed an elongated molecule of 126 Å in length and the presence of three bacterial Ig-like domains, each of 120 amino acids in length (Fig. 2B), similar to invasin (14). Adopting the domain nomenclature of Hamburger et al. (14), numbering the invasin domains D1 to D5 from the N to the C termini, the comparison with invasin (Protein Data Bank identification [PDB ID] 1CWV) showed that 241 out of the 319 Cα atoms in the FdeCB fragment aligned with invasin (PDB_ID 1CWV) with a root mean square deviation (RMSD) of 2.67 Å. This corresponds to alignment with domains D2 to D4 of invasin (Fig. 2C and D). Enteropathogenic E. coli intimin (PDB_ID 1F00) aligns with the FdeCB structure for 139 out of the 282 Cα atoms with an RMSD of 2.9 Å (Fig. 2C and D). In the C-terminal portion of both invasin and intimin, a domain D5, which is a C-type lectin-like moiety that is closely connected to the D4 domain, is present. In both intimin and invasin, this domain plays an important role in bacterial binding to their respective receptors. Sequence analysis and modeling of the FdeC structure revealed no evidence of a C-type lectin domain (Fig. 2D).
Recombinant FdeCAB binds to human epithelial cells.
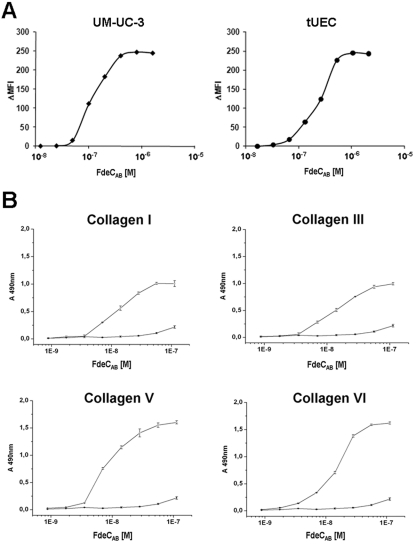
Based on the presence of structural features typical of bacterial surface determinants interacting with host cells, we evaluated the intrinsic capacity of FdeC to bind to human bladder (UM-UC-3) and urethral epithelial (tUEC) cells. We used an antibody-based indirect immunofluorescence detection system supported by fluorescence-activated cell sorting (FACS) analysis (see Materials and Methods) to compare the adhesive properties of the two soluble fragments obtained, FdeCAB and FdeCB. Recombinant FdeCAB bound to both UM-UC-3 and tUEC cells (Fig. 3A), reaching a plateau at a concentration of ≈10−6 M. The binding constant (Kd) value was calculated as the FdeCAB concentration resulting in saturation of 50% of the putative receptors present on the cells and was estimated to be on the order of ≈10−7 M. Recombinant FdeCAB also bound to ovary (CHO), cervix (HeLa), and kidney (Vero) epithelial cell lines (see Fig. S2 in the supplemental material). In contrast, FdeCB did not bind to human cells (data not shown), suggesting the importance of the A region in maintaining the molecule in a functional conformation. Epithelial cells produce a specific array of extracellular matrix (ECM) proteins that have a fundamental role in maintaining tissue integrity. To investigate the propensity of FdeCAB to also interact with selected ECM components, we performed in vitro binding assays using fibrinogen, fibronectin, laminin, and various types of collagen as target molecules. Binding was detected using polyclonal anti-FdeC antiserum and quantified by enzyme-linked immunoabsorbent assay (ELISA). Recombinant FdeCAB showed a pronounced dose-dependent binding to collagen types I, III, V, and VI, with an estimated Kd on the order of ≈10−8 M (Fig. 3B). No binding was observed to other ECM compounds, such as fibrinogen, fibronectin, laminin, and collagen IV (data not shown).
FIG 3 .
Binding of recombinant FdeC to epithelial cells and ECM. (A) UM-UC-3 and tUEC cells were incubated for 1 h at 4°C with increasing concentrations of recombinant FdeCAB. Binding of FdeCAB to cells was assessed by flow cytometry, and data are shown as mean fluorescence intensity (MFI) values plotted in the saturation curves. (B) Binding of increasing concentrations of FdeCAB to collagen I, III, V, and VI (open circles). BSA is used as a negative control (solid circles). Binding was quantified by ELISA. Points represent the means (error bars show standard errors of the means) of measurements made in triplicate.
Expression of FdeC is induced by interaction with host cells.
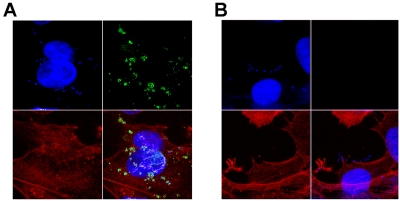
Since several attempts to reproduce urinary tract or intestinal conditions in vitro did not result in native FdeC expression on the bacterial surface (see the supplemental material), we hypothesized that expression of FdeC may be triggered upon contact with host cells. Using confocal immunofluorescence microscopy, we examined the expression of FdeC in ExPEC strain IHE3034 (Fig. 4A) following contact with human uroepithelial cells. We observed that FdeC was expressed by the bacterial population that was tightly associated with the plasma membrane of the mammalian cell, as visualized by F-actin staining. Similar results were obtained using the uropathogenic strain CFT073 (data not shown). No fluorescent signal was observed when infections were carried out using the fdeC deletion mutant strains (Fig. 4B). These data suggest that FdeC surface expression may depend on sensing or binding to host cells. To address the kinetics of FdeC expression during interaction with host cells, we performed infection experiments and compared the appearances of the FdeC signals on the bacterial surface at 15, 30, 45, 60, 90, 120, 180, and 240 min. Confocal imaging analysis of IHE3034, 536, and CFT073 strains in contact with uroepithelial cells revealed that the switching of FdeC expression did not occur earlier than 60 min from bacterial loading and that the proportion of bacteria expressing the protein remained constant up to 4 h (data not shown). FdeC deletion mutant strains were used as internal controls. Quantitative association assays were performed using IHE3034, 536, and CFT073 strains. Infection of UM-UC-3 cells at different time points (ranging from 15 min to 4 h) revealed no significant differences in the CFU counts between wild-type and respective isogenic fdeC mutant strains (data not shown). These data suggest that in this in vitro system, constitutively expressed adhesins mask the contribution of FdeC to bacterial adhesion.
FIG 4 .
Confocal analysis of FdeC expression in contact with cultured epithelial cells. Monolayers of UM-UC-3 cells were infected with IHE3034 (A) and the respective fdeC knockout mutant strain (B). At the end of the infection, the samples were fixed and stained for confocal immunofluorescence microscopy. FdeC was detected using specific antibodies and visualized using a fluorescent secondary antibody (green). DAPI was used to stain the host cell nuclei and visualize bacteria (blue). Cellular actin was stained with fluorescent phalloidin (red).
Constitutive expression of FdeC results in bacterial aggregation.
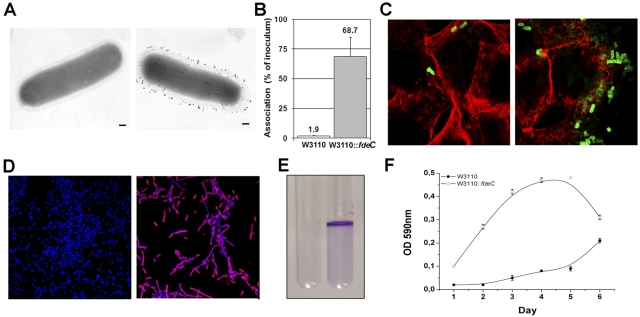
To better evaluate the contribution of FdeC to adhesion, we engineered the well-characterized, weakly adhesive E. coli K-12 strain W3110 (naturally devoid of the fdeC gene) for constitutive surface expression of FdeC (W3110::fdeC). Localization of the protein on the surface of the W3110::fdeC strain was confirmed by electron microscopy analysis (Fig. 5A). Assays of the adherence of W3110 wild-type and W3110::fdeC strains to human bladder UM-UC-3 cells, quantified using viable CFU counts (Fig. 5B) and by confocal microscopy (Fig. 5C), clearly showed the strong contribution of FdeC in the adherence of E. coli to epithelial cells. However, gentamicin protection assays reveal a low invasion of UM-UC-3 cells by the W3110::fdeC strain (data not shown). The use of E. coli K-12 and its derivatives as a model to assess bacterial attachment to solid surfaces has been established previously (15). By comparing the W3110 wild-type and W3110::fdeC strains, we observed a strong propensity of adhering bacteria to form large aggregates on glass slides (Fig. 5D). ExPEC species are known to use a number of surface-associated determinants to form in vivo macrocommunities, often defined as biofilms (16). In particular, several studies have indicated the important role of biofilms in UTIs, notably in catheterized patients (17, 18). Using a previously established protocol for determining the propensity of E. coli strains to form aggregates in vitro (19), we demonstrated that, compared to the wild-type control, the W3110::fdeC strain is able to strongly interact with solid surfaces and produce macrocomplexes when grown in polystyrene tubes (Fig. 5E). The capacity of the W3110::fdeC strain to form bacterial aggregates was monitored up to 6 days and quantified by crystal violet staining (Fig. 5F).
FIG 5 .
Constitutive surface exposure of FdeC in strain W3110::fdeC induces the formation of bacterial aggregates. (A) Immunogold transmission electron microscopy of FdeC surface localization in W3110 wild-type (left panel) and W3110::fdeC (right panel) strains. Fixed bacteria were incubated with an anti-FdeC serum and then with the secondary antibodies labeled with 10-nm gold particles. Bars, 200 nm. (B) Association rate of W3110 and W3110::fdeC with UM-UC-3 cells. The W3110 (white bar) and the W3110::fdeC (gray bar) strains were used to infect UM-UC-3 monolayers for 3 h at a multiplicity of infection of 10:1. Viable cell-associated bacteria were quantified after cell lysis. Results are expressed as a percentage of the inoculum. The data displayed illustrate the results of three independent experiments, including standard deviations. (C) Confocal microscopy analysis of UM-UC-3 cells infected with W3110 wild-type (left panel) and W3110::fdeC (right panel) strains. The W3110 strain and its derivative were detected using a polyclonal rabbit anti-E. coli K-12 antibody (green fluorescence). Actin was stained in red using phalloidin. (D) Confocal images of W3110 wild-type (left panel) and W3110::fdeC (right panel) strains adhering to glass slides reveal the propensity of the FdeC-expressing strain to form aggregates. Bacteria were localized with DAPI (blue), and FdeC was detected using the anti-FdeC serum and a fluorescent secondary antibody (red). (E) In vitro microcolony formation by W3110 (left) and W3110::fdeC (right) strains grown in LB medium at RT in polystyrene tubes. The macroscopic aggregates were visualized by crystal violet staining. (F) Kinetics of microcomplex formation at up to 6 days. Quantitative crystal violet staining was used to measure the level of the aggregation for W3110::fdeC in comparison with the W3110 wild-type strain. Data represent means and standard deviations of three independent measurements.
FdeC contributes to colonization of the urinary tract.
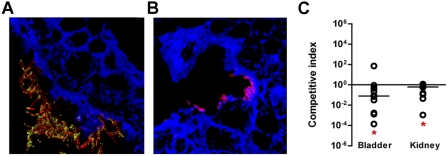
In order to determine if FdeC plays a role in virulence, we performed an independent challenge experiment in mice infected with wild-type E. coli 536 and its isogenic fdeC mutant strain. As shown in Fig. 6A, confocal immunofluorescence microscopy of paraffin-embedded bladder sections revealed that FdeC is specifically expressed by wild-type bacteria closely associated with the bladder tissue and that the number of bacteria was significantly higher than that in tissue derived from mice inoculated with the fdeC mutant strain (Fig. 6B). Imaging data were representative of a qualitative imaging analysis of 10 bladder sections that collectively revealed reduced bladder colonization by the mutant strain. To evaluate the contribution of FdeC to bacterial fitness in vivo during colonization of the urinary tract, female CBA/J mice were transurethrally inoculated with a 1:1 ratio of E. coli 536 and the fdeC, kanamycin-resistant, knockout mutant. At 48 h postinoculation, bacteria from tissue homogenates were prepared in the presence of 0.1% saponin to separate eventual aggregates. Live bacteria were enumerated by viable counts. Bacterial fitness was determined as competitive index (CI) for cochallenge data. We observed that deletion of fdeC from strain 536 caused a significant reduction in fitness during UTI in both the bladder and the kidney (Fig. 6C). The significant difference between the mutant and wild-type colonization levels indicated that FdeC may have an important role in E. coli fitness during an ascending urinary tract infection.
FIG 6 .
Role of FdeC in vivo in a model of ascending UTI. Confocal imaging analysis of FdeC expression in bladder from mice independently infected with UPEC wild-type strain 536 (A) and the isogenic fdeC mutant strain (B). Tissue sections embedded in paraffin were stained with WGA conjugated with Alexa Fluor 647 (blue), while UPEC strain 536 was detected using a polyclonal serum raised against inactivated whole bacteria (red). FdeC was stained in green. The images are representative of multiple observations, from at least 10 sections. (C) Cochallenge experiments using the 536 wild-type strain and the fdeC isogenic knockout mutant. In vivo competitive indices (CI) were determined following cochallenge infections of female CBA/J mice with a 1:1 ratio of wild-type 536 and fdeC knockout mutant strains. At 48 h postinoculation, bacteria were enumerated by plating serial dilutions from tissue homogenates. The CI from each tissue was determined by dividing the mutant-to-wild-type ratio of CFU/g by the mutant-to-wild-type ratio of CFU from the mixed inoculum. Each dot represents bladders and kidneys from an individual animal. Bars indicate the median CI. Significant differences in colonization (*) (P values, <0.05) were determined by the Wilcoxon signed rank test. A CI of <1 indicates a fitness defect.
Vaccination with recombinant FdeC confers protection against ExPEC during experimental UTI.
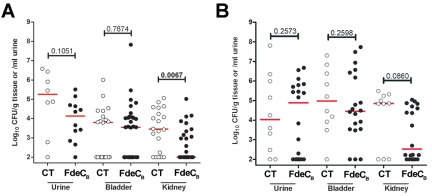
Since FdeC (ECOK1_0290) was among the nine antigens shown to be protective in a mouse model of sepsis (10) and shown here to contribute to virulence, we decided to test whether the antigen could also induce protection in the murine model of ascending UTI. Towards this end, FdeCB was mixed with the adjuvant cholera toxin (CT) at a ratio of 10:1 (antigen to CT) and groups of mice (n = 30) were intranasally inoculated with either the antigen-CT mixture or CT alone. Following primary immunization (day 0) and booster doses (days 7 and 14), animals were transurethrally challenged with the UPEC strain 536 and protection was assessed at 48 h postinfection by determining CFU in the urine, bladder, and kidneys. FdeCB induced significant protection in the kidney with a 1.5-log reduction in median CFU/g (P value = 0.0067) (Fig. 7A). In addition, 17/30 vaccinated mice had undetectable levels of bacteria in the kidney (≤102 CFU/g tissue). We also observed a strong trend in reduction of kidney colonization after challenge with the UPEC strain CFT073, with a 2.5-log reduction in median CFU/g (Fig. 7B). In this case, 50% of mice had undetectable levels of bacteria (≤102 CFU/g tissue) in the kidney. This demonstrated that mucosal immunization in the nares generates a protective effect at distal sites. These data strongly support the use of FdeCB as a component in a vaccine able to prevent all ExPEC-associated diseases.
FIG 7 .
FdeCB prevents the spread of UPEC infection into the kidney. Mice were immunized intranasally with a 10:1 ratio of FdeCB to CT containing 100 µg of FdeCB or with 10 µg CT alone (mock immunized). After two boosts of 25 µg antigen (10:1 ratio of antigen to CT) or CT alone given on days 7 and 14, the individual mice were transurethrally challenged with 108 CFU of UPEC strain 536 (A) and CFT073 (B). After 48 h, bladder and kidneys were harvested and homogenized. Bacteria in urine and in the tissue homogenates were enumerated by plating serial dilutions. Symbols represent CFU/g tissue or CFU/ml urine of individual mice, and bars indicate median values. P values were determined using the nonparametric Mann-Whitney significance test.
DISCUSSION
The development of an efficacious, broadly cross-protective vaccine against most E. coli pathotypes would have significant public health and economic benefits, considering the increasing antibiotic resistance among E. coli strains and the associated mortality, morbidity, and lost productivity. We have recently compared available genome sequences of different E. coli strains, and by using a subtractive reverse vaccinology approach, we identified a number of antigens common to different pathotypes, capable of inducing protection in a mouse model of sepsis, thus proposing their use for the development of a universal E. coli vaccine (10). Bioinformatic analysis of one of these antigen candidates, here named FdeC, revealed an interesting structural similarity with invasin and intimin, bacterial virulence factors known to bind the host cells. In particular, the FdeC region B (FdeCB) is predicted to have the same arrangement of four bacterial immunoglobulin folds as does the Y. pseudotuberculosis invasin structure. The X-ray structure of FdeCB confirmed this structural homology, with the truncated form of FdeCB (678 to 991 amino acids [aa]) sharing both the immunoglobulin fold arrangement and the quaternary structure of invasin (14).
Since the fdeC gene is present in different E. coli pathotypes and has structural similarity to other pathogenic adhesion molecules, we hypothesize that FdeC could contribute to virulence, as an adhesin essential for infection in the host. In UPEC strains, for instance, adhesion is vitally important to overcome host defenses in the urinary tract and resist the flushing effect of the urine. The role of FdeC in E. coli colonization of host tissues was supported not only by in vivo evidence showing decreased bacterial fitness of the fdeC mutant strain during experimental urinary tract infection but also by the fact that the recombinant FdeC binds with strong affinity to several epithelial cell lines and that its surface expression on the bacteria is host cell contact dependent. In addition, its capacity to bind different collagen types, including type V and VI (both widely expressed in the interstitial space of kidney and bladder), strongly supports the hypothesis for a specific role of FdeC during bacterial colonization.
Genome comparison of the loci containing the fdeC gene in different E. coli strains revealed that it is present as a pseudogene in the nonpathogenic K-12 E. coli strains, such as MG1655, W3110, and DH10B. However, in human commensal E. coli isolates, HS and SE11, and in many fecal strains, the full-length fdeC is present, indicating that the protein might be functional and provide a general advantage during host colonization. For example, E. coli HS has been shown to stably colonize the human gastrointestinal tract but cause no detectable disease or adverse effects (20). On the other hand, strain SE11, a commensal E. coli isolate from the feces of a healthy human adult, evolved to acquire and accumulate the functions advantageous for stable colonization of intestinal cells (21). It possesses more genes involved in adhesion than does the K-12 strain MG1655, which is no longer able to colonize the human gut.
In general, the biogenesis of bacterial adhesive structures is a tightly controlled process. The cost in energy and other resources required for expression of an adhesin must be balanced with any potential benefits that a particular adhesive organelle might provide to a bacterium. Variable expression of different adhesive organelles, such as fimbriae, may allow the bacterium to alter its binding characteristics in response to environmental changes encountered within a host during the course of an infection (22). In this context, although FdeC may share functional homology with other adhesins localized on the E. coli surface, its host cell-dependent expression suggests a specific role during colonization. Nevertheless, we also observed that in vitro constitutive expression of the protein is associated with an increased aggregative phenotype. We therefore suggest that the expression of FdeC at the site of colonization may contribute to the formation of macrocolonies, a phenotype often associated with E. coli infections. This is in agreement with our observation that fdeC mutant bacteria are attenuated for bladder colonization during cochallenge experiments, confirming that FdeC is a virulence factor in mice. In fact, in vivo competition experiments between wild-type uropathogenic E. coli and its fdeC mutant strain revealed that the loss of FdeC caused a significant fitness defect during extraintestinal colonization.
We recently described FdeC (ECOK1_0290) as a promising vaccine candidate against ExPEC infections (10). In vivo protection was evaluated in mice by subcutaneous injection of the purified recombinant antigen followed by intraperitoneal challenge with the E. coli pathogenic strain IHE3034. Using this model, we found a good correlation between mortality and the bacterial counts in the blood, with a significant, although rather low, protective efficacy compared to the control group. Based on the functional properties of FdeC, we hypothesized that the antigen may exert a more efficacious protective role in an experimental UTI model. Indeed, we found that intranasal immunization with FdeC provided considerable protection against experimental infection with two different UPEC strains. Of interest, site-specific protection was observed, as mice immunized with the FdeC protein were significantly protected from strain 536 colonization only in the kidney, with a >1.5-log reduction in the median CFU/g tissue and more than 50% of mice having undetectable levels of bacteria in that organ. Surprisingly, despite the significant protection in the kidney, immunization with the FdeC protein did not reduce the level of bacteria able to colonize the bladder.
The evidence reported in this study indicates that functional characterization of potential vaccine candidates, and determination of their role in pathogenesis, may be pivotal to understanding the basis for inducing protective immunity during specific stages of disease and support the development of more efficacious vaccines.
MATERIALS AND METHODS
Ethics statement.
All procedures were conducted according to protocol 08999 approved by the University Committee on the Care and Use of Animals at the University of Michigan Medical School. The approved procedures are in compliance with university guidelines, state and federal regulations, and the standards of the Guide for the Care and Use of Laboratory Animals.
Bacterial strains and culture conditions.
ExPEC strain IHE3034 (serotype O18: K1:H7), isolated in Finland in 1976 from a case of human neonatal meningitis (23); RS218 (serotype O18:K1:H7); 536 (serotype O6:K15:H31) (24); CFT073 (serotype O6:K2:H1) (25); MG1655 (K-12) (26); and W3110 (K-12) (27) were used for comparative genome analysis. Bacteria were cultured in Luria-Bertani (LB) liquid medium or agar plates at 37°C. E. coli DH5α was used for cloning purposes, and E. coli BL21(λDE3) was used for expression of His-tagged fusion proteins. The clones carrying a specific cassette conferring antibiotic resistance were grown in the presence of ampicillin (100 µg/ml) or kanamycin (50 µg/ml).
Crystallization and data collection.
Crystallization experiments were performed in Greiner 96-well low-profile sitting-drop plates. Initial screen conditions consisted of a sparse matrix of 480 conditions set up at 4°C and 20°C (28, 29). The reservoir volume was 50 µl, and the drop consisted of 200 nl protein with an equal volume of precipitant. Promising protein crystals took over 4 months to grow under a condition containing 20% polyethylene glycol 6000 (PEG 6000) in 0.1 M Bicine buffer at pH 9.0. Crystals were looped and chilled at liquid nitrogen temperature. Cryoprotection was achieved via the addition of 20% glycerol solution to the mother liquor prior to harvesting the crystal. Data were collected at the Advanced Light Source (ALS; Berkeley) on beamline 5.0.2. Data were reduced using HKL2000 and the CCP4 suite (30). Data reduction statistics are presented in Table 2.
TABLE 2 .
Crystallographic and refinement statistics
| Parameter | FdeC |
|---|---|
| Protein | |
| Space group | P21212 |
| No. of molecules in ASUd | 1 |
| Unit cell (Å) | 44.6, 150.93, 47.40 |
| Wavelength (Å) | 0.9202 |
| Resolution (Å) | 50.0-1.9 |
| Total no. of unique reflections | 24,734 |
| Completeness, % (highest shell) | 95.6 (67.0) |
| R merge, % (highest shell) | 0.084 (0.634) |
| Highest-resolution shell, Å | 1.97–1.90 |
| Mean I/σ (I) (highest shell) | 23.9 (1.7) |
| Refinement | |
| No. of references, working set | 23,413 |
| No. of references, test set | 1,265 |
| Rcryst (Rfreea)b (%) | 17.1 (20.5) |
| RMSD bonds (Å) | 0.017 |
| RMSD angles (°) | 1.44 |
| Avg B (Å)2 | 26.27 |
| ESUe based on Rfree (Å)c | 0.135 |
Rfree = as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
R factor = Σ|Ii − <Ii>| |/Σ|Ii|, where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Estimated overall coordinate error.
ASU, asymmetric unit.
ESU, estimated standard uncertainty.
Structure solution and refinement.
The structure was solved by molecular replacement using the program Phaser (31) and an ensemble of individual immunoglobulin domains from the homologues invasin and intimin (PDB IDs 1CWV and 1F00, respectively) as probe molecules with all data to 2.5 Å. Full-length probes of invasin and intimin failed to produce expectable solutions, in initial runs. In all the positions and orientations of three immunoglobulin domains found via this method, packing considerations made it clear that the fourth domain, believed to be in the construct, was missing in the crystal structure, which contained only 36% solvent with the 3 domains fitted. Iterative rounds of building and refinement with Buster (32) and Coot (33) produced a structure whose refinement converged with excellent geometry (data and refinement statistics are shown in Table 2). Superposition with structural homologues was performed with Coot and SSM (33, 34). All crystallographic manipulations other than those stated used the CCP4 package (30).
Binding assay using purified protein.
Approximately 2 × 105 cells were incubated with increasing concentrations of recombinant protein for 1 h at 4°C. Cells were then incubated with rabbit polyclonal anti-FdeC serum. Subsequently, cells were incubated with a goat anti-rabbit IgG fluorescein isothiocyanate (FITC)-conjugated antibody (Jackson ImmunoResearch Laboratories). FACS acquisition was performed using a fluorescence-activated cell sorter (Becton Dickinson). Data were analyzed using FlowJo software.
Confocal microscopy analysis of E. coli strains adhering to eukaryotic cells and tissues.
Epithelial cells were grown to confluence and infected with E. coli strains at a multiplicity of infection of 10:1 for different time points. Samples were washed and fixed in 3% paraformaldehyde (PFA) for 15 min at room temperature (RT). After 2 h of blocking in 1% bovine serum albumin (BSA), the rabbit anti-FdeC serum was used to detect the protein, using a donkey anti-rabbit IgG Rhodamine Red X-conjugated antibody as secondary antibody. Alexa Fluor dye-labeled phalloidins (Invitrogen) were used to stain the actin cytoskeleton according to the manufacturer’s instructions. Bacterial and cellular DNA were stained with 4′,6-diamidino-2-phenylindole (DAPI). The ProLong Gold reagent (Invitrogen) was used as a liquid mountant. The W3110 strain was detected using the polyclonal rabbit anti-E. coli K-12 (Dako) and the Alexa-Fluor 488 goat anti-rabbit IgG secondary antibody. The analysis was done with a Zeiss LSM 710 laser scanning microscope. Confocal imaging on tissues was performed on bladder sections derived from ExPEC-infected animals. In detail, the bladder was cut transversely, preserved in 10% formalin (pH 7.2), and embedded in paraffin. Tissue sections were cut and mounted onto slides. Samples were dewaxed and subjected to antigen retrieval before the immunofluorescence labeling. Staining of the sections was done as described for in vitro staining, except for tissues that were labeled with wheat germ agglutinin (WGA) (Alexa Fluor 647 conjugated; Invitrogen).
Vaccination in a murine model of ascending UTI.
Female CBA/J mice, 6 to 8 weeks old, were transurethrally inoculated as previously described (35). Purified antigen was mixed with cholera toxin (CT) (Sigma) at a ratio of 10:1. The vaccine was administered intranasally in a total volume of 20 µl/animal (10 µl/nare). Animals received a primary dose on day 0 of 100 µg antigen (containing 10 µg CT) or 10 µg CT alone. Two boosts of 25 µg antigen (mixed with 2.5 µg CT) or 2.5 µg CT alone were given on days 7 and 14, and mice were challenged on day 21. E. coli CFT073 and 536 suspensions in phosphate-buffered saline (PBS) (50 µl/mouse) were delivered transurethrally using a sterile 0.28-mm-inner-diameter polyethylene catheter connected to an infusion pump (Harvard Apparatus), with a total inoculum of 108 CFU/mouse. For determination of CFU, organs were aseptically removed from euthanized animals at 48 h postinoculation and homogenized in PBS with a GLH homogenizer (Omni International). Bacteria in tissue homogenates were enumerated by being plated on LB agar containing 0.5 g/liter NaCl using an Autoplate 4000 spiral plater (Spiral Biotech), and CFU were determined using a QCount automated plate counter (Spiral Biotech). Blood was collected as necessary from anesthetized mice by an infraorbital bleed using 1.1- to 1.2-mm Micro-Hematocrit capillary tubes (Fisher), and serum was separated using Microtainer serum separator tubes (Becton Dickinson). The animals were ≤15 weeks old at the conclusion of all experiments.
Protein structure accession number.
The structure has been deposited in the structure database of the Genomics Institute of the Novartis Research Foundation (San Diego, CA), and entry code in the Protein Data Bank (PDB) is 4E9L.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB.
FdeC is not expressed under experimental in vitro conditions. Download Text S2, DOCX file, 0.1 MB.
Region encompassing the fdeC gene in non-ExPEC pathotype strains. Download Figure S1, TIF file, 5.5 MB.
Representative flow cytometry profiles of the binding of 100 µg/ml of FdeCAB to CHO, HeLa, and Vero cell lines. Filled gray histograms represent the binding to control cells incubated with the anti-FdeC antiserum followed by incubation with the fluorescently conjugated secondary antibody. Download Figure S2, DOCX file, 1.4 MB.
ACKNOWLEDGMENTS
This work was supported by internal funding from Novartis Vaccines and Diagnostics.
We are grateful to Marina Cerquetti (Istituto Superiore di Sanità, Rome) for kindly providing ExPEC clinical isolates and to Lothar H. Wieler (Freie Universität, Berlin), Ulrich Vogel (Universität Würzburg), and Karin Schnetz (Universität Köln) for providing other E. coli isolates. We thank Angela Spagnuolo for technical assistance in the septic mouse model, Anna Rita Taddei for contributing to the electron microscopy studies, and Paolo Ruggiero and Laura Pancotto for assistance in preparation of sections from tissue samples. We are indebted with gratitude to Kate Seib for critical reading of the manuscript.
The crystallization experiments were conducted at beamline 5.0.2 of the Advanced Light Source (ALS). The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division of the U.S. Department of Energy, under contract no. DE-AC03-76SF00098, at Lawrence Berkeley National Laboratory. We thank all of the staff of these beamlines for their continued support.
Footnotes
Citation Nesta B, et al. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3(2):e00010-12. doi:10.1128/mBio.00010-12.
REFERENCES
- 1. Bielaszewska M, et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 2. Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754 [DOI] [PubMed] [Google Scholar]
- 3. Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449–456 [DOI] [PubMed] [Google Scholar]
- 4. Gransden WR, Eykyn SJ, Phillips I, Rowe B. 1990. Bacteremia due to Escherichia coli: a study of 861 episodes. Rev. Infect. Dis. 12:1008–1018 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L, Foxman B. 2003. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front. Biosci. 8:e235–e244 [DOI] [PubMed] [Google Scholar]
- 6. Mysorekar IU, Hultgren SJ. 2006. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 103:14170–14175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winberg J, Bergström T, Jacobsson B. 1975. Morbidity, age and sex distribution, recurrences and renal scarring in symptomatic urinary tract infection in childhood. Kidney Int. Suppl. 4:S101–S106 [PubMed] [Google Scholar]
- 8. Ruben FL, et al. 1995. Clinical infections in the noninstitutionalized geriatric age group: methods utilized and incidence of infections. The Pittsburgh Good Health Study. Am. J. Epidemiol. 141:145–157 [DOI] [PubMed] [Google Scholar]
- 9. Kucheria R, Dasgupta P, Sacks SH, Khan MS, Sheerin NS. 2005. Urinary tract infections: new insights into a common problem. Postgrad. Med. J. 81:83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moriel DG, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Isberg RR, Voorhis DL, Falkow S. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769–778 [DOI] [PubMed] [Google Scholar]
- 12. Luo Y, et al. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073–1077 [DOI] [PubMed] [Google Scholar]
- 13. Chen Q, Savarino SJ, Venkatesan MM. 2006. Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152:1041–1054 [DOI] [PubMed] [Google Scholar]
- 14. Hamburger ZA, Brown MS, Isberg RR, Bjorkman PJ. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291–295 [DOI] [PubMed] [Google Scholar]
- 15. Lehti TA, et al. 2010. Mat fimbriae promote biofilm formation by meningitis-associated Escherichia coli. Microbiology 156:2408–2417 [DOI] [PubMed] [Google Scholar]
- 16. Ulett GC, Mabbett AN, Fung KC, Webb RI, Schembri MA. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153:2321–2331 [DOI] [PubMed] [Google Scholar]
- 17. Trautner BW, Darouiche RO. 2004. Role of biofilm in catheter-associated urinary tract infection. Am. J. Infect. Control 32:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ha US, Cho YH. 2006. Catheter-associated urinary tract infections: new aspects of novel urinary catheters. Int. J. Antimicrob. Agents 28:485–490 [DOI] [PubMed] [Google Scholar]
- 19. Bomchil N, Watnick P, Kolter R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rasko DA, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oshima K, et al. 2008. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 15:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Snyder JA, et al. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73:7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Achtman M, et al. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hacker J, et al. 1985. Cloning and characterization of genes involved in production of mannose-resistant, neuraminidase-susceptible (X) fimbriae from a uropathogenic O6:K15:H31 Escherichia coli strain. Infect. Immun. 47:434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mobley HL, et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 27. Hayashi K, et al. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:20060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page R, et al. 2003. Shotgun crystallization strategy for structural genomics: an optimized two-tiered crystallization screen against the Thermotoga maritima proteome. Acta Crystallogr. D Biol. Crystallogr. 59:1028–1037 [DOI] [PubMed] [Google Scholar]
- 29. Stevens RC, et al. 2002. An approach to rapid protein crystallization using nanodroplets. J. Appl. Crystallogr. 35:278–281 [Google Scholar]
- 30. Collaborative Computational Project , Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 31. McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458–464 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 35. Hagberg L, et al. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuenne CT, Ghai R, Chakraborty T, Hain T. 2007. GECO—linear visualization for comparative genomics. Bioinformatics 23:125–126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB.
FdeC is not expressed under experimental in vitro conditions. Download Text S2, DOCX file, 0.1 MB.
Region encompassing the fdeC gene in non-ExPEC pathotype strains. Download Figure S1, TIF file, 5.5 MB.
Representative flow cytometry profiles of the binding of 100 µg/ml of FdeCAB to CHO, HeLa, and Vero cell lines. Filled gray histograms represent the binding to control cells incubated with the anti-FdeC antiserum followed by incubation with the fluorescently conjugated secondary antibody. Download Figure S2, DOCX file, 1.4 MB.