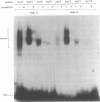
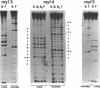
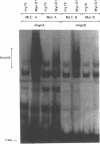
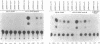Abstract
A potent muscle-specific enhancer element, originally described in the rat myosin light chain (MLC) 1/3 locus located downstream of the coding region, is found in an analogous position in the human MLC1/3 gene. When linked to a CAT reporter gene and transfected into muscle or non-muscle cells, the human MLC enhancer directs high levels of muscle-specific gene expression from homologous or heterologous promoters, irrespective of position or orientation relative to the CAT transcription unit. A significant degree of sequence homology (over 85%) in the 3'-flanking regions of the two MLC genes is restricted to a 200 bp sequence which lies approximately 1.5 kb downstream of the polyadenylation site in both species. The human enhancer sequence includes binding sites for human myogenic determination factors containing a common basic helix-loop-helix motif, and it can be trans-activated to varying degrees in non-muscle cells by these factors. This study establishes the MLC enhancer as an evolutionarily conserved, integral component of the MLC1/3 locus which constitutes a novel target for the action of myogenic determination factors.
Full text
PDF


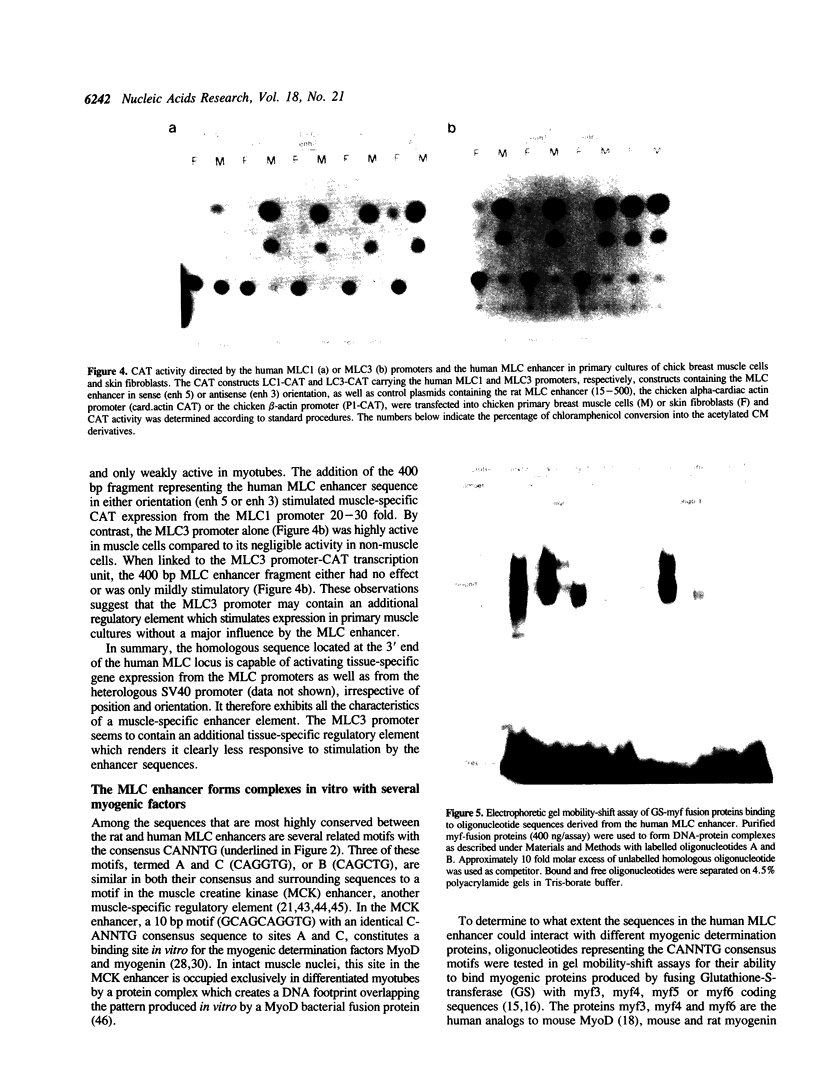
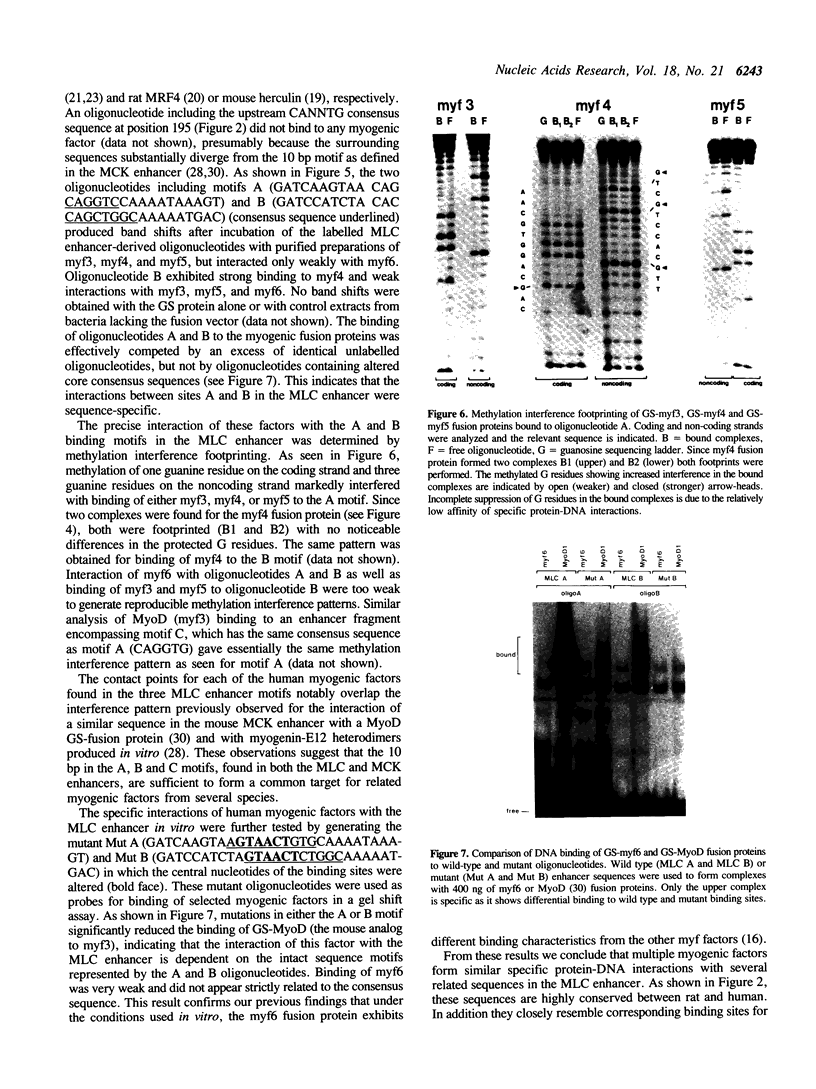



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billeter R., Quitschke W., Paterson B. M. Approximately 1 kilobase of sequence 5' to the two myosin light-chain 1f/3f gene cap sites is sufficient for differentiation-dependent expression. Mol Cell Biol. 1988 Mar;8(3):1361–1365. doi: 10.1128/mcb.8.3.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Bober E., Buschhausen-Denker G., Kohtz S., Grzeschik K. H., Arnold H. H., Kotz S. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 1989 Dec 1;8(12):3617–3625. doi: 10.1002/j.1460-2075.1989.tb08535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Bober E., Winter B., Rosenthal N., Arnold H. H. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 1990 Mar;9(3):821–831. doi: 10.1002/j.1460-2075.1990.tb08179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H. H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989 Mar;8(3):701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Tannich E., Buschhausen-Denker G., Arnold H. H. Promoter upstream elements of the chicken cardiac myosin light-chain 2-A gene interact with trans-acting regulatory factors for muscle-specific transcription. Mol Cell Biol. 1989 Jun;9(6):2513–2525. doi: 10.1128/mcb.9.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. J., Olson E. N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990 Apr;4(4):582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- Buskin J. N., Hauschka S. D. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol Cell Biol. 1989 Jun;9(6):2627–2640. doi: 10.1128/mcb.9.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubas P., Robert B., Garner I., Buckingham M. A comparison between mammalian and avian fast skeletal muscle alkali myosin light chain genes: regulatory implications. Nucleic Acids Res. 1985 Jul 11;13(13):4623–4643. doi: 10.1093/nar/13.13.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Ernst H., Wentworth B., Nadal-Ginard B., Rosenthal N. A muscle-specific enhancer is located at the 3' end of the myosin light-chain 1/3 gene locus. Genes Dev. 1988 Dec;2(12B):1779–1790. doi: 10.1101/gad.2.12b.1779. [DOI] [PubMed] [Google Scholar]
- Edlund T., Walker M. D., Barr P. J., Rutter W. J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5' flanking elements. Science. 1985 Nov 22;230(4728):912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- Edmondson D. G., Olson E. N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989 May;3(5):628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- Gauthier G. F., Lowey S. Distribution of myosin isoenzymes among skeletal muscle fiber types. J Cell Biol. 1979 Apr;81(1):10–25. doi: 10.1083/jcb.81.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett L. A., Kelvin D. J., Sternberg E. A., Olson E. N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989 Nov;9(11):5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Kiledjian M., Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990 Jan 26;247(4941):467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Horlick R. A., Benfield P. A. The upstream muscle-specific enhancer of the rat muscle creatine kinase gene is composed of multiple elements. Mol Cell Biol. 1989 Jun;9(6):2396–2413. doi: 10.1128/mcb.9.6.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes J. B., Chamberlain J. S., Buskin J. N., Johnson J. E., Hauschka S. D. Transcriptional regulation of the muscle creatine kinase gene and regulated expression in transfected mouse myoblasts. Mol Cell Biol. 1986 Aug;6(8):2855–2864. doi: 10.1128/mcb.6.8.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lohse P., Arnold H. H. The down-regulation of the chicken cytoplasmic beta actin during myogenic differentiation does not require the gene promoter but involves the 3' end of the gene. Nucleic Acids Res. 1988 Apr 11;16(7):2787–2803. doi: 10.1093/nar/16.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S., Risby D. Light chains from fast and slow muscle myosins. Nature. 1971 Nov 12;234(5324):81–85. doi: 10.1038/234081a0. [DOI] [PubMed] [Google Scholar]
- Miner J. H., Wold B. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1089–1093. doi: 10.1073/pnas.87.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Paterson B., Strohman R. C. Myosin synthesis in cultures of differentiating chicken embryo skeletal muscle. Dev Biol. 1972 Oct;29(2):113–138. doi: 10.1016/0012-1606(72)90050-4. [DOI] [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
- Quitschke W. W., DePonti-Zilli L., Lin Z. Y., Paterson B. M. Identification of two nuclear factor-binding domains on the chicken cardiac actin promoter: implications for regulation of the gene. Mol Cell Biol. 1989 Aug;9(8):3218–3230. doi: 10.1128/mcb.9.8.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes S. J., Konieczny S. F. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989 Dec;3(12B):2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- Robert B., Daubas P., Akimenko M. A., Cohen A., Garner I., Guenet J. L., Buckingham M. A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell. 1984 Nov;39(1):129–140. doi: 10.1016/0092-8674(84)90198-3. [DOI] [PubMed] [Google Scholar]
- Rosenthal N., Kornhauser J. M., Donoghue M., Rosen K. M., Merlie J. P. Myosin light chain enhancer activates muscle-specific, developmentally regulated gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7780–7784. doi: 10.1073/pnas.86.20.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R. K., Sarkar S. Correlation between the protein and mRNA levels for myosin light chains and tropomyosin subunits during chick fast muscle development in vivo. FEBS Lett. 1982 Nov 22;149(1):22–28. doi: 10.1016/0014-5793(82)81063-6. [DOI] [PubMed] [Google Scholar]
- Roy R. K., Sreter F. A., Sarkar S. Changes in tropomyosin subunits and myosin light chains during development of chicken and rabbit striated muscles. Dev Biol. 1979 Mar;69(1):15–30. doi: 10.1016/0012-1606(79)90271-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel U., Arnold H. H. Identification of the functional promoter regions in the human gene encoding the myosin alkali light chains MLC1 and MLC3 of fast skeletal muscle. J Biol Chem. 1989 Sep 25;264(27):16109–16117. [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sternberg E. A., Spizz G., Perry W. M., Vizard D., Weil T., Olson E. N. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988 Jul;8(7):2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler E. E., Periasamy M., Strehler-Page M. A., Nadal-Ginard B. Myosin light-chain 1 and 3 gene has two structurally distinct and differentially regulated promoters evolving at different rates. Mol Cell Biol. 1985 Nov;5(11):3168–3182. doi: 10.1128/mcb.5.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988 Oct 21;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Thayer M. J., Tapscott S. J., Davis R. L., Wright W. E., Lassar A. B., Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989 Jul 28;58(2):241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- Trask R. V., Strauss A. W., Billadello J. J. Developmental regulation and tissue-specific expression of the human muscle creatine kinase gene. J Biol Chem. 1988 Nov 15;263(32):17142–17149. [PubMed] [Google Scholar]
- Wang Y., Xu H. P., Wang X. M., Ballivet M., Schmidt J. A cell type-specific enhancer drives expression of the chick muscle acetylcholine receptor alpha-subunit gene. Neuron. 1988 Aug;1(6):527–534. doi: 10.1016/0896-6273(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Sassoon D. A., Lin V. K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989 Feb 24;56(4):607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- Yutzey K. E., Kline R. L., Konieczny S. F. An internal regulatory element controls troponin I gene expression. Mol Cell Biol. 1989 Apr;9(4):1397–1405. doi: 10.1128/mcb.9.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]