Abstract
TFIIH is a multifunctional RNA polymerase II general initiation factor that includes two DNA helicases encoded by the Xeroderma pigmentosum complementation group B (XPB) and D (XPD) genes and a cyclin-dependent protein kinase encoded by the CDK7 gene. Previous studies have shown that the TFIIH XPB DNA helicase plays critical roles not only in transcription initiation, where it catalyzes ATP-dependent formation of the open complex, but also in efficient promoter escape, where it suppresses arrest of very early RNA polymerase II elongation intermediates. In this report, we present evidence that ATP-dependent TFIIH action in transcription initiation and promoter escape requires distinct regions of the DNA template; these regions are well separated from the promoter region unwound by the XPB DNA helicase and extend, respectively, ≈23–39 and ≈39–50 bp downstream from the transcriptional start site. Taken together, our findings bring to light a role for promoter DNA in TFIIH action and are consistent with the model that TFIIH translocates along promoter DNA ahead of the RNA polymerase II elongation complex until polymerase has escaped the promoter.
TFIIH is a multifunctional RNA polymerase II general initiation factor that includes two DNA helicases encoded by the Xeroderma pigmentosum complementation group B (XPB) and D (XPD) genes, as well as a cyclin-dependent protein kinase encoded by the CDK7 gene (1). Previous studies have shown that the TFIIH XPB DNA helicase functions at multiple steps to promote efficient transcription initiation and promoter escape by RNA polymerase II. The TFIIH XPB DNA helicase catalyzes ATP(dATP)-dependent formation of the open complex before synthesis of the first phosphodiester bond of nascent transcripts (2), and it is required to suppress premature arrest of very early RNA polymerase II elongation intermediates at promoter-proximal sites ≈10–12 bp downstream of the transcriptional start site before their escape from the promoter (3–5).
In a previous study, we identified a requirement in transcription initiation and promoter escape by RNA polymerase II for promoter DNA extending ≈23–50 bp downstream from the transcriptional start site (6). In that study, we showed that synthesis of the first phosphodiester bond of nascent transcripts by RNA polymerase II requires promoter DNA extending ≈23–39 bp downstream from the transcriptional start site and that efficient promoter escape by the enzyme requires promoter DNA extending ≈39–50 bp downstream from the transcriptional start site. That study, however, did not identify which of the general initiation factors require downstream promoter DNA during these stages of transcription.
In this report, we present direct biochemical evidence that TFIIH requires downstream promoter DNA for its action in transcription initiation and promoter escape by RNA polymerase II. In addition, we show that TFIIH function in synthesis of the first phosphodiester bond of nascent transcripts and in promoter escape requires distinct regions of DNA downstream of the transcriptional start site. These regions of DNA are well separated from the region unwound by the XPB DNA helicase during formation of the open complex (2, 7–9) and extend, respectively, ≈23–39 and ≈39–50 bp downstream from the transcriptional start site. Taken together, our findings are consistent with the model that TFIIH translocates along promoter DNA ahead of the RNA polymerase II elongation complex until polymerase has escaped the promoter, and they provide a means of reconciling two recently proposed models (10–12) for the mechanism of TFIIH action in ATP(dATP)-dependent formation of the open complex and promoter escape.
Materials and Methods
DNA Templates.
A 444-bp duplex DNA template containing adenovirus major-late (AdML) promoter sequences from positions −50 to +10 was prepared by PCR, with M13 mp19-AdML as a template. The primers were 5′-GACGGCCAGTGAATTCGA-3′ and 5′-CCAGCGTGGACCGCTTGC-3′. The resulting DNA fragment, which contains sequences that extend 77 bp upstream and 367 bp downstream from the transcriptional start site, was gel purified before use in transcription reactions. The Ad(−9/−1) and Ad(−9/+9) templates were prepared as described (3, 5).
RNA Polymerase II and Transcription Factors.
RNA polymerase II (13) and TFIIH [rat d, TSK SP-5-PW fraction (14)] were purified as described from rat liver nuclear extracts. Recombinant yeast TBP (15, 16) and TFIIB (17) were expressed in Escherichia coli and purified as described. Recombinant TFIIE was prepared as described (18), except that the 56-kDa subunit was expressed in E. coli BL21(DE3)-pLysS. Recombinant TFIIF was purified as described (19) from E. coli JM109(DE3) coinfected with M13 mpET-RAP30 and M13 mpET-RAP74.
Transcription Assays.
Preinitiation complexes were assembled on 20 ng of the indicated DNA template at 30°C by a 45-min preincubation of 30-μl reaction mixtures containing 20 mM Hepes-NaOH (pH 7.9), 20 mM Tris⋅HCl (pH 7.9), 50 mM KCl, 4 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.5 mg/ml BSA, 2% (wt/vol) polyvinyl alcohol, 6% (vol/vol) glycerol, ≈50 ng of recombinant yeast TBP, ≈10 ng of recombinant TFIIB, ≈20 ng of recombinant TFIIF, ≈20 ng of recombinant TFIIE, ≈150 ng of highly purified TFIIH, and ≈0.01 units of RNA polymerase II. Transcription reactions were performed in the presence of the ribonucleoside triphosphates and for the times indicated in the figure legends. Reactions were stopped by the addition of 15 μl of 0.1 M EDTA, followed by the addition of 55 μl of 10 M urea, 0.025% bromophenol blue, and 0.025% xylene cyanole. Reaction mixtures were heated at 90°C for 5 min, and RNA transcripts were separated by electrophoresis in 25% acrylamide, 3% bisacrylamide gels containing 0.5× TBE buffer (1× TBE = 89 mM Tris/89 mM boric acid/2.0 mM EDTA, pH 8.0) and 5.0 M urea. Radioactive RNA transcripts were visualized by autoradiography.
Results
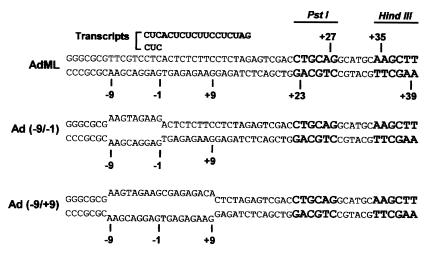
To investigate the possibility that TFIIH function in transcription initiation and promoter escape requires downstream promoter DNA, we took advantage of the artificial AdML promoter derivatives Ad(−9/−1) and Ad(−9/+9), which contain premelted DNA from positions −9 to −1 and positions −9 to + 9 relative to the normal AdML transcriptional start site. Previous studies have shown (i) that TFIIH and an ATP(dATP) cofactor are dispensable for initiation by RNA polymerase II from the Ad(−9/−1) promoter but are required for efficient promoter escape (3), and (ii) that TFIIH and an ATP(dATP) cofactor are not required for either transcription initiation or promoter escape by RNA polymerase II from the Ad(−9/+9) promoter (5). As illustrated in Fig. 1, these AdML promoter derivatives direct synthesis of identical transcripts by RNA polymerase II and have restriction sites, which are conveniently located for assessment of the contribution of downstream promoter DNA to TFIIH function in transcription initiation, inasmuch as they can be cleaved by PstI and HindIII at sites 23 and 39 bp downstream from the AdML transcriptional start site.
Figure 1.
Structures of the transcriptional start sites of the duplex AdML, Ad(−9/−1), and Ad(−9/+9) promoters. The bottom (coding) strands of all three templates are identical. PstI and HindIII restriction sites are indicated in bold. The sequences of transcripts initiated with CpU and synthesized in abortive initiation and promoter escape assays are shown above the DNA templates.
In our experiments, transcription by RNA polymerase II was carried out in a transcription system reconstituted with recombinant TBP, TFIIB, TFIIE, TFIIF, and purified polymerase and TFIIH from rat liver. Promoter-specific initiation was assayed by measuring synthesis of abortive, dinucleotide-primed trinucleotide transcripts. As shown previously, RNA polymerase II will use dinucleotides to prime synthesis of promoter-specific transcripts (20–24). Transcription initiation by RNA polymerase II from the AdML promoter can be primed by a variety of dinucleotides complementary to template DNA surrounding the transcriptional start site (20). In our experiments, synthesis of the first phosphodiester bond of nascent transcripts was assayed by measuring the synthesis of trinucleotide transcripts in reactions containing the initiating dinucleotide CpU and [α-32P]CTP, which support synthesis by polymerase of radioactively labeled CpUpC transcripts intiated at a position 3 bp upstream from the normal AdML transcriptional start site (Fig. 1).
Promoter escape by RNA polymerase II was assayed by measuring successful synthesis of 18 nucleotide transcripts in reactions containing the initiating dinucleotide CpU, ATP, UTP, [α-32P]CTP and the RNA chain-terminating nucleotide 3′-O-methylguanosine triphosphate (3′-O-MeGTP), which prevents most transcription beyond the first G residue of the nascent transcript. In previous studies (3, 25) we observed that very early RNA polymerase II elongation intermediates that have synthesized transcripts shorter than ≈10 nucleotides are prone to premature arrest at promoter-proximal sites ≈10–12 bp downstream from the transcriptional start site, either in the absence of TFIIH or an ATP(dATP) cofactor or in the presence of ATPγS, a potent inhibitor of the TFIIH XPB DNA helicase. In contrast, further transcript elongation by very early RNA polymerase II elongation intermediates that have successfully synthesized transcripts extending to the U or A residue immediately preceding the G residue at position +15 requires neither TFIIH nor an ATP(dATP) cofactor and is not inhibited by ATPγS. Furthermore, digestion of the duplex AdML DNA template with restriction enzymes that cleave the template ≤39 nucleotides downstream from the transcriptional start site before assembly of the preinitiation complex was shown to result in the arrest of RNA polymerase II at promoter-proximal sites, whereas digestion of the DNA template with the same restriction enzymes after polymerase had successfully synthesized ≈14 nucleotide transcripts did not prevent further elongation of transcripts by the enzyme (6). Accordingly, we operationally define early RNA polymerase II elongation intermediates that have synthesized ≈14-nucleotide or longer transcripts as those that have successfully escaped the promoter.
TFIIH Action in Transcription Initiation and Promoter Escape Requires Distinct Regions of Downstream Promoter DNA.
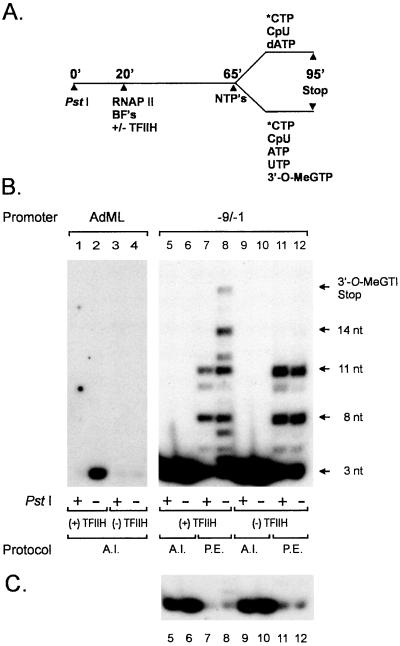
As discussed above, we previously observed that promoter DNA downstream of the PstI site at +23 in the duplex AdML promoter (Fig. 1) was essential for TFIIH-dependent synthesis by RNA polymerase II of the first phosphodiester bond of nascent transcripts (6). To investigate the possibility that TFIIH function in this process requires promoter DNA downstream of the PstI site, we asked whether the requirement for downstream DNA in the synthesis of the first phosphodiester bond of nascent transcripts is lost when transcription reactions are carried with the premelted Ad(−9/−1) promoter, which does not require TFIIH or an ATP(dATP) cofactor. Transcription reactions were carried out according to the protocol diagrammed in Fig. 2A. As shown in the control reactions of Fig. 2B, lanes 1–4, and consistent with our previous results, synthesis of abortive CpU-primed trinucleotide transcripts from the duplex AdML promoter is strictly dependent on TFIIH (Fig. 2, lanes 2 and 4); in addition, synthesis of abortive CpU-primed trinucleotide transcripts depends strongly on promoter DNA downstream of +23, because synthesis of trinucleotide transcripts is completely inhibited by digestion of the template with PstI before transcription reactions. In contrast, synthesis of abortive CpU-primed trinucleotide transcripts from the premelted Ad(−9/−1) promoter, in the presence or absence of TFIIH, was largely unaffected by PstI cleavage of the DNA template before transcription reactions (Fig. 2C; compare lanes 5, 6, 9, and 10). These findings indicate that promoter DNA downstream of +23 is not essential for assembly of a transcriptionally competent preinitiation complex, but is very likely required for TFIIH function in synthesis of the first phosphodiester bond of nascent transcripts.
Figure 2.
Downstream DNA is dispensable for transcription initiation by RNA polymerase II under conditions that bypass the requirement for TFIIH in initiation. (A) Schematic diagram of reaction protocol. BF's, basal initiation factors; *CTP, [α-32P]CTP; NTP's, ribonucleoside triphosphates; RNAPII, RNA polymerase II. (B) AdML (lanes 1–4) or Ad(−9/−1) (lanes 5–12) DNA templates were incubated for 20 min at 30°C with or without 15 units of PstI. Preinitiation complexes were assembled, with or without TFIIH, on templates as described in Materials and Methods. Abortive initiation assays (A.I.) were performed with 5 μM dATP, 200 μM CpU, and 0.5 μM [α-32P]CTP (3000 Ci/mmol). Promoter escape assays (P.E.) were performed with 5 μM ATP, 200 μM CpU, 0.5 μM [α-32P]CTP, 100 μM UTP, and 150 μM 3′-O-MeGTP. (C) Shorter exposure of the lower portion of the gel shown in B to allow visualization of abortively initiated trinucleotide transcripts in lanes 5–12.
We previously observed that downstream promoter DNA is required for efficient promoter escape during transcription initiated from the duplex AdML DNA template (6). To determine whether downstream promoter DNA is also required for promoter escape during transcription from the premelted Ad(−9/−1) promoter, transcription reactions were carried out with the Ad(−9/−1) promoter in the presence of the initiating dinucleotide CpU, ATP, UTP, [α-32P]CTP and the RNA chain-terminating nucleotide 3′-O-MeGTP. In the presence of TFIIH, the undigested Ad(−9/−1) promoter supported promoter escape, as evidenced by the formation of 3′-O-MeGTP-terminated transcripts (Fig. 2B, lane 8). In the absence of TFIIH, promoter escape by RNA polymerase II was suppressed, resulting in synthesis by polymerase of ≈11-nucleotide or shorter RNA transcripts (lane 12). In contrast, the PstI-digested Ad(−9/−1) promoter did not support synthesis by RNA polymerase II of transcripts longer than ≈11 nucleotides in either the presence or absence of TFIIH (lanes 7 and 11). Taken together, these findings indicate that very early RNA polymerase II elongation intermediates are capable of synthesizing short transcripts in the absence of promoter DNA downstream of +23, but that these elongation intermediates suffer premature arrest at the same promoter-proximal sites as those intermediates transcribing in the absence of TFIIH (compare lanes 7 and 12). Therefore, TFIIH and promoter DNA downstream of +23 are likely to function at a very similar stage during promoter escape.
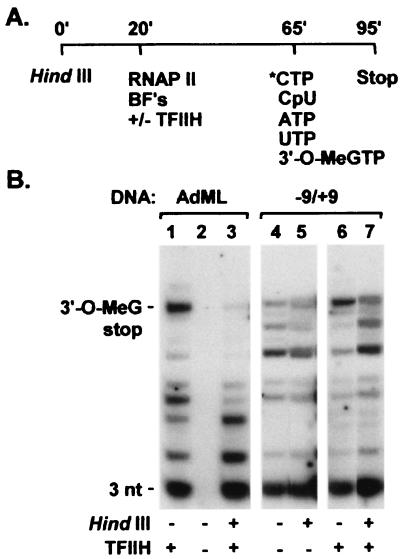
Our observation that premature arrest of early RNA polymerase II elongation intermediates occurs at the same promoter-proximal sites in the absence of either downstream DNA or TFIIH suggested that TFIIH function in promoter escape might depend on downstream DNA. To address this possibility, we asked whether removal of downstream DNA affects the efficiency of promoter escape by RNA polymerase II when transcription reactions are carried out with the premelted Ad(−9/+9) promoter, which supports both synthesis of the first phosphodiester bond of nascent transcripts and promoter escape in the absence of TFIIH and/or an ATP(dATP) cofactor (5). Transcription reactions were carried out according to the protocol diagrammed in Fig. 3A. As shown in Fig. 3B and consistent with our previous results, efficient synthesis by RNA polymerase II of 15-nucleotide-long, 3′-O-MeG-terminated transcripts from the duplex AdML promoter is strictly dependent on TFIIH; in addition, synthesis by polymerase of 18-nucleotide-long, 3′-O-MeG-terminated transcripts depends strongly on downstream DNA, because synthesis of these transcripts is completely inhibited by digestion of the DNA template with HindIII at position +39 before transcription reactions. In contrast, both transcription initiation and promoter escape occur at the premelted Ad(−9/+9) promoter even in the absence of TFIIH. Notably, TFIIH-independent initiation and promoter escape are largely unaffected by HindIII cleavage of the DNA template before transcription reactions. Taken together, these findings argue that TFIIH action in efficient promoter escape by RNA polymerase II depends on promoter DNA downstream of +39.
Figure 3.
Downstream DNA is dispensable for promoter escape by RNA polymerase II under conditions that bypass the requirement for TFIIH in promoter escape. (A) Schematic diagram of reaction protocol. (B) AdML (lanes 1–3) or Ad(−9/+9) (lanes 4–7) DNA templates were incubated for 20 min at 30°C with or without 5 units of HindIII before the assembly of preinitiation complexes. Promoter escape assays were performed, with or without TFIIH, as described in the legend to Fig. 2.
Sarkosyl Bypasses the Requirements for Both TFIIH and Downstream DNA in Promoter Escape by RNA Polymerase II.
Low concentrations of the detergent Sarkosyl (typically 0.015–0.025%) have been shown to inhibit assembly of RNA polymerase II preinitiation complexes but to have little effect on promoter-specific initiation or elongation by preassembled preinitiation complexes, whereas Sarkosyl concentrations of ≈0.05% or more have been shown to inhibit initiation, but to have little effect on subsequent transcript elongation (26–29). As a consequence, Sarkosyl has been widely used in studies of the mechanism of promoter-specific transcription to limit initiation events to a single round, as well as to prepare “washed elongation complexes” for studies of transcript elongation (e.g., refs. 30 and 31) and to prepare initiation complexes for cross-linking studies (12).
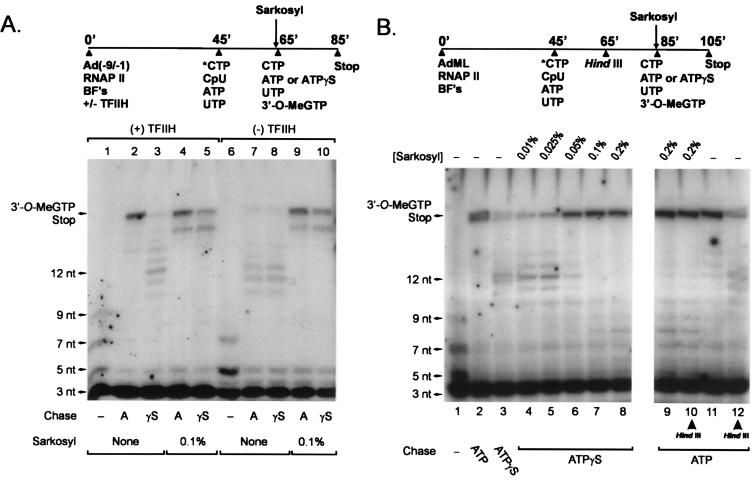
In the course of experiments investigating the mechanism of promoter escape by RNA polymerase II, we discovered that addition of Sarkosyl to very early RNA polymerase II elongation intermediates effectively relieves the requirements for TFIIH and an ATP(dATP) cofactor in promoter escape. Transcription reactions were carried out according to the pulse–chase protocol diagrammed in Fig. 4A. Preinitiation complexes were assembled on the premelted Ad(−9/−1) promoter, in the presence or absence of TFIIH. Short radioactive transcripts were synthesized by incubating preassembled preinitiation complexes with 200 μM initiating dinucleotide CpU, 5 μM ATP, 0.5 nM UTP, and 0.5 μM [α-32P]CTP. Under these conditions, transcripts with a maximum length of ≈7–9 nucleotides are synthesized by RNA polymerase II (lanes 1 and 6); notably, a significant fraction of transcripts synthesized in these reactions are abortively initiated trinucleotide transcripts, which cannot be chased into longer transcripts. The stably initiated transcripts were then chased into longer products by the addition of 200 μM CTP, 100 μM UTP, and 150 μM 3′-O-MeGTP, with or without 100 μM ATP or ATPγS. Consistent with previous results (3, 25), synthesis by RNA polymerase II of 18 nucleotide, 3′-O-MeG-terminated transcripts depended on the presence of both ATP and TFIIH and was inhibited by ATPγS (lanes 2, 3, 7, and 8). In contrast, when 0.1% Sarkosyl was included in the chase phase of the reaction, the majority of short transcripts were chased into 16- to 18-nucleotide transcripts, independently of TFIIH and in the presence of ATP or ATPγS (lanes 4, 5, 9, and 10).
Figure 4.
The requirements for TFIIH, an ATP(dATP) cofactor, and downstream DNA in promoter escape by RNA polymerase II are bypassed after treatment of very early elongation intermediates with Sarkosyl. (A) Transcription reactions were performed with the premelted Ad(−9/−1) promoter, with a pulse–chase protocol (diagrammed at the top of the figure) to test the requirement for TFIIH specifically during promoter escape. Preinitiation complexes were assembled on the Ad(−9/−1) promoter, with or without TFIIH, as described in Materials and Methods. Short RNA transcripts were synthesized during a 20-min incubation at 30°C with 5 μM ATP, 200 μM CpU, 0.5 μM [α-32P]CTP, and 0.5 nM UTP. Where indicated, Sarksoyl was added to a final concentration of 0.1% (wt/vol). One minute later, short transcripts were chased into longer transcripts in the presence of 100 μM ATP or 100 μM ATPγS and 200 μM CTP, 100 μM UTP, and 150 μM 3′-O-MeGTP. (B) Preinitiation complexes were assembled on the duplex AdML DNA template as described in Materials and Methods. After synthesis of short radioactively labeled transcripts according to the procedure described for A, reaction mixtures were incubated for 20 min with or without 5 units of HindIII. Sarkosyl was added to reaction mixtures at the concentrations indicated in the figure, and short transcripts were chased into longer transcripts as described in the experiment in A.
Sarkosyl relieved ATPγS-induced arrest of early RNA polymerase II elongation intermediates at concentrations as low as 0.05% (Fig. 4B, lanes 6–8), whereas lower Sarkosyl concentrations of 0.01–0.025% had little effect on ATPγS-induced arrest of early elongation intermediates. The reactions shown in Fig. 4B were performed with the duplex AdML DNA template; thus, Sarkosyl is able to relieve the block to promoter escape not only on premelted DNA templates, as shown in Fig. 4A, but also on duplex DNA templates. Notably, Sarkosyl treatment also relieves the requirement for downstream DNA in promoter escape, inasmuch as early RNA polymerase II elongation intermediates efficiently escape the promoter on DNA templates digested with HindIII at +39, in the presence but not the absence of 0.2% Sarkosyl (Fig. 4B; compare lanes 9–12). The observations that Sarkosyl relieves ATPγS-induced arrest of early RNA polymerase II elongation intermediates as well as the requirements for TFIIH and downstream DNA in promoter escape provide further support for the model that downstream DNA is required for TFIIH action in promoter escape.
Discussion
In previous studies (3, 4, 6, 25), efficient promoter escape by RNA polymerase II was shown to depend strongly on the TFIIH XPB DNA helicase, an ATP(dATP) cofactor, and a region of promoter DNA extending ≈39–50 bp downstream from the transcriptional start site. In addition, we showed that transcription initiation by RNA polymerase II depends on a region of promoter DNA extending ≈23–39 bp downstream from the transcriptional start site when initiation depends on TFIIH and an ATP(dATP) cofactor.
In this report, we have shown that downstream promoter DNA is not required for transcription by RNA polymerase II under conditions that bypass the requirements for TFIIH and an ATP(dATP) cofactor in initiation, efficient promoter escape, or both processes, arguing that downstream promoter DNA is essential for TFIIH action in both processes. First, we observe that downstream promoter DNA is not required for transcription initiation by RNA polymerase II when reactions are carried out with the premelted Ad(−9/−1) promoter, which supports transcription initiation but not efficient promoter escape in the absence of TFIIH and an ATP(dATP) cofactor. Second, we observe that removal of downstream promoter DNA does not affect the efficiency of TFIIH-independent promoter escape by RNA polymerase II when reactions are carried out with the premelted Ad(−9/+9) promoter, which supports both transcription initiation and efficient promoter escape in the absence of TFIIH and an ATP(dATP) cofactor. In additional experiments, we have shown that treatment of very early RNA polymerase II elongation intermediates with the detergent Sarkosyl bypasses the requirement for TFIIH, an ATP(dATP) cofactor, and downstream promoter DNA in efficient promoter escape by polymerase.
Based on our findings, which argue that TFIIH action in the synthesis of the first phosphodiester bond of nascent transcripts and in promoter escape requires distinct downstream promoter regions well separated from the region unwound by the XPB DNA helicase (2, 7–9), we propose that TFIIH, and perhaps its XPB DNA helicase subunit, has a DNA binding domain that is distinct from the helicase catalytic site and that binds DNA downstream of the transcriptional start site in the initiation complex. Furthermore, we propose that, after initiation, this TFIIH DNA binding domain translocates along promoter DNA ahead of the RNA polymerase II elongation complex until the completion of promoter escape. This model is consistent with results of several other studies of the structure and function of TFIIH and the RNA polymerase II initiation complex. DNase I footprinting analysis revealed that addition of a protein fraction containing TFIIE, TFIIF, and TFIIH to promoter-bound complexes that include RNA polymerase II, TBP, and TFIIB results in specific protection of promoter DNA between positions +20 and +30 (32). Results of two-dimensional electron crystallography performed on yeast RNA polymerase II transcription complexes suggest that TFIIE binds to a polymerase domain that contacts downstream DNA (33–36); because TFIIE and TFIIH bind specifically to one another (37, 38), these findings suggest that TFIIH might be similarly positioned. Results of two recent cross-linking studies are also consistent with the idea that TFIIH makes protein–DNA contacts downstream of the RNA polymerase II initiation complex during transcription initiation and, by extension, during promoter escape (11, 12). Although results of these studies differ in many respects, they each provide evidence that, in both closed and open RNA polymerase II initiation complexes, the TFIIH XPB DNA helicase makes protein–DNA contacts with promoter DNA downstream of the transcriptional start site, between positions +10 and +20, according to the findings of Kim et al. (12), and between positions +10 and +38, according to findings of Douziech et al. (11). Finally, it is intriguing that the recently reported structures of yeast and mammalian TFIIH have revealed that they are ring-like molecules with a central hole of sufficient size to accommodate double-stranded DNA (39, 40). In light of this structural information, it is tempting to speculate that TFIIH possesses a sliding clamp-like DNA binding domain that could mediate its translocation along promoter DNA ahead of the RNA polymerase II elongation complex.
What is the downstream DNA-dependent function of TFIIH in promoter escape? Results of previous experiments suggest that a TFIIF activity required for transcription initiation presents an impediment to efficient promoter escape and is at least partly responsible for inducing the premature arrest of very early RNA polymerase II elongation intermediates (5). Together with results of cross-linking experiments suggesting that TFIIF and TFIIE promote tight wrapping of DNA around RNA polymerase II by making contacts in the initiation complex with promoter DNA upstream of the TATA box and downstream of the transcriptional start site (10, 11, 41), this observation raises the possibility that protein–DNA contacts between promoter DNA and TFIIF and TFIIE might present an impediment to promoter escape. Based on our observations (i) that, like TFIIH action in formation of the open complex, TFIIH action in promoter escape by RNA polymerase II requires the XPB DNA helicase activity, an ATP(dATP) cofactor, and downstream DNA and (ii) that the requirement for the TFIIH XPB DNA helicase, an ATP(dATP) cofactor, and downstream DNA in transcription initiation and promoter escape by RNA polymerase II is lost when transcription is carried out with the Ad(−9/+9) template, which contains a premelted region extending to +9, it is reasonable to propose that TFIIH XPB helicase could suppress arrest simply by unwinding promoter DNA downstream of the transcriptional start site. Indeed, the regions of the TFIIF RAP30 and TFIIE small subunits required for transcription initiation by RNA polymerase II in vitro have been shown to include double-stranded DNA binding domains (42–44); thus, unwinding of downstream promoter DNA by the TFIIH XPB DNA helicase could relieve a TFIIF- and TFIIE-induced impediment to promoter escape by disrupting interactions of TFIIF and TFIIE with double-stranded DNA downstream of the transcriptional start site. Further experiments will be required, however, to test this hypothesis.
Finally, our findings provide a way to explain the different results obtained in cross-linking studies performed by Kim et al. (12) and by Douziech et al. (11) and used to propose conflicting models for how TFIIH promotes unwinding of promoter DNA. Kim et al. observed (i) that the TFIIH XPB DNA helicase subunit is the only TFIIH subunit that efficiently cross-links to promoter DNA, (ii) that the XPB subunit is only efficiently cross-linked to promoter DNA downstream of the transcriptional start site, and (iii) that only the TFIIH XPB DNA helicase and some RNA polymerase II subunits were reproducibly cross-linked to downstream promoter DNA (12). Based on their findings, Kim et al. proposed that the TFIIH XPB DNA helicase does not catalyze formation of the open complex by a conventional DNA helicase mechanism, but rather functions as a “molecular wrench” that melts promoter DNA surrounding the transcriptional start site by binding to and rotating downstream promoter DNA relative to rotationally fixed upstream DNA. As described above, Douziech et al. observed that the TFIIH XPB DNA helicase subunit cross-links not only to promoter DNA downstream of the transcriptional start site, but also to promoter DNA upstream of the transcriptional start at a position near −5 and upstream of the TATA box (11). Taken together with their evidence that TFIIE, TFIIF, and RNA polymerase II subunits crosslink to promoter DNA both upstream and downstream of the transcriptional start site, this finding led Douziech et al. to propose that protein–DNA contacts between the promoter and RNA polymerase II, TFIIE, TFIIF, and TFIIH induce tight wrapping of DNA around the initiation complex and destabilize the DNA helix near position −5, giving the TFIIH XPB DNA helicase access to a single-stranded region of DNA to initiate unwinding by a conventional DNA helicase mechanism.
Until recently, it has been difficult to reconcile the results of Douziech et al. (11) and Kim et al. (12). It is noteworthy, however, that, unlike the experiments of Douziech et al., the cross-linking experiments of Kim et al. (12) were performed with Sarkosyl-treated transcription complexes. Our observation that Sarkosyl treatment of very early RNA polymerase II elongation intermediates relieves the TFIIF-induced impediment to efficient promoter escape and, in so doing, the requirement for TFIIH and an ATP(dATP) cofactor in this process suggests that Sarkosyl may disrupt functionally important protein–DNA contacts within the initiation complex. These results may explain the failure by Kim et al. to detect crosslinking of the TFIIH XPB DNA helicase in the vicinity of the transcription start site and of TFIIF and TFIIE to downstream promoter sequences (12) and raise the possibility that the model of Douziech et al. (11) may more accurately describe molecular events that occur during TFIIH-dependent formation of the open complex.
Acknowledgments
We thank Kristen Maslonka for expert technical assistance and Qin Yan for a gift of premelted promoter DNA templates. Work in the authors' laboratories is supported by National Science Foundation Grant MCB-9817004 and the Oakland University Research Excellence Program in Biotechnology (A.D.) and by National Institutes of Health Grant R37 GM41628 and funds provided to the Oklahoma Medical Research Foundation by the H. A. and Mary K. Chapman Charitable Trust (R.C.C. and J.W.C.). J.W.C. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- 3′-O-MeGTP
3′-O-methylguanosine triphosphate
- AdML
adenovirus major-late
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Svejstrup J Q, Vichi P, Egly J M. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 2.Tirode F, Busso D, Coin F, Egly J M. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 3.Dvir A, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreland R J, Tirode F, Yan Q, Conaway J W, Egly J M, Conaway R C. J Biol Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- 5.Yan Q, Moreland R J, Conaway J W, Conaway R C. J Biol Chem. 1999;274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 6.Dvir A, Tan S, Conaway J W, Conaway R C. J Biol Chem. 1997;272:28175–28178. doi: 10.1074/jbc.272.45.28175. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Carey M, Gralla J D. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 8.Holstege F C P, van der Vliet P C, Timmers H, Th M. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 9.Holstege F C P, Fiedler U, Timmers H, Th M. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert F, Douziech M, Forget D, Egly J M, Greenblatt J, Burton Z F, Coulombe B. Mol Cell. 1998;2:342–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douziech M, Coin F, Chipoulet J M, Arai Y, Ohkuma Y, Egly J M, Coulombe B. Mol Cell Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T K, Ebright R H, Reinberg D. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 13.Serizawa H, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaway R C, Reines D, Garrett K P, Powell W, Conaway J W. Methods Enzymol. 1996;273:194–207. doi: 10.1016/s0076-6879(96)73020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt M C, Kao C C, Pei R, Berk A J. Proc Natl Acad Sci USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conaway J W, Hanley J P, Garrett K P, Conaway R C. J Biol Chem. 1991;266:7804–7811. [PubMed] [Google Scholar]
- 17.Tsuboi A, Conger K, Garrett K P, Conaway R C, Conaway J W, Arai N. Nucleic Acids Res. 1992;20:3250. doi: 10.1093/nar/20.12.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Nature (London) 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 19.Tan S, Conaway R C, Conaway J W. BioTechniques. 1994;16:824–828. [PubMed] [Google Scholar]
- 20.Samuels M, Fire A, Sharp P A. J Biol Chem. 1984;259:2517–2525. [PubMed] [Google Scholar]
- 21.Luse D S, Jacob G A. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 22.Jiang Y, Yan M, Gralla J D. J Biol Chem. 1995;270:27332–27338. doi: 10.1074/jbc.270.45.27332. [DOI] [PubMed] [Google Scholar]
- 23.Jacob G A, Luse S W, Luse D S. J Biol Chem. 1991;266:22537–22544. [PubMed] [Google Scholar]
- 24.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 25.Dvir A, Conaway R C, Conaway J W. J Biol Chem. 1996;271:23352–23356. doi: 10.1074/jbc.271.38.23352. [DOI] [PubMed] [Google Scholar]
- 26.Hawley D K, Roeder R G. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 27.Hawley D K, Roeder R G. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 28.Cai H, Luse D S. J Biol Chem. 1987;262:298–304. [PubMed] [Google Scholar]
- 29.Conaway R C, Conaway J W. J Biol Chem. 1988;263:2962–2968. [PubMed] [Google Scholar]
- 30.Izban M G, Luse D S. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 31.Marshall N F, Price D H. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buratowski S, Hahn S, Guarente L, Sharp P A. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 33.Leuther K K, Bushnell D A, Kornberg R D. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 34.Poglitsch C L, Meredith G D, Gnatt A L, Jensen G J, Chang W H, Fu J, Kornberg R D. Cell. 1999;98:791–798. doi: 10.1016/s0092-8674(00)81513-5. [DOI] [PubMed] [Google Scholar]
- 35.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 36.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 37.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 38.Bushnell D A, Bamdad C, Kornberg R D. J Biol Chem. 1996;271:20170–20174. doi: 10.1074/jbc.271.33.20170. [DOI] [PubMed] [Google Scholar]
- 39.Chang W H, Kornberg R D. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 40.Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly J M. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 41.Coulombe B, Burton Z F. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan S, Garrett K P, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1994;91:9808–9812. doi: 10.1073/pnas.91.21.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan S, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1995;92:6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda M, Watanabe Y, Okamura H, Hanaoka F, Ohkuma Y, Nishimura Y. EMBO J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]