Abstract
The regulation of ion channel activity by specific lipid molecules is widely recognized as an integral component of electrical signaling in cells1,2. In particular, phosphatidylinositol 4,5-bisphosphate (PIP2), a minor yet dynamic phospholipid component of cell membranes, is known to regulate many different ion channels3–7. PIP2 is the primary agonist for classical inward rectifier (Kir2) channels, through which this lipid can regulate a cell’s resting membrane potential2,7–9. However, the molecular mechanism by which PIP2 exerts its action is unknown. Here we present the x-ray crystal structure of a Kir2.2 channel in complex with a short-chain (dioctanoyl) derivative of PIP2. We found that PIP2 binds at an interface between the transmembrane domain (TMD) and the cytoplasmic domain (CTD). The PIP2 binding site consists of a conserved non-specific phospholipid binding region (RWR) in the TMD and a specific phosphatidylinositol binding region in the CTD. Upon PIP2 binding a flexible expansion linker contracts to a compact helical structure, the CTD translates 6 Å and becomes tethered to the TMD, and the inner helix gate begins to open. In contrast, the small anionic lipid dioctanoyl glycerol pyrophosphatidic acid (PPA) also binds to the non-specific TMD region, but not to the specific phosphatidylinositol region, and thus fails to engage the CTD or open the channel. Our results show how PIP2 can control the resting membrane potential through a specific ion channel receptor-ligand interaction that brings about a large conformational change, analogous to neurotransmitter activation of ion channels at synapses.
PIP2 influences the metabolic state of cells by at least three distinct pathways (figure S1a,b); first, as the prototypical second messenger being cleaved into diacyl glycerol (DAG) and inositol triphosphate (IP3)10,11, second, as a localization signal targeting soluble proteins to the plasma membrane12–14, and third, as a signaling molecule capable of agonizing an ion channel2,8,15,16. This latter role, in which an ion channel is activated by PIP2, was first discovered in 1998 when Hilgemann and colleagues showed that PIP2 acted alone to open a Kir channel8.
Figure 1a, b shows the influence of PIP2 on the function of Kir2.2 from chicken. Following excision of an inside-out membrane patch from a Xenopus oocyte expressing Kir2.2 channels, initially large inward K+ currents diminish over time. The diminution occurs because PIP2 is depleted from the membrane’s inner leaflet8. The K+ currents can be restored partially by exposing the cytoplasmic face of the patch to the short-chain derivative of PIP2 in a dose-dependent manner17,18 (figure 1b). PIP2 is the primary agonist for classical inward rectifier (Kir2) channels, through which this lipid can regulate a cell’s resting membrane potential. Here we use x-ray crystallography to understand the mechanism by which PIP2 opens a Kir2 channel.
Figure 1. Effect of a short chain PIP2 on Kir2.2.
a, Endogenous PIP2 depletion causes “run down” of Kir2.2 channels in an excised inside out patch from Xenopus oocytes as shown by the three macroscopic current traces recorded with a voltage ramp from−80 to +80 mV immediately (green), 30 min (blue), and 50 min (black) after patch excision. b, The short chain PIP2 added to the bath solution (solid line with concentration indicated below) 30 min after patch excision partially rescued Kir2.2 channel activity. The bath was then perfused (dashed line) at time 40 min with ~1 ml/min bath solution for 3 min. c, X-ray crystal structures of apo (left, pdb 3JYC) and PIP2 bound (right, pdb 3SPI) Kir 2.2 tetramer (grey α-carbon traces) viewed from the side with the extracellular solution above. The lipid bilayer boundaries are shown as grey bars. Four PIP2 molecules are shown as sticks and colored according to atom type: carbon, yellow; phosphorous, orange; and oxygen, red. One PIP2 molecule in the same orientation as in figure 2a is outlined by a black box. Upon PIP2 binding the flexible linker between CTD and TMD consisting of two strands (highlighted green for one subunit, dotted line indicating disordered region in the crystal structure) form helical structures, and the CTD translates towards the TMD by 6 Å. A set of reference atoms (Asp72 and Lys220 α-carbons) are highlighted as blue spheres in each structure.
Kir2.2 is a tetrameric ion channel comprised of a TMD, which forms the prototypic potassium selective pore, and a large CTD, which characterizes all Kir channels19 (figure 1c). We determined the structures of wild type Kir2.2 from chicken (with disordered segments of the N- and C-termini truncated) in the presence of the short-chain derivative of PIP2 at 3.3 Å resolution (figure 1c). We also determined the structures of two point mutants, I223L and R186A, in the presence of PIP2 at 3.0 Å and 2.6 Å resolution, respectively (Table S1). These mutants were studied because they are described in the literature as altering the apparent affinity for PIP29,20. All three channels have overall similar structures and taken together enhance our knowledge of the chemical details through which PIP2 binds to and modifies the channel’s structure and function (figure S2a,b). Electron density maps are of high quality for the entire protein and strong density for the three phosphates in PIP2, observed in all three structures, allowed accurate placement of the ligand (figure S3a). Furthermore, the glycerol backbone of PIP2 is well ordered and easily placed in the higher resolution structures, along with 4–6 carbons of the lipid acyl chains (figure S3a). One PIP2 molecule binds to each of the four channel subunits (figure 1c).
PIP2 binds at the interface between the TMD and CTD and produces a large conformational change in Kir2.2 (figure 1c). The entire CTD translates 6 Å towards the TMD in association with the formation of two new helices, an N-terminal extension of the ‘interfacial’ helix and a ‘tether’ helix at the C-terminus of the inner helix (figure 2a and S3b). The 6 Å translation of the CTD is reflected in a compression along the c axis of the unit cell (Table S1). The protein conformational changes position amino acids that form the binding site for the 4′, 5’′ phosphate-substituted inositol head group of PIP2.
Figure 2. PIP2 binding site.
a, A detailed view of the PIP2 binding site is shown in the same orientation as outlined in figure 1c. Helices (shown as ribbon) from different subunits are colored orange and cyan interior. Residues hydrogen bonded (dashed lines) to PIP2 are colored green, and residues stabilizing the PIP2 binding site in the CTD while lacking direct contact are colored blue. All side chains are shown as sticks. PIP2 is shown as sticks and colored according to atom type: carbon, yellow; phosphorous, orange; and oxygen, red. b, An amino acid sequence alignment of selected eukaryotic Kirs showing residues predicted from literature (blue outline) and not predicted (purple outline) to interact with PIP29,28. Residues with direct bonding interactions to PIP2 and with a structural role are highlighted with green and blue shade, respectively. The two residues serving as the inner helix gate are also highlighted with grey shade.
The PIP2 binding site comprises amino acids from two main structural regions of the channel. The acyl chains, glycerol backbone, and 1′ (phosphodiester) phosphate of PIP2 interact with the TMD, while the inositol head group makes interactions with the CTD (figure 2a and S4). In detail, the acyl chains insert into the membrane layer where they interact with hydrophobic amino acids on both the inner and outer helices, while the 1′ phosphate makes interactions with amino acids forming the sequence RWR (amino acids 78–80 in Kir2.2) (figure 2b). This sequence is conserved (as RWR or KWR) among many different Kir channels, and the reason for this conservation is made clear by the PIP2 complex: the RWR sequence is located at the N-terminus of the outer helix and forms a binding site in which the 1′ phosphate caps the helix and is cradled by main chain amide nitrogen atoms and the guanidinium groups from the two arginine residues (figure 2a and S4). The tryptophan residue appears to anchor the end of the outer helix at the membrane interface and also interact with one of the acyl chains. With the acyl chains, glycerol backbone and 1’ phosphate of the lipid molecule contacting the TMD, the inositol ring of the head group is oriented towards the CTD, where the 4′ and 5′ phosphates are positioned to interact directly with Lys183, Arg186, Lys188 and Lys189 (figure 2a and S4). The latter two positively charged amino acids are located on the tether helix, whose structure is induced by the binding of PIP2. Other amino acids on the tether helix, including Arg190, participate in the formation of a hydrogen-bonding network that appears to strengthen the interaction between the tether helix and other regions of the CTD, especially the N-terminal extension of the interfacial helix, whose structure is also induced by the binding of PIP2 (figure 2a and S4). A sequence alignment shows that the amino acids binding to PIP2 are highly conserved among the large family of inward rectifier K+ channels (figure 2b). Because all members of this ion channel family appear to be regulated by PIP2 (some in concert with other ligands such as ATP or G-proteins)16, we anticipate that the PIP2 site described here will be observed in many other inward rectifiers.
The detailed chemical properties of the PIP2 binding site suggest that the TMD region should bind to any lipid that contains a glycerol backbone, acyl chains and a 1′ phosphate, whereas the CTD should provide the specificity for the inositol phosphate head group. Moreover, because the head group region of the binding site is formed only after the conformational changes occur in the channel, we would predict that a glycerol phospholipid without an inositol head group would bind to the TMD but not induce the conformational changes. We tested this prediction by determining a 2.45 Å resolution crystal structure of Kir2.2 in the presence of a short-chain derivative of pyrophosphatidic acid (PPA), which contains as a head group only phosphoric acid instead of the 4′, 5′ phosphate-substituted inositol ring (Table S1). This lipid is bound to the TMD in a manner almost identical to PIP2, however, the head group does not interact with the CTD and the protein conformational changes induced by PIP2 do not occur (figure 3a–c). This finding is compatible with recent functional studies showing that small head group anionic lipids failed to activate Kir channels in the absence of PIP221.
Figure 3. Conserved non-specific lipid binding site in Kirs.

a, A grey α-carbon representation of Kir2.2 tetramer in complex with PPA, a small anionic lipid lacking an inositol ring. PPA bound Kir2.2 assumes a closed conformation similar to apo (pdb 3JYC) with the flexible linker elongated and the CTD unengaged. The four PPA molecules are shown as sticks and colored according to atom type: carbon, yellow; phosphorous, orange; and oxygen, red. b, A close-up view of the PPA binding site. PPA contacts Kir at the cytoplasmic end of the outer helix making strong interactions with the guanidiniums of R78 and R80 and the backbone amide nitrogens of the helix turn; similar to the interactions of the 1′ phosphate of PIP2. However, residues (blue sticks) for interacting with the PIP2 inositol 4′,5′ phosphate remain distant to the lipid binding site; R186 orients with its side chain pointing towards the ion conduction pathway. c, Superposition of PPA (colored the same as in panel a) and PIP2 (grey). The position of the 1′ phosphate in PIP2 is between the pyrophosphate of PPA.
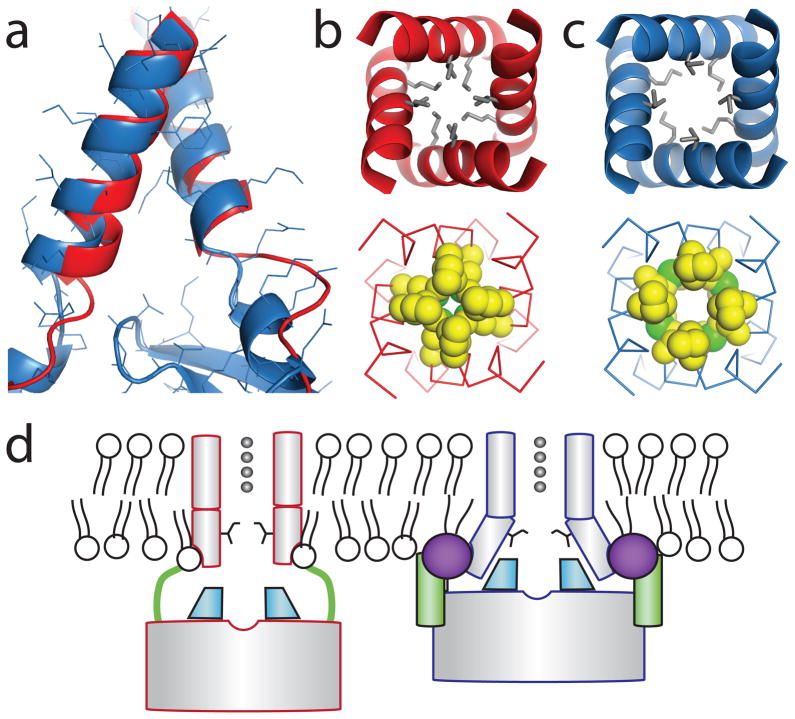
We wish to understand how the PIP2 induced conformational changes relate to channel activity. Comparison of the inner helix gate in the PIP2 and PPA complexes shows that the interaction of the CTD with the TMD induced by PIP2 is key to opening the gate (figure 4a–c). In the PPA complex, in which the CTD is extended away from the TMD, the gate in the TMD is tightly closed (4.9 Å at Ile177), whereas in the PIP2complex the inner helices have begun to separate (6.3 Å). The separation of helices comes about as a result of a slight splaying, but more significantly a rotation of the inner helices, which moves hydrophobic amino acid side chains away from the ion pathway (figure 4a–c). Opening of the inner helix gate to approximately 6.3 Å (approximately 5 Å diameter between van der Waal surfaces of carbon atoms at the narrowest region) is probably still insufficient to permit ion conduction, but the gate is clearly on the way to an open conformation. A previously published study of prokaryotic Kir channels proposed that interactions between the TMD and CTD of those channels influence the distribution of ions in the selectivity filter22. We observe no such influence of the CTD on ions in the filter in the high quality electron density maps in our analysis of Kir2.2 (figure S5a–c). In the present study, the data support a simple allosteric mechanism of gating control by the signaling lipid PIP2, in which the lipid mediates docking of the CTD to the TMD and opening of the inner helix gate, as depicted (figure 4d).
Figure 4. A proposed mechanism of Kir2.2 activation by PIP2.
a, Superposition of the TMD inner helices of the PIP2 bound (blue ribbon) and apo (red ribbon ) Kir2.2 structures. PIP2 binding results in a splaying of the helices near the helix bundle activation gate. b and c, comparison of the inner helix bundle gate in PPA bound Kir2.2 (b) and PIP2 bound Kir2.2 (c) viewed from the extracellular side. Side chains of the residues in the bundle crossing are represented as either grey sticks or space filling CPK models (carbon, yellow; and sulfur, green). d, A proposed mechanism for Kir2.2 activation by PIP2. PIP2(purple sphere) binds at an interface between the TMD (grey cylinder) and the CTD (grey rectangle) and induces a large conformational change: a flexible linker (green line) contracts to a compact helical structure (green cylinder), the CTD translates towards and becomes tethered to the TMD, the G-loop (cyan wedge) inserts into the TMD, and the inner helix activation gate opens.
The ion pathway in Kir channels has a second constriction formed by the G-loop, at the apex of the CTD. This loop in some instances is thought to function as a gate, referred to as the G-loop gate23. In Kir2.2 the conformation of the G-loop is altered by PIP2, either directly through the binding of PIP2 or indirectly through the docking of the CTD to the TMD when PIP2 binds (figure S6a,b). But it appears that PIP2 does not control the G-loop gate to a large extent, because in both conformations this gate is open (smallest diameter 7.8 Å) (figure S6a). The mutation I223L affects the conformation of the G-loop gate in a manner that might explain the apparent increased affinity for PIP2 20. In this mutant, although the CTD does not bind to the TMD and the tether and interfacial helices do not form, the G-loop adopts its PIP2 bound conformation (figure S6c,d). It is thus possible that the mutant favors PIP2 binding by tending to favor the PIP2-bound configuration prior to PIP2 binding.
The membrane lipid PIP2 plays a central role in cell signaling through three distinct pathways (figure S1a). In one of these pathways PIP2 acts directly on specific ion channels to regulate their activity. PIP2 is the primary agonist for Kir2 channels, which control the resting membrane potential in many cells. Since this discovery fifteen years ago, this form of ion channel regulation has been a topic of intense study. The crystal structures presented here reveal the mechanism of PIP2 activation of Kir2 channels. PIP2 binds to a lipid-binding site at the membrane’s inner leaflet, and through specific interactions between the 4’, 5’ inositol phosphate head group and the channel a large conformational change occurs, initiating pore opening.
Methods Summary
Chicken Kir2.2 with a C-terminal GFP and a 1D4 epitope was expressed in Pichia and purified in n-decyl-β-D-maltopyranoside (DM, Anatrace) by 1D4 antibody affinity chromatography followed by PreScission protease cleavage and gel filtration19. Purified protein was concentrated to 9 mg/ml and mixed with freshly prepared dioctanoyl PIP2 (10 mM stock in water) or dioctanoyl PPA (100 mM stock in water) to a final concentration of 0.6–1 mM and 5 mM, respectively. Crystals, diffracting between 2.45 and 3.3 Å, were obtained from a 200 nL hanging drop with 4 mM DM, 20 mM DTT, 3 mM TCEP, 0.5 M KCl, and PEG 400 or PEG 4000 as a precipitant and cryo-protected in reservoir solution containing 25% glycerol. Phases were obtained by molecular replacement with apo Kir2.2 (pdb 3JYC) using MolRep24 in the CCP4 suite25. The models were built in Coot26 and refined in Phenix27 to Rfree of 0.22 to 0.28. Complete crystallographic data and refinement statistics are shown in the online supplementary information.
Electrophysiology experiments were conducted using patch-clamp on Xenopus oocytes expressing wt Kir2.2. Briefly, oocytes were injected with 50 nL (~2 mg/ml) cRNA and used for patch recording after 2–3 days. Large pipette tips with typical resistance of 0.4–0.9 M Ω were used.
Methods
Cloning, expression, and purification
Kir2.2 from chicken with a GFP and a 1D4 epitope at the C-terminus was expressed in Pichia and purified in n-decyl-β-D-maltopyranoside (DM, Anatrace) by 1D4 antibody affinity chromatography followed by PreScission protease cleavage and gel filtration on a superdex 200 column as previously described19. Purified protein was concentrated to 9 mg/ml. For crystallization trials of PIP2-Kir2.2 channel complex, freshly prepared PIP2 (10 mM stock in water) was added to the concentrated protein at a final concentration of 0.6–1 mM lipid and 8 mg/ml protein and incubated for about an hour before setting up trays. For the crystallization trials of the PPA-Kir2.2 channel complex, 5 mM PPA (100 mM stock in water) was used.
Structure determination
Co-crystals of Kir2.2 with PIP2 or PPA were obtained from a 200 nL (100:100 nL protein:reservoir mixture) hangingdrops. The protein buffer solution contained 4 mM DM, 20 mM DTT, 3 mM TCEP, 150 mM KCl, and 20 mM Tris-HCl pH 8.0. Reservoir solution yielding the best diffracting crystals contained 0.3–0.6 M KCl, 50 mM HEPES (pH 6.5–7.5) plus 10–20% PEG 400 (w/v) or 3–8% PEG 4000 (w/v). Diamond-shaped crystals, 150 to 350 microns in the longest dimension, grew within 48 hrs at 4°C. The crystals were cryo-protected in reservoir solution plus 25–30% (v/v) glycerol (5% increment steps) and flash frozen in liquid nitrogen. Diffraction data were collected at beamlines X29 and X25 (Brookhaven NSLS). Crystals with PIP2 or PPA diffracted to 2.6–3.3 Å or 2.45 Å respectively. The crystals all belong to I4 space group with one subunit in the asymmetric unit. Phases were obtained by molecular replacement from apo Kir2.2 (pdb 3JYC) using MolRep24 in the CCP4 suite25. The models were built in Coot26 and refined in Phenix27 to an Rfree of 0.22 to 0.28. There are no Ramachandran outliers (97% most favored, 3% allowed). Complete crystallographic data and refinement statistics are shown in the supplementary Table S1. The PIP2 bound model contains residues from 42–369. In the PPA bound structure, part of the interfacial helix is disordered and the final model contains residues 42–62 and 70–369. Waters were added with ARP/wARP29 in the CCP4 suite25 and manually adjusted in the 2.45 and 2.6 Å structures. Figures were made with PYMOL30.
Electrophysiology
cRNA of chicken Kir2.2 was made from NdeI linearized Kir2.2 in the pGEM vector19 using the Amplicap T7 RNA kit (Epicentre Biotechnologies). Xenopus oocytes were prepared as described19 and injected with 50 nL of cRNA 12–20 hours later. All recordings were made with patch-clamp in inside-out configuration 2–3 days after injection. Injected oocytes were treated with ND96 (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 50 μg/ml gentamycin, pH 7.6 with NaOH) plus 200 mM NaCl for 5–10 min and the vitelline membrane was removed prior to seal formation. On cell membrane seals were formed using pipettes with typical resistance of 0.4–0.9 MΩ and large inside-out patches were excised with currents ranging from 0.5 to 5 nA and seals from 0.4 to 1 GΩ. The bath solution contained 130 mM KCl, 5 mM HEPES, 5 mM K2EDTA, pH 7.4 with KOH. The pipette solution contained 140 mM KCl, 5 mM HEPES, 0.3 mM CaCl2, 1 mM MgCl2, pH 7.4 with KOH. For PIP2 rescuing experiments described in figure 1b, 10 mM dioctanoyl PIP2 prepared in water was added to the bath solution and mixed by pipetting.
All patch-recordings were made with a voltage ramp from +80 to −80 mV in 10 sec duration under the control of an Axopatch 200B amplifier, Digidata 1440A analogue-to-digital converter and pClamp10.1 software (Axon Instruments, Inc). For figure 1b, the voltage ramp was repeated every 30 seconds after patch excision and the amount of current at +70 mV was plotted against the time (immediately after excision: time = 0). Figure 1a and 1b were made with Igor Pro (Wavemetrics, Lake Oswego, OR, USA).
Supplementary Material
Acknowledgments
We thank staff members at NSLS X29 and X25, Brookhaven National Laboratory for beamline assistance, members of the Gadsby lab (Rockefeller University) for help in Xenopus oocyte preparation, R. Molday (University of British Columbia) for providing the anti-1D4 tag cell line, and members of the MacKinnon laboratory for helpful suggestions. R.M. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Supplementary Information: is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: S.B.H purified and crystallized the protein; collected, processed and refined crystallographic data, and performed electrophysiology experiments. X.T. aided in experimental design and provided assistance in all aspects of the project. R.M. designed the study, and analyzed data. All authors wrote and discussed the manuscript.
Atomic coordinates and structure factors for the reported crystal structures have been deposited into the Protein Data Bank with accession codes 3SPI (wt/PIP2), 3SPC (wt/PPA), 3SPH ( I223L/PIP2), 3SPJ (I223L/apo), 3SPG (R186A/PIP2).
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interest.
References
- 1.Dart C. Lipid microdomains and the regulation of ion channel function. J Physiol. 588:3169–3178. doi: 10.1113/jphysiol.2010.191585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilgemann DW, Feng S, Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE. 2001;2001:re19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- 3.Suh BC, Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Fujiwara Y, Kubo Y. Regulation of the desensitization and ion selectivity of ATP-gated P2X2 channels by phosphoinositides. J Physiol. 2006;576:135–149. doi: 10.1113/jphysiol.2006.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logothetis DE, Jin T, Lupyan D, Rosenhouse-Dantsker A. Phosphoinositide-mediated gating of inwardly rectifying K(+) channels. Pflügers Archiv : European journal of physiology. 2007;455:83–95. doi: 10.1007/s00424-007-0276-5. [DOI] [PubMed] [Google Scholar]
- 6.Vaithianathan T, et al. Direct regulation of BK channels by phosphatidylinositol 4,5-bisphosphate as a novel signaling pathway. The Journal of general physiology. 2008;132:13–28. doi: 10.1085/jgp.200709913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamper N, Shapiro MS. Regulation of ion transport proteins by membrane phosphoinositides. Nat Rev Neurosci. 2007;8:921–934. doi: 10.1038/nrn2257. [DOI] [PubMed] [Google Scholar]
- 8.Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 9.Lopes CMB, et al. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 2002;34:933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 10.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 11.Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 13.Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 14.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science (New York, NY) 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilgemann DW, Ball R. Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science (New York, NY) 1996;273:956–959. doi: 10.1126/science.273.5277.956. [DOI] [PubMed] [Google Scholar]
- 16.Stanfield PR, Nakajima S, Nakajima Y. Constitutively active and G-protein coupled inward rectifier K+ channels: Kir2.0 and Kir3.0. Rev Physiol Biochem Pharmacol. 2002;145:47–179. doi: 10.1007/BFb0116431. [DOI] [PubMed] [Google Scholar]
- 17.Rohács T, et al. Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:745–750. doi: 10.1073/pnas.0236364100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enkvetchakul D, Jeliazkova I, Nichols CG. Direct modulation of Kir channel gating by membrane phosphatidylinositol 4,5-bisphosphate. The Journal of biological chemistry. 2005;280:35785–35788. doi: 10.1074/jbc.C500355200. [DOI] [PubMed] [Google Scholar]
- 19.Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science (New York, NY) 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, He C, Yan X, Mirshahi T, Logothetis DE. Activation of inwardly rectifying K+ channels by distinct PtdIns(4,5)P2 interactions. Nature cell biology. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 21.Cheng WW, D’Avanzo N, Doyle DA, Nichols CG. Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys J. 2011;100:620–628. doi: 10.1016/j.bpj.2010.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke OB, et al. Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels. Cell. 2010;141:1018–1029. doi: 10.1016/j.cell.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Pegan S, et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nature neuroscience. 2005;8:279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 24.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 25.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie LH, John Sa, Ribalet B, Weiss JN. Activation of inwardly rectifying potassium (Kir) channels by phosphatidylinosital-4,5-bisphosphate (PIP2): interaction with other regulatory ligands. Progress in biophysics and molecular biology. 2007;94:320–335. doi: 10.1016/j.pbiomolbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 30.Delano WL. Delano Scientific. Palo Alto, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.