Summary
Polycomb group (PcG) proteins are essential for accurate axial body patterning during embryonic development. PcG-mediated repression is conserved in metazoans and is targeted in Drosophila by Polycomb response elements (PREs). However, targeting sequences in humans have not been described. While analyzing chromatin architecture in the context of human embryonic stem cell (hESC) differentiation, we discovered a 1.8kb region between HOXD11 and HOXD12 (D11.12) that is associated with PcG proteins, becomes nuclease hypersensitive, and then shows alteration in nuclease sensitivity as hESCs differentiate. The D11.12 element repressed luciferase expression from a reporter construct and full repression required a highly conserved region and YY1 binding sites. Furthermore, repression was dependent on the PcG proteins BMI1 and EED and a YY1-interacting partner, RYBP. We conclude that D11.12 is a Polycomb-dependent regulatory region with similarities to Drosophila PREs, indicating conservation in the mechanisms that target PcG function in mammals and flies.
Introduction
Proper embryonic development requires an orchestration of precise temporal and spatial gene expression patterns. Polycomb repressive complexes PRC2 and PRC1 act as gene-specific epigenetic silencers throughout development. Conservation of Polycomb-mediated silencing across metazoans underlies its importance; disruption of this controlled and complex phenomenon often leads to gross abnormalities along the anterior-posterior axis. Initial insights into how Polycomb-Group (PcG) complexes affect development were observed in Drosophila (reviewed in (Grimaud et al., 2006), (Schwartz and Pirrotta, 2007)), where extensive genetic analysis over the past sixty years has shown that the PcG system is required to maintain differentiated states.
In mammals, PcG genes are essential for proper differentiation and development. For example, in mice defects in a central PRC1 component, Bmi1, display homeotic transformations in axial segmentation (van der Lugt et al., 1996); many of the segmentation defects were suppressed when crossed with mice bearing mutations in Mll, a trxG protein (Hanson et al., 1999). PcG proteins are required for the maintenance of pluripotency in mouse and human embryonic stem cells (hESCs) and are found at the promoters of many genes involved in differentiation (Boyer et al., 2006), (Lee et al., 2006). They maintain the silenced state of genes through cell divisions although commitment to a new cell fate may result in loss of their association with the promoters of upregulated genes (Bracken et al., 2006).
The mammalian PRC1 and PRC2 complexes are comprised of evolutionarily conserved proteins that combine to create a repressed state. The core components of the mammalian PRC2 complex are SUZ12, EED, EZH2, and RBAP48/46 (reviewed by (Simon and Kingston, 2009)). The mammalian and Drosophila PRC1 complexes form around a core of four proteins; many sub-complexes of PRC1 exist in mammals which include core proteins from the CBX family (CBX2, 4, 6, 7, or 8), BMI1, RING1, and PH. Mechanistically, the PRC2 complex methylates histone H3 at lysine 27 converting it to a tri-methylated state (H3K27me3), which is believed to play a key role in regulating PRC1-mediated repression complexes (Simon and Kingston, 2009). In vitro, physical compaction of nucleosomal arrays occurs in the presence of the core PRC1 complex (Francis et al., 2004) and in vivo data suggest that a looping of chromatin partitions the silenced genes away from activating factors (Tiwari et al., 2008) (Kahn et al., 2006). PRC1-family complexes can also ubiquitylate histone H2A (Cao et al., 2005; Kallin et al., 2009) and have been proposed to impede transcriptional elongation (Stock et al., 2007). A third PcG complex is the Drosophila PHO-RC complex, which has sequence specific DNA-binding capability and is involved in targeting PcG function (Oktaba et al., 2008).
A central question in PcG function revolves around the multiple mechanisms required for appropriate targeting. In Drosophila, homeotic genes contained within the Antennapedia and Bithorax complexes are repressed by PcG proteins. DNA sequences within these complexes, called Polycomb Response Elements (PREs), target the repression machinery via binding by several different sequence-specific binding factors. PREs are relatively large and complex regions that can be located tens of kilobases from the homeotic genes they regulate. Indeed, chromatin immunoprecipitation (ChIP) of PcG proteins Polycomb (PC) and Polyhomeotic (PH) from Drosophila embryos show that a majority of binding occurred between 2kb to 40kb away from the nearest promoter (Negre et al., 2006). PcG protein binding is developmentally regulated; differences in binding are observed between embryo and adult chromatin and large-scale studies differ in specifics of binding patterns, presumably because Drosophila cell lines reflecting different stages of development were used (Negre et al., 2006; Schwartz et al., 2006; Tolhuis et al., 2006). Genome-wide identification of PcG binding sites was not sufficient to identify PREs and some known PREs were not targeted. Another approach using a prediction algorithm based upon the frequency of known DNA binding motifs yielded some targets that did not show repression in transgenic studies (Ringrose and Paro, 2007; Ringrose et al., 2003). This approach might have been limited by the fact that binding sites for these proteins do not show perfect overlap with PRE elements. The protein most consistently associated with PRE function in Drosophila is the PcG protein PHO (Brown et al., 2003; Brown et al., 1998; Wang et al., 2004). PHO binding sites, however, are not sufficient to define a PRE.
PREs in Drosophila tend to be conspicuously depleted of nucleosomes (Mohd-Sarip et al., 2006; Muller and Kassis, 2006; Papp and Muller, 2006), although the nucleosomes surrounding the PRE are enriched in H3K27me3 (Schwartz et al., 2006). At several PREs in the Drosophila homeotic cluster, nuclease-hypersensitive sites correlated with peaks of H3.3 localization (Mito et al., 2007). Enrichment of H3.3 at these PREs suggests that there is continual nucleosome disruption to keep cis-acting elements accessible. How the binding sites within the largely non-nucleosomal PRE and the surrounding methylated nucleosomes coordinate to target PcG function is a matter for ongoing debate (Ringrose et al., 2004);(Wang et al., 2004);(Kahn et al., 2006).
Targeting of PcG function in mammals is not as well understood as it is in flies. The mammalian homolog of PHO, YY1, is a candidate targeting factor (Wang et al., 2004). Studies involving YY1 in the context of PcG-mediated repression are complicated by the fact that YY1 interacts with many regulatory proteins in various cell types. YY1 interacts with PcG proteins EED and BMI1 in separate complexes and colocalizes in the trunks of E12.5 mouse embryos upstream of the repressed Hoxa5 and Hoxc8 genes (Kim et al., 2006). Another YY1-interacting protein, RYBP, directly interacts with the PRC1 components Ring1A, Ring1B, and M33, and has demonstrated repressive activity in a transcriptional reporter assay (Garcia et al., 1999). Therefore, RYBP may link YY1 with the PcG system. Interestingly, aside from YY1 there are no known mammalian homologs of the most commonly found PRE-binding Drosophila proteins; GAF, Pipsqueak, and Zeste. One clue as to how PcG proteins are targeted is that there is a high correlation between localization of PRC2 components and CpG islands, suggesting the possibility that these islands influence recruitment (Ku et al., 2008). It is likely that PcG recruitment in mammals, as in flies, requires many components that are not yet understood.
Two recent studies address PcG targeting in mice. The discovery that a large inversion of sequences caused mis-expression of the MafB gene during mouse development led to the search for possible PcG targeting sequences near the inversion (Sing et al., 2009). A minimal 3kb fragment from this region has PRE function in flies and confers repression on a reporter gene in mouse embryos that was abrogated when the fragment was excised. This segment, called PRE-kr, was bound by PcG proteins in cultured cells, repressed activity in a PcG-dependent manner and thus has been proposed to constitute a mouse PRE. A second report characterized the deregulation of gene expression and changes in histone modifications within the mouse Hoxd cluster in a mutant carrying an inversion separating Hoxd11 to Hoxd13 from the rest of the locus by 3Mb (Soshnikova and Duboule, 2009). Of particular relevance to PcG function and to the element examined in our study, during early development, Hoxd11 and Hoxd12 remained silent in the inversion mutant while their wild-type littermates exhibited normal upregulated expression of these genes. Intriguingly, several unexpected peaks of H3K4me3 and loss of H3K27me3 were observed in wild-type mice, including a region between Hoxd11 and Hoxd12. These data suggest that potential regulatory region(s) in which PcG and trxG complexes mediate these histone modifications exist in the distal part of the Hoxd cluster.
To characterize PcG function during differentiation of human cells, we used the pluripotent H1 and H9 hESC lines and their derivatives to examine changes in chromatin structure and epigenetic modifications throughout the HOX clusters. We found a region of high chromatin plasticity between HOX genes HOXD11 and HOXD12. This region possesses several characteristics consistent with an involvement in PcG targeting, including nucleosome depletion, high sequence conservation across species, YY1 binding sites and GC-rich sequences. Functionally, this region is sufficient to target PcG function to a reporter gene in differentiated hESCs. We report that this repressive ability can be heritably transmitted through differentiation into another cell type in a PcG-dependent manner.
Results
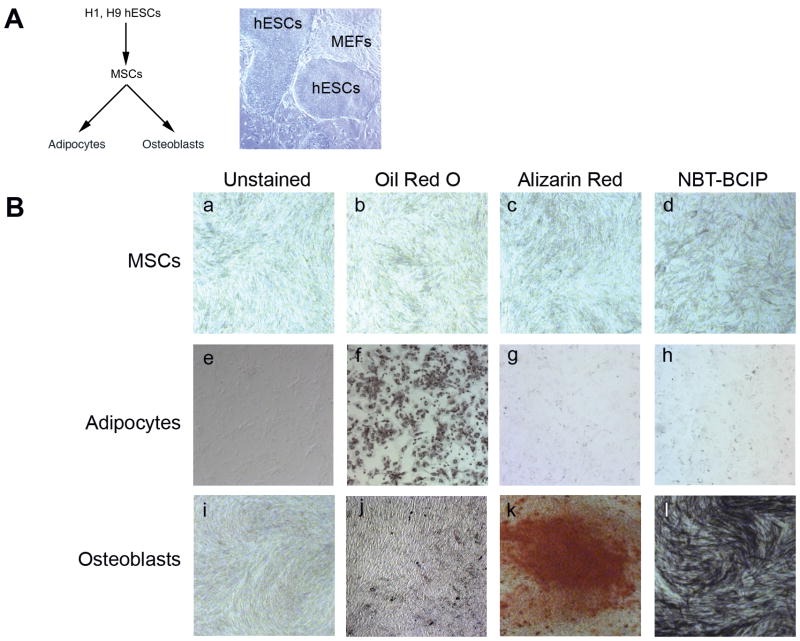
To analyze changes in the structure of chromatin at the HOX clusters, we first established conditions that would enable us to isolate sufficient numbers of cells in defined states of differentiation. The starting population of hESCs had the characteristic clustered morphology (Figure 1A) and expressed the pluripotency markers OCT4, SOX2, and NANOG, as well as TERT, the enzymatic component of the telomerase complex (Figure S1). These cells were differentiated into mesenchymal stem cells (MSCs) and expressed a panel of MSC markers (Figure S1). The multipotent MSCs were differentiated into either adipocytes or osteoblasts as described previously (Barberi et al., 2005). The adipocytes stained positively for lipid accumulation (Figure 1B, f) but not for osteoblast markers (Figure 1C, g and h). The osteoblasts had an elongated morphology, and stained positively for calcium deposition and for alkaline phosphatase activity (panels k, l) but not for lipid accumulation (panel j). The MSCs stained negatively for all of these cell markers and did not express significant levels of the adipocyte or osteoblast-associated genes (Figure 1B, a–d, and Figure S1).
Figure 1. Lineage commitment from pluripotent cells to differentiated cell types.
(A) Differentiation schematic; H9 hESCs cultured with MEFs (B) MSCs (panels a–d: unstained, treated with Oil Red O, Alizarin Red, and NBT-BCIP); Adipocytes, day 14 (panels e–h: unstained, treated with Oil Red O, Alizarin Red, and NBT-BCIP); and Osteoblasts, day 28 (panels i–l: unstained, treated with Oil Red O, Alizarin Red, and NBT-BCIP). See Figure S1 for qRT-PCR results.
We used quantitative RT-PCR to measure changes in expression for a panel of genes across HOXA, HOXB, HOXC, and HOXD clusters as cells underwent differentiation. When the hESCs were differentiated into MSCs, most of the twenty surveyed HOX genes remained unchanged in expression (Table S1) while HOXA13, HOXB1, HOXD10, HOXD12, and HOXD13 were silenced. HOXA4 was upregulated when the cells differentiated from hESC to MSCs. When the MSCs were differentiated into adipocytes, HOXC10 was upregulated and HOXA9, HOXA10, and HOXD1 were silenced. In the osteoblasts, HOXA10, HOXB2, HOXB3 increased expression, while HOXA1 was silenced. We conclude that regulated changes to the HOX loci occurred in our cultured hESC cell-based system. We used this controlled cell culture system of differentiation to search for potential regulatory elements through analysis of chromatin structural changes.
Characterization of chromatin regulation in HOX clusters
We examined chromatin changes during differentiation to identify potential regulatory regions. Sites with high levels of enrichment of the PcG proteins BMI1 and SUZ12 and H3K27me3 were identified by ChIP using a high-density tiled microarray (Kharchenko et al., 2008) covering the four HOX clusters. Such sites were observed in the HOX clusters in the different ES-derived cell types (Figure S2), as has previously been studied by others (Squazzo, 2006). The same array format was employed to detect micrococcal nuclease (MNase) sensitive sites by hybridizing mononucleosome-sized DNA fragments following digestion (Dennis et al., 2007; Kharchenko et al., 2008). We looked for intergenic regions that might predict nuclesome-free regions (NFRs) or areas with low nucleosome occupancy, as determined by MNase hypersensitivity, and correlated that with enrichment of PcG proteins and H3K27me3, as these are all features associated with PREs in Drosophila. In this study, we focus upon one sequence with these characteristics, an intergenic region between HOXD11 and HOXD12.
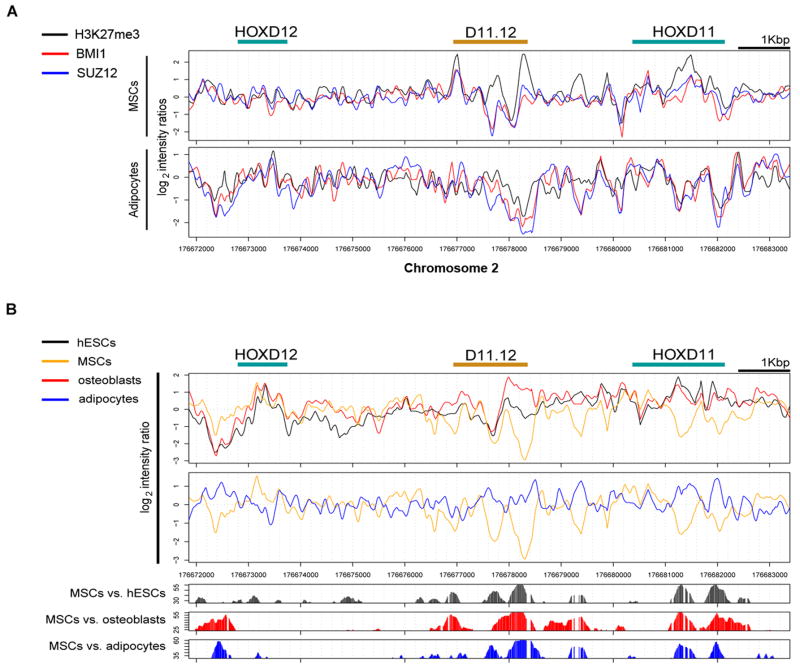
Figure 2A shows the normalized location analysis results from ChIP-chip experiments in MSCs and in adipocytes for the PcG proteins BMI1 and SUZ12 and for H3K27me3. In MSCs, a region between HOXD11 and HOXD12 (Figure 2, bracket) showed peaks corresponding to occupancy of H3K27me3, BMI1 and SUZ12 at the boundaries (Figure 2A, top). These peaks were not observed in a similar analysis of adipocytes (Figure 2A, bottom). In Figure 2B, the plots display the comparisons of MNase mapping data of all four cell types for the region of the HOXD cluster containing HOXD11 through HOXD13. Statistically significant differences in the MNase profiles between the different cell types are found throughout the entire region. Prominent differences are illustrated by a statistical comparison of MSCs with hESCs, osteoblasts and adipocytes, with the extent of difference indicated by the height of each bar (Figure 2B, bottom). We were intrigued that a region between HOXD11 and HOXD12 showed not only flanking peaks of PcG binding and H3K27 methylation but also significant changes in nucleosome occupancy as cells differentiated. In particular, this region appears low in nucleosome occupancy in MSCs (yellow line, Figure 2B), the same stage of differentiation where this area is flanked by high H3K27 methylation and PcG protein occupancy. These are characteristics associated with Drosophila PREs, so we chose to pursue a functional analysis of this region, which is referred to below as D11.12.
Figure 2. ChIP-chip and MNase hypersensitivity results.
(A) Summary of location analyses across HOXD13 to HOXD10 for H3K27me3, BMI1, and SUZ12 in MSCs. Log2 ratios were normalized and sliding windows of 20 bp were used to plot the values. (B) Nucleosomal mapping of MNase-sensitive sites. Top graphs: plots of normalized log2 ratios from MNase-digested chromatin for hESCs (black), MSCs (orange), osteoblasts (red), and adipocytes (blue). X-axis represents 21kb along chromosome 2 along the HOXD cluster. The bracket points to a large region of difference between HOXD11 and HOXD12. Bottom graphs: Plots of the statistically significant differences of the log2 ratios of the MNase profiles between the cells. Figure S2 displays data across the HOX clusters. Figure S3 displays MNase sensitivity data of D11.12.
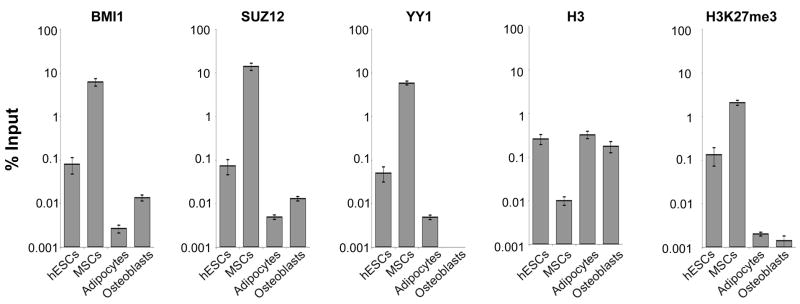
We characterized further the MNase sensitivity of D11.12. For MNase mapping of nucleosome occupancy, mononucleosome-sized DNA is isolated and hybridized to arrays. If a span of DNA is hypersensitive to cleavage, then no signal will be observed, even with digestion by the lowest amounts of MNase. Alternatively, the DNA might exist within a compacted structure that is resistant to MNase cleavage and thus not form mononucleosome-sized fragments. This would also score (in this instance, artifactually) as high in MNase sensitivity. The raw nucleosome mapping data indicates the region of apparent MNase sensitivity (Figure S3). To validate that D11.12 was sensitive to MNase digestion and not a region that was resistant, we performed a Southern blot after treating native MSC chromatin with a range of MNase concentrations (Figure S3). The signal disappeared even with the lightest MNase digestions. As another independent method of analysis, we sonicated formaldehyde-crosslinked MSCs and used ChIP with an antibody that recognizes H3 regardless of modification status. The association of H3 with D11.12 was one or two orders of magnitude less than the levels of H3 associated with this region in the hESCs, adipocytes, and osteoblasts (Figure 6). The relative levels of enrichment observed by ChIP-qPCR agree with the comparisons of MNase protection from our mapping experiments. These results indicate that D11.12 in MSCs is a nuclease sensitive region depleted for histone H3.
Figure 6. PcG proteins are enriched at the endogenous D11.12 in different cell types.
ChIP-qPCR results of BMI1, SUZ12, YY1, H3, and H3K27me3 at the endogenous D11.12 region in MSCs. Results are represented as % input and were not normalized against histone H3. Data are represented as mean +/−SEM.
Analysis of D11.12 regulatory function
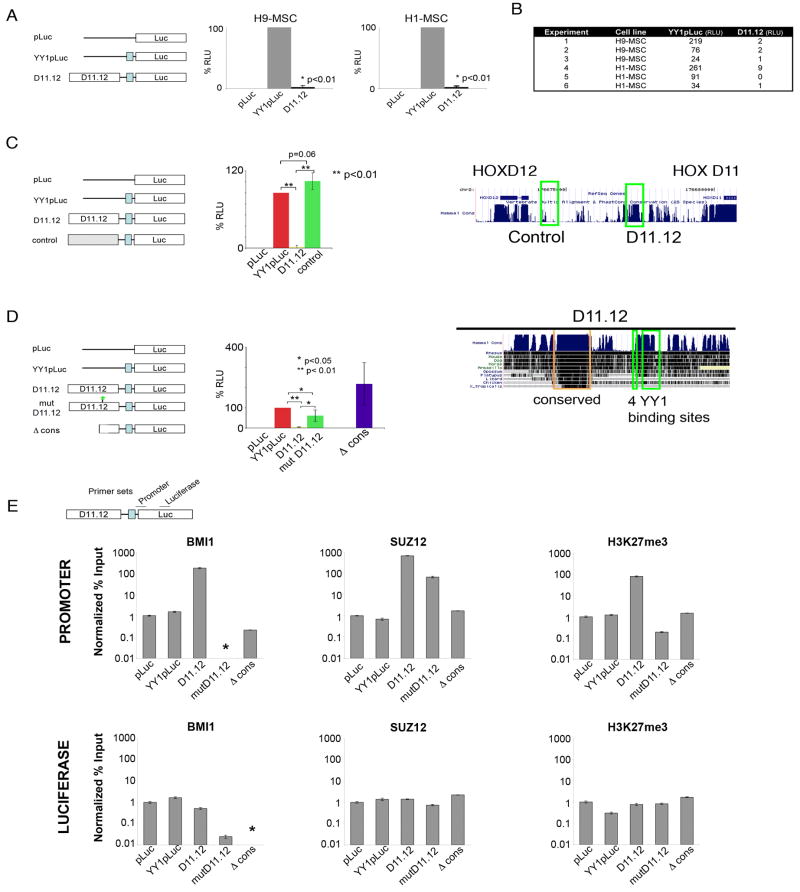
The association of D11.12 with PcG proteins and MNase hypersensitivity suggested that this region might serve a silencing function, since these are characteristics found in Drosophila PREs (Muller and Kassis, 2006) (Ringrose and Paro, 2007) (Henikoff et al., 2009). PREs in Drosophila have the ability to repress heterologous genes in transgenic assays and are PcG-dependent (Cavalli and Paro, 1998) (Dejardin et al., 2005) (Sengupta et al., 2004). To determine whether D11.12 could repress gene activity in cultured human cells, a transient luciferase assay was developed (Figure 3). The MSCs are amenable to nucleofection with high efficiency and grow rapidly in tissue culture. A parental luciferase construct containing an upstream thymidine kinase (TK) promoter (pLuc) showed minimal luciferase activity that was close to the measured background. To augment the activity of this reporter, we used a construct containing multiple binding sites for YY1, which functions as an activator in this context, immediately upstream of the TK promoter (YY1pLuc). This construct showed a high level of luciferase activity (Figure 3A). When D11.12 was placed upstream of the YY1 enhancer (D11.12), the luciferase activity was reduced to less than 5% of the activity of YY1pLuc (Figure 3A). Repression of luciferase activity was observed in both H1 and H9 hESC-derived MSC lines. Results from individual experiments, without normalization to YY1pLuc, are shown to underscore the reproducibility and extent of the repression observed mediated by D11.12 (Figure 3B).
Figure 3. D11.12 represses luciferase activity in MSCs.
(A) Constructs are displayed on the left: the firefly luciferase construct (pLuc), with an upstream YY1 enhancer (YY1pLuc), and with a 1.8kb region between HOXD11 and HOXD12 upstream of YY1pLuc (D11.12). Luciferase measurements of the co-transfected firefly luciferase and renilla luciferase constructs (n=3, each cell line). Data are represented as mean +/−SEM. (B) Chart of the individual lucifease results, presented as RLU (n=6). (C) Luciferase activity of a control region located between HOXD11 and HOXD12 (left panel). The UCSC Genome Browser map shows the locations of the control region and D11.12 and the level of mammalian conservation of sequence (right panel). (D) Luciferase activity of D11.12 having mutated YY1 binding sites (mutD11.12) or deletion of the conserved region (Δ cons) (left panel). The UCSC Genome Browser map depicts the degree of conservation across 10 other vertebrate species (right panel) with the conserved region (orange) and the 4 YY1 binding sites (green). (E) ChIP-qPCR of BMI1, SUZ12, and H3K27me3. ChIP results are displayed as ratios of the % input(IP)/% input (histone H3). These templates had H3 levels that were consistently 5–10 fold lower in the luciferase gene than for the promoter, perhaps indicative of differences in nucleosome occupancy across these transiently transfected templates. (*) indicates values lower than 0.01% input. Data are represented as mean +/−SEM.
To control for whether repression is specific to D11.12, a construct was made that contains a fragment similar in size to D11.12 from genomic sequences also located between HOXD11 and HOXD12, but approximately 3kb away from D11.12 (Figure 3C, right panel). This region was not associated with PcG proteins in our initial ChIP-chip experiments and was not MNase hypersensitive (Figure 2). There was no significant difference between the luciferase activity of YY1pLuc and the control region (Figure 3C, left panel), suggesting that the repression of luciferase activity observed with D11.12 was localized and specific.
Analysis of the D11.12 sequence showed two notable features: a cluster of predicted YY1 binding sites and a highly-conserved region (Figure 3D, right panel). YY1 is a DNA binding protein that can recruit PcG proteins for transcriptional repression (Atchison et al., 2003). To determine whether the YY1 binding sites contribute to the repressive function of D11.12, four predicted binding sites, two of which match the extended PHO-binding motif (Oktaba et al. 2008), were mutated. The minimal YY1 binding site, GCCAT, was mutated in each case such that GCC was substituted with ATT (‘mutD11.12’). Repressive activity was mostly lost with mutD11.12, but not completely as its luciferase activity remained consistently lower than the activity from YY1pLuc. The partial repressive activity might be due to the binding of other factors to D11.12. The second notable feature of D11.12 is a 237 bp region that has a high degree of similarity across many vertebrates as evolutionarily distant as X. tropicalis. Notably, when this region is deleted from D11.12 (Δcons), there is a complete loss of D11.12 repressive activity (Figure 3D). Therefore, the conserved region appears to be required for repression of luciferase expression.
If the repression conferred by D11.12 is PcG-mediated, we would expect recruitment of PcG proteins to constructs that contained D11.12. We performed quantitative ChIPs (ChIP-qPCR) to determine whether PcG proteins were associated with the transfected DNA (Figure 3E). The regions that were tested covered the promoter/5′end of the luciferase gene (Promoter) and a site 1kb within the luciferase gene (Luciferase). The Promoter contains the TK promoter and the 5′ end of the luciferase gene as the region closest to D11.12 that could be amplified specifically on the transfected templates. We observed an enrichment of BM11 and SUZ12 as well as H3K27me3 at the promoter of the D11.12 construct and not in the pLuc or YY1pLuc constructs that lack D11.12 (Figure 3E). There were limited differences in the body of the luciferase gene. Interestingly, although SUZ12 was enriched at the promoter on mutD11.12, albeit to a lower extent than on D11.12, BMI1 could not be detected. Lower levels of H3K27me3 were detected at the promoter as well. These results, along with the partial depression of luciferase activity, suggest that the YY1 sites might be important in establishing a stable repressed state through the activities associated with BMI1. When the conserved region was deleted, BMI1, SUZ12, and H3K27m3 could no longer be detected in association with the promoter or inside the luciferase gene. The conserved region appears to be required for the recruitment and/or stability of both PRC2 and PRC1 to confer repression.
Silencing function is lost following PcG knockdown
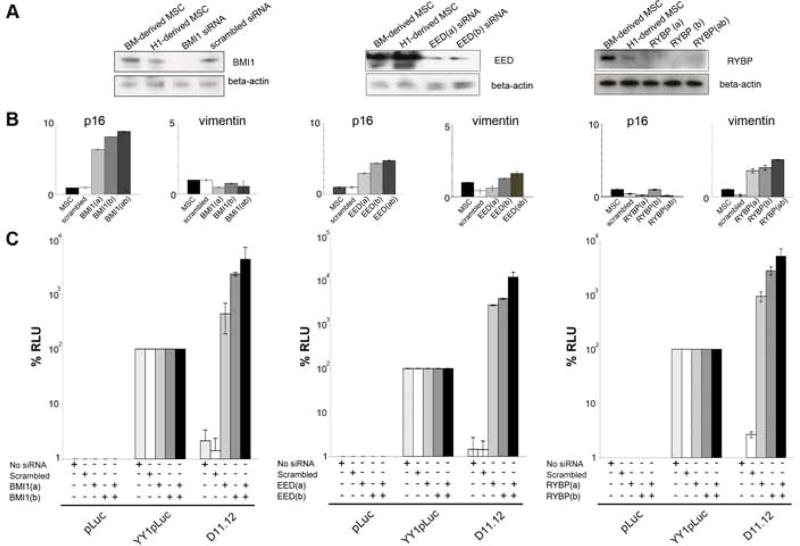
Based upon the association of BMI1 and SUZ12 with D11.12, we wanted to determine whether the repressive activity of this element was dependent upon the complexes that contain these proteins, PRC1 and PRC2. siRNA lentiviruses were used to target BMI1 (in PRC1) and EED (in PRC2). To knock down BMI1 expression in MSCs, two lentiviruses containing siRNA targeted to distinct regions of the BMI1 mRNA were used individually and in combination (referred to as BMI1(a), BMI1(b), and BMI1 (ab)). The same strategy was used to knock down EED expression in MSCs. The loss of BMI1 and EED were confirmed by western blots (Figure 4A). We infected cells with a control scrambled lentivirus construct and observed no change in BMI1 or EED expression. We surveyed HOX gene expression in the BMI1 knockdown cells and HOXD11, HOXD12, and HOXD13 were upregulated while HOXD1 and HOXD4, and HOXD10 remained unchanged (Table S2). As another functional readout, we found that p16 of the INK4A/ARF locus, a well-studied target repressed by BMI1 in dividing cells (Bruggeman et al., 2005), was upregulated in the BMI1 knockdown cells and to a lesser extent in the EED knockdown cells (Figure 4B). The expression changes in p16 in BMI1 and EED knockdown cells are similar to what was observed in hematopoietic cells (Lessard et al., 1999). We conclude that the lentiviral constructs are able to provide effective knockdown of these two PcG proteins.
Figure 4. The repressive activity of D11.12 is dependent on BMI1, EED, and RYBP.
Western blots of BMI1, EED, and RYBP knockdown in MSCs. Western blots of beta-actin were used to demonstrate equal loading of samples. (B) qRT-PCR results p16/Arf and vimentin in the 3 different BMI1 knockdown cells (Bmi1(a), Bmi1(b), Bmi1(ab), EED knockdown cells (Eed(a), Eed(b), Eed(ab)), and RYBP knockdown cells (RYBP(a), RYBP(b), RYBP(ab)). (C) Luciferase activity of pLuc, YY1pLuc, and D11.12 in the scrambled, BMI1, SUZ12, and RYBP knockdown cells. Data are represented as mean +/−SEM. See Figure S4 for HOXD11-D13 expression levels in the knockdown cells.
We tested the impact of PcG protein knockdown on repression conferred by D11.12 and measured luciferase levels. The D11.12 element repressed function in uninfected cells and those infected with the control lentivirus. Surprisingly, this repression by D11.12 was not only alleviated in the BMI1 or EED knockdown cells, but luciferase activity was nearly two orders of magnitude higher than YY1pLuc activity (Figure 4C). It appears as though D11.12 also contains sequences that lead to activation following depletion of PcG proteins.
Although we observed in the transient transfection assays that the YY1 sites are required for the recruitment of BMI1 and for the full repression of luciferase activity (Figure 3), we could not investigate the effect of YY1 in D11.12 repression by knockdown strategies because YY1 elements were also present in the enhancer region of the reporter construct thereby complicating interpretation. To address the role of the YY1 at D11.12 indirectly, RYBP was targeted for lentiviral knockdown, as it interacts with YY1 and has repressive activity in reporter assays suggesting a possible PcG connection (Garcia et al., 1999). The two lentiviruses independently and in combination effectively knocked down the RYBP protein (Figure 4A). Vimentin mRNA levels increased from an already expressed state in the MSCs although this was not known previously to be a target of RYBP, while p16 levels remained unchanged (Figure 4B). HOXD11, HOXD12, and HOXD13 were upregulated in the RYBP knockdown cells (Figure S4). In the absence of RYBP, D11.12 activated luciferase activity beyond the levels observed with YY1pLuc similar to that seen in the BMI1 knockdown cells (Figure 4C). We conclude that RYBP functions in the repression conferred by the D11.12 element, which extends previous studies showing repression by a GAL4-RYBP construct (Bejarano et al., 2005). Taken together, we conclude from these knockdown experiments that the repressive function of D11.12 is PcG-dependent.
Repressive activity of ectopic D11.12 is maintained following differentiation
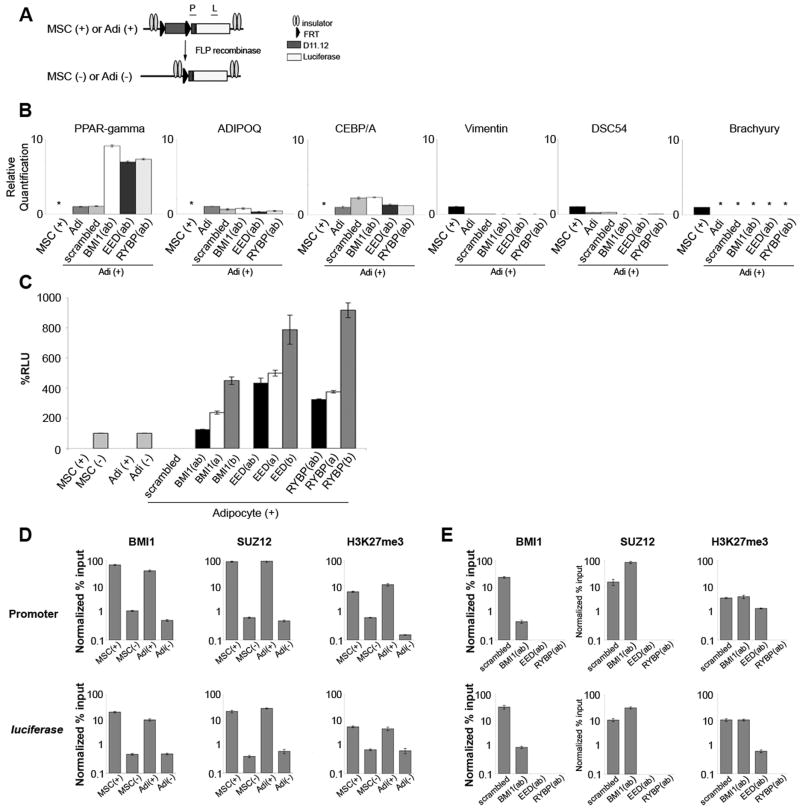
To determine if D11.12 behaves as a PcG-responsive repressor following integration into the genome, we generated MSCs that carries a stably integrated D11.12 construct (Figure 5A). Two beta-globin insulators flank the D11.12 construct to prevent position effects of neighboring elements following integration. In addition, FRT sites flanking D11.12 were inserted with the purpose of being able to excise D11.12 element. A LacZ gene and a drug selection marker on a second plasmid were integrated into the genome in parallel. ChIP-qPCR analyses on the cells carrying the D11.12 element demonstrated enrichments for BMI1, SUZ12, and H3K27me3 with the promoter and with the luciferase gene (Figure 5D). FLP recombinase was nucleofected to excise the D11.12 region and positively transfected cells were selected by dsRED expression. Thus, we could compare constructs with or without D11.12, MSC(+) and MSC(−) respectively, minimizing the possibility of effects from variables such as copy number and integration site. After approximately 14 doublings, the MSC(−) cells were analyzed by ChIP-qPCR for BMI1, SUZ12, and H3K27me3 which were not enriched in the promoter region or the luciferase gene compared to the MSC(+) results (Figure 5D). Concomitantly, luciferase readings were measured and the results were normalized with beta-galactosidase activity. The removal of D11.12 from the construct resulted in the upregulation of luciferase activity (Figure 5C). Thus the presence of D11.12 is required for the maintenance of repression in stably integrated constructs.
Figure 5. D11.12 repression is maintained when MSCs are differentiated into adipocytes.
(A) D11.12 construct in the stable cell lines. FRT sites flank D11.12 and insulators flank the D11.12 construct. (B) qRT-PCR from adipocytes (Adi(+)) derived from MSCs carrying the stably integrated D11.12 luciferase construct and from BMI1, SUZ12, and RYBP knockdown Adi(+) cells shows PPAR-gamma, AdipoQ, CEBP-alpha, vimentin, DSC54, and brachyury were similarly expressed in the adipocytes and Adi(+) cells. (*) were not detectable. (C) Luciferase results from MSC(+), MSC(−), Adi(+) and Adi(−) cells normalized by beta-galactosidase measurements. Adi(+) cells infected with scrambled, BMI1, SUZ12, and RYBP lentiviruses were assayed in parallel. Data are represented as mean +/−SEM (D) Bar graphs represent results from ChIP-qPCR experiments for MSC(+), MSC(−), Adi(+), and Adi(−) cells for BMI1, SUZ12, H3, and H3K27me3 at the promoter and the luciferase gene. ChIP results represented as Normalized % Input were normalized for the % input of histone H3. (E) ChIP-qPCR results for Bmi1, Suz12, and H3K27me3 from Adi(+) cells and lentiviral control, BMI1, SUZ12, and RYBP knockdown derivatives. Data are represented as mean +/−SEM.
One hallmark of the Drosophila PcG system is the ability to maintain repression as the embryo develops. This cellular memory is critical to advance the differentiation of cells along prescribed pathways. In order to address whether the repressive activity of D11.12 is maintained following differentiation, we differentiated the MSC(+) and MSC(−) cells into adipocytes, namely Adi(+) and Adi(−) cells. To verify differentiation, we tested Adi(+) cells for adipocyte markers, which were upregulated as expected, and for MSC markers, which were downregulated or undetectable by qRT-PCR (Figure 5B). Osteoblast markers were undetectable in the MSC(+) and Adi(+) cells (data not shown). Luciferase activity from the Adi(+) and Adi(−) cells showed that repression was observed only when D11.12 was present (Figure 5C). The Adi(−) cells displayed luciferase activity similar to the levels observed in the MSC(−) cells suggesting that the differentiation process itself has no effect on transcription or repression on the luciferase construct. Importantly, BMI1, SUZ12, and H3K27me3 remained associated with the promoter and the luciferase gene in the Adi(+) cells (Figure 5D). The Adi(−) cells showed no enrichment of BMI1, SUZ12, and H3K27me3 from either region (Figure 5D). Thus, the association of the PRC1 and PRC2 proteins and the luciferase activity is maintained when the human MSCs are differentiated into adipocytes in culture. We conclude that, like a Drosophila PRE, D11.12 can maintain a repressive state as a cell differentiates.
To determine whether the maintenance of repression in differentiated adipocytes requires PcG proteins, we used a lentivirus knockdown scheme as described above for MSCs. The loss of BMI1, EED or RYBP in the Adi(+) cells led to an upregulation of luciferase activity (Figure 5C). In addition, the knockdown of BMI1 resulted in the loss of BMI1 at D11.12 at the promoter and luciferase gene (Figure 5E). Interestingly, SUZ12 was enriched in the BMI1 knockdown cells but whether this is physiological or whether the antibody was better able to access SUZ12 without PRC1 components nearby cannot be discriminated. H3K27me3 levels did not increase despite the greater enrichment of SUZ12 and stayed similar to levels observed in the control-infected Adi(+) cells. In the EED knockdown cells, there was a complete loss of BMI1 and SUZ12 at the promoter and the luciferase gene (Figure 5E). H3K27me3 levels were reduced but remained above background levels. It is possible that while SUZ12 and BMI1 may no longer be present on D11.12, the H3K27me3 mark might persist until a demethylase removes it. In the RYBP knockdown Adi(+) cells, in addition to the loss of BMI1 and SUZ12 from the promoter and luciferase gene, there is a complete loss of the H3K27me3 mark. If RYBP is involved in the stability of PRC2 and/or PRC1 on D11.12, then the loss of the H3K27me3 mark could be explained by the inability of PRC2 to be recruited to D11.12. We conclude that the stably repressed transgene in differentiated adipocytes requires the function of PcG proteins.
Endogenous D11.12 through differentiation
The data above demonstrate that the D11.12 element is able to function as a PcG-dependent repressor when placed in a reporter construct and integrated into differentiating hESCs. This prompted us to examine the association of key components of the proposed PcG targeting and repression system on this element at the endogenous locus as hESCs differentiated. Expression from this region of the HOXD locus is repressed in the cells under study. Specifically, low levels of HOXD10, HOXD12, and HOXD13 mRNA are detectable in hESCs but are silenced when the cells differentiate into MSCs (Table S1). They continue to be repressed as the cells differentiate into adipocytes and osteoblasts. HOXD11 is not expressed in hESCs and remains silent throughout differentiation into the three cell types.
ChIP was performed for BMI1, SUZ12, YY1, H3, and H3K27me3 at the endogenous D11.12 locus and levels were quantified using qPCR. In the undifferentiated hESCs, we observed an association of the PcG proteins at D11.12 (Figure 6). However, we did not observe a hypersensitivity to MNase in these cells (Figure 2B). When the cells were differentiated into MSCs, D11.12 had a slightly greater association with Bmi1 and Suz12, MNase sensitivity, and a lower enrichment of H3. Notably, there was not a high level of H3K27me3 enrichment. There remains a debate as to whether H3K27me3 marks are in or are adjacent to PREs (Kahn et al., 2006; Ringrose et al., 2004; Wang et al., 2004). When cells are differentiated, BMI1 and SUZ12 remain present at D11.12 but at lower levels, and the H3K27me3 mark decreases. Levels of PcG proteins at PREs are known to change across differentiated cell states in Drosophila (Negre et al., 2006; Schwartz et al., 2006; Tolhuis et al., 2006). We conclude that the endogenous D11.12 locus is occupied by PcG proteins in its natural location and that the extent of association changes during differentiation.
To test whether PcG-associated DNA binding proteins are present at the endogenous D11.12, we observed YY1 binding as measured by ChIP in the hESCs, MSCs, and adipocytes. The presence of YY1 at D11.12, combined with the results from the mutD11.12 experiments, suggest that YY1 participates at D11.12 to confer PcG-mediated repression. Interestingly, in the osteoblasts, YY1 was not found to be associated with D11.12 despite the presence of BMI1 and SUZ12. It is possible that other unidentified proteins have stabilized the PRC complexes, or that they are maintained in the absence of the initial signal. Alternatively, the chromatin structure at D11.12 in the osteoblasts is in a conformation that prevents its detection.
Discussion
The D11.12 element has several characteristics of a Drosophila PRE, indicating that there is conservation of the mechanisms that target PcG function. The multiple components that combine to make a functional PRE in Drosophila are diverse and still not fully understood. While the study of mammalian PREs is in its infancy, there is reason to think that, like Drosophila, multiple components might contribute to function. We observe roles in D11.12 for a hyperconserved region, for YY1 and the interacting protein RYBP, and suggest that an NFR is also central to function.
We initially focused on D11.12 as playing a potential regulatory role due to its depletion in nucleosome occupancy in MSCs, a level of depletion that changes during differentiation (Figure 2B). It is intriguing and somewhat counter-intuitive that sequences associated with recruiting the PcG system are nucleosome depleted. Most characterized activities of the PRC1 and PRC2 families in vitro, including histone methylation, histone ubiquitylation, and chromatin compaction, involve nucleosomes. However, several studies have directly examined depletion of nucleosomes on Drosophila PREs and their association with PcG proteins (Kahn et al., 2006; Mishra et al., 2001; Mohd-Sarip et al., 2006; Papp and Muller, 2006). Dynamic accessibility of protein-binding sequences might be important for recruiting PcG complexes in vivo (reviewed in (Muller and Kassis, 2006). Recent studies suggest that in addition to nucleosome depletion, high levels of histone replacement could be observed where PcG and trxG binding sites exist (Henikoff et al., 2009; Mito et al., 2007). This suggests that PRE sequences in flies might be open and dynamic, consistent also with proposals that RNA production from these regions might be important for function (reviewed in (Schmitt and Paro, 2006)). We find that D11.12 is nuclease-sensitive and associated with the PcG proteins BMI1 and SUZ12. Nucleosome depletion might therefore play a key role mechanistically in establishing the ability to recruit PcG function to a region of the genome, explaining the apparent conservation of this feature between Drosophila and humans.
To date, there is only one known human DNA-binding protein, YY1, which has homology to one of the Drosophila proteins which functions to recruit PcG proteins at PREs. Several lines of evidence suggest that YY1 is important to D11.12 function, consistent with previous proposals based upon both functional studies and homology to PHO (Atchison et al., 2003; Srinivasan et al., 2005). It is important to note that while YY1 appears central to D11.12 function, it is unlikely that this protein (or any protein) is generally required for mammalian PRE function. In mice, the PRE-kr has a single YY1 binding site as determined by sequence analysis (Sing et al., 2009), however this YY1 binding site is not conserved in the homologous human sequence and no other apparent YY1 binding sites are present. The contribution of the YY1 binding site at the PRE-kr was not examined. We note that in reporter constructs containing D11.12, mutation of the YY1 binding sites impacts binding of BMI1, a PRC1 component, but has little impact on binding of SUZ12, a PRC2 component (Figure 3). This is consistent with models in which PRC2 is recruited prior to PRC1, and suggests that different components of D11.12 might be involved differentially in recruitment of these two complexes. YY1 interacts with RYBP, which in turn interacts with three PRC1 proteins, RING1A, RING1B and CBX2. Thus, at D11.12, YY1 might be involved primarily in PRC1 recruitment.
A highly conserved region within D11.12, which shares sequence homologies to organisms as evolutionarily different as zebrafish, is essential for repressive function. This 237 bp conserved region was required for the recruitment of both PRC1 and PRC2 components and for full repression of the reporter gene. In a search for potential regulatory sequences in the Hoxd cluster, Duboule and colleagues made knockout mice deleted of highly conserved sequences, amongst them the conserved sequence in D11.12 studied here (Beckers and Duboule, 1998). Transgenic studies determined that deletion of this conserved region impacted hoxd11 and hoxd12 expression, however knockout mice with this region deleted displayed no gross phenotype. This lack of gross phenotype might reflect redundancy in either Hox protein function or in regulatory elements with the entire hoxd cluster. These previous data are consistent with this conserved region having the potential to contribute to regulation in mice; further analysis is needed to determine whether there are contributions of the other nearby elements to function of D11.12 in the genomic context. The mouse PRE-kr element contains a conserved 450 bp sequence within the functionally defined 3kb fragment. Comparison of the conserved regions of D11.12 and PRE-kr using the TRANSFAC database revealed only conserved GAGA factor binding sites, a site defined in Drosophila that has no known binding protein in mammals. Interestingly both conserved region sequences were predicted to form NFRs when analyzed by the nucleosome occupancy feature at the UCSC Genome Browser.
The D11.12 element also contains a CpG island. We have not tested whether this is important to D11.12 function, in part because it is surrounded by key functional elements (namely, the YY1 binding sites and the conserved element), making interpretation of any deletion effect problematic. This element might contribute to the nucleosome-free nature of D11.12, as CpG islands in other areas have been shown to form nucleosomes poorly thereby generating low nucleosome occupancy (Ramirez-Carrozzi et al., 2009). It has previously been noted that there is a high correlation of PcG binding sites with CpG islands (Ku et al., 2008), leading to the proposal that these elements might be a key determinant of PRE function in mammals.
The D11.12 sequence behaves as a strong activating sequence in cells when PcG proteins are knocked down. These knockdowns therefore change the expression from the D11.12 reporter construct by several orders of magnitude in MSCs. A loss of association of the PcG proteins with the D11.12 construct in these cells might allow for the recruitment of activating factors. In Drosophila there is precedent for the same sequence being involved in repression and activation, as PRE elements overlap with Trithorax response elements involved in maintaining activation (Papp and Muller, 2006). It is possible that there is association of trxG components with D11.12 when PcG components have been removed.
A key aspect of PcG function is to maintain repression of genes as cells differentiate. It is not clear to what extent PRE sequences, as opposed to other aspects of PcG function, are required for this heritable repression. We showed that repression of an integrated reporter is maintained when MSCs are differentiated into adipocytes. In its natural location, D11.12 remains associated with PcG proteins in adipocytes, although to a lesser degree than in MSCs. In Drosophila, it is known that PcG association can be plastic during differentiation and can be impacted by local activators (Beuchle et al., 2001; Schmitt et al., 2005). A test for whether D11.12 is required for embryonic development will require that the homologous mouse sequence function in this manner, as this type of experiment would require a genetically tractable model system.
Experimental Procedures
Tissue culture
H1 and H9 hESCs (WA01 and WA09, WiCell) were maintained in hESC medium.containing 8ng/ml bFGF (Millipore). hESCs were differentiated into MSCs following a protocol described by Seda Tigli (Seda Tigli et al., 2009). MSCs were then maintained in MSCGM (Lonza).
For the generation of the stable transgenic D11.12 MSCs, cells were nucleofected with the MSC nucleofection kit (Amaxa) with the D11.12 derived from the parental luciferase constructs (Panomics) and the pVitro1 neomycin/LacZ construct (InVivoGen). Cells were placed under G418 drug selection and grown in culture for 2 months. To remove transgenic D11.12, FLP recombinase (Invitrogen) that was placed into a pIRES2-DsRed2 vector (Clontech) was transiently nucleofected into the cells. Transfected cells were sorted by DsRed to produce MSC(−) cells. The MSC(+) and (−) cells were differentiated into adipocytes.
Lentiviral siRNA knockdown
293FT cells were transfected with Lipofectamine2000 using siRNA constructs for BMI1, EED, and RYBP (Sigma). Supernatants containing virus were used to infect cells. Infected cells were selected by puromycin resistance for at least 14 days.
Expression analyses
Total RNA was isolated from cells using Trizol (Invitrogen). RNA was treated with DNaseI (Roche) before converting to double-stranded cDNA using Superscript II (Invitrogen). Inventoried primers for qRT-PCR (Applied Biosystems) and used in the Applied Biosystems 7500 System.
ChIP-chip and ChIP-qPCR
ChIPs were performed for 2 or more biological replicates. Cells were either pretreated with detergent for pre-extraction of proteins or directly crosslinked. The ChIP protocol provided by Agilent was followed. The following antibodies were used: anti-BMI1 (Kingston lab), SUZ12 (Abcam), H3 (Abcam), H3K27me3 (Abcam), YY1 (Santa Cruz). After purification, the DNA was amplified with the WGA2 Kit (Sigma) for 2 rounds. Nimblegen custom tiled microarrays were used for the mapping experiments. Q-PCR was used to analyze ChIP DNA in triplicate. For the stable and transient ChIPs, the % input of the IP (after subtraction of the rabbit IgG control IP) was normalized to the % input of the histone H3 IP. For the endogenous locus, % input was determined as above without normalization.
Western blots
Lysates were prepared using RIPA buffer and protease inhibitor cocktails (Roche) and probed with BMI1, EED, Beta-Actin (Abcam) or the RYBP antibody (Millipore) at 1:1000 and the secondary-HRP antibodies (Amersham) were at 1:10,000.
Luciferase Assay
The parental pTranslucent (pLuc, YY1pLuc) firefly luciferase constructs were used. D11.12 was inserted immediately upstream of the YY1 enhancer of the YY1pLuc construct. The Renilla luciferase plasmid (pRL-TK)(Promega) was used as the assay control. Site-directed mutagenesis of the D11.12 construct was done with the Quick-change Mutagenesis Kit (Stratagene).
Cells were co-nucleofected with a firefly luciferase plasmid and the control plasmid at a ratio of 10:1. 48h post-nucleofection, both luciferases were measured with the Dual-Luciferase Reporter Assay System (Promega). For beta-galactosidase readings, the NovaBright system was used (Invitrogen). The Monolight 3010 (Pharmigen) luminometer was used for all readings.
To account for variability between experiments, which is common when using transfection protocols, expression from the experimental construct was first normalized to pRL-TK. The RLU were further normalized by setting the value obtained with the pLuc construct to 0% RLU and that obtained with YY1pLuc to 100% RLU.
Accession Numbers
Microarray data are deposited in the NCBI GEO database with series accession number GSE19046.
Supplementary Material
Acknowledgments
We thank SK Bowman and MD Simon for discussions and critical reading of the manuscript. Funding for this work for REK and CJW was provided by the NIH (GM43901 and HG003141.) CJW was supported by NRSA GM072265. PJK and PVK were supported by NIH grant GM082798.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. Embo J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2:e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers J, Duboule D. Genetic analysis of a conserved sequence in the HoxD complex: regulatory redundancy or limitations of the transgenic approach? Dev Dyn. 1998;213:1–11. doi: 10.1002/(SICI)1097-0177(199809)213:1<1::AID-AJA1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Bejarano F, Gonzalez I, Vidal M, Busturia A. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech Dev. 2005;122:1118–1129. doi: 10.1016/j.mod.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Dejardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, Locker D, Cavalli G. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2005;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- Dennis JH, Fan HY, Reynolds SM, Yuan G, Meldrim JC, Richter DJ, Peterson DG, Rando OJ, Noble WS, Kingston RE. Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome Res. 2007;17:928–939. doi: 10.1101/gr.5636607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. Embo J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, Korsmeyer SJ. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci U S A. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- Kallin EM, Cao R, Jothi R, Xia K, Cui K, Zhao K, Zhang Y. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 2009;5:e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ. Nucleosome positioning in human HOX gene clusters. Genome Res. 2008;18:1554–1561. doi: 10.1101/gr.075952.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Paylor SW, Magnuson T, Schumacher A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development. 2006;133:4957–4968. doi: 10.1242/dev.02677. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 1999;13:2691–2703. doi: 10.1101/gad.13.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Mol Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Paro R. RNA at the steering wheel. Genome Biol. 2006;7:218. doi: 10.1186/gb-2006-7-5-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Seda Tigli R, Ghosh S, Laha MM, Shevde NK, Daheron L, Gimble J, Gumusderelioglu M, Kaplan DL. Comparative chondrogenesis of human cell sources in 3D scaffolds. J Tissue Eng Regen Med. 2009;3:348–360. doi: 10.1002/term.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta AK, Kuhrs A, Muller J. General transcriptional silencing by a Polycomb response element in Drosophila. Development. 2004;131:1959–1965. doi: 10.1242/dev.01084. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Pan X, Atchison ML. Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J Cell Biochem. 2005;96:689–699. doi: 10.1002/jcb.20562. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.