Abstract
Mitochondrial protein hyperacetylation is a known consequence of sustained ethanol consumption and has been proposed to play a role in the pathogenesis of alcoholic liver disease (ALD). The mechanisms underlying this altered acetylome, however, remain unknown. The mitochondrial deacetylase sirtuin 3 (SIRT3) is reported to be the major regulator of mitochondrial protein deacetylation and remains a central focus for studies on protein acetylation. To investigate the mechanisms underlying ethanol-induced mitochondrial acetylation, we employed a model for ALD in both wild-type (WT) and SIRT3 knockout (KO) mice using a proteomics and bioinformatics approach. Here, WT and SIRT3 KO groups were compared in a mouse model of chronic ethanol consumption, revealing pathways relevant to ALD, including lipid and fatty acid metabolism, antioxidant response, amino acid biosynthesis and the electron-transport chain, each displaying proteins with altered acetylation. Interestingly, protein hyperacetylation resulting from ethanol consumption and SIRT3 ablation suggests ethanol-induced hyperacetylation targets numerous biological processes within the mitochondria, the majority of which are known to be acetylated through SIRT3-dependent mechanisms. These findings reveal overall increases in 91 mitochondrial targets for protein acetylation, identifying numerous critical metabolic and antioxidant pathways associated with ALD, suggesting an important role for mitochondrial protein acetylation in the pathogenesis of ALD.
INTRODUCTION
Histone acetylation has been implicated in a host of diseases, ranging from inflammation to cancer.1–4 Until recently, little has been known about the role of acetylation on non-core histone proteins and it remains to be determined exactly how many proteins are regulated through acetylation. Recent studies, however, suggest that over 20% of mitochondrial proteins contain this post-translational modification (PTM).5 Protein acetylation has been shown to affect a plethora of cellular pathways including lipid and amino acid metabolism, oxidative stress, the urea cycle and cellular respiration.6–13 While the past decade has provided much insight into regulatory mechanisms of protein acetylation, the biochemical consequences and resulting physiological disease states due to altered acetylation remain unclear. Much like other PTMs, altered acetylation has been implicated in numerous disease states including cardiovascular disease, cancer, ageing, metabolic disorders and alcoholic liver disease (ALD).14–17
Protein acetylation occurs on N-ε-lysine residues and has been shown to affect protein structure, function and activity.18–20 Currently, little is known about the mechanisms involved in the acetylation of mitochondrial proteins, although current theories assume the existence of mitochondrial acetyltransferases which carry out these acetylation reactions as well as mechanisms of auto-acetylation.21 The mechanisms surrounding the removal of these PTMs, however, is well characterized and has become a focus for many researchers over the past decade. The deacetylation of N-ε-lysine residues is carried out by a family of proteins known as histone deacetylases (HDACs).22 Specifically, the HDAC class III enzymes known as Sirtuins (SIRT) are a subset of NAD+-dependent, zinc-requiring deacetylase enzymes which are known to regulate nuclear, cytosolic and mitochondrial proteins.23 Currently, the SIRT family of proteins contains 7 isoforms, identified as SIRT1-7; most notably, SIRT3-5 are located within the mitochondria and likely regulate most, if not all, protein function modulated through deacetylation; however, few direct relationships between protein acetylation and function are known.5, 24 As evidenced by publications, from 2000–2005 a mere 310 publications were present on sirtuins, whereas from 2006–2011 1322 independent publications were reported (search term “Sirtuin”, PubMed). These numbers alone provide insight into this rapidly expanding field of research.
Recent reports have described increased mitochondrial protein acetylation in rodent models of chronic ethanol consumption.25, 26 Mitochondrial dysregulation is a well-documented factor in the development and progression of ALD and protein acetylation provides a potential mechanism contributing to the observed dysfunction.27–30 Through the use of genetically modified mice, SIRT3 has been demonstrated to be the major regulating enzyme of mitochondrial protein acetylation and implicates SIRT3 in a number of disease states resulting, in part, from perturbations in mitochondrial processes.31 Given the dependence of SIRTs on NAD+ for activity, disease states, such as ALD, associated with altered metabolic NAD+/NADH ratios are a prime target for investigation. Importantly, decreases in cytosolic NAD+ are a known consequence of chronic ethanol ingestion through the oxidative metabolism of large quantities of ethanol.32, 33 These reducing equivalents are then shuttled into the mitochondria through the malate-aspartate shuttle leading to a shift in mitochondrial redox status. The oxidative metabolism of ethanol-derived acetaldehyde also utilizes mitochondrial NAD+-dependent aldehyde dehydrogenase 2 (ALDH2), further exacerbating mitochondrial NAD+ depletion.34, 35 A significant shift in the balance of NAD+/NADH during chronic ethanol consumption likely plays a role in modulating SIRT activity and may contribute to the observed increases in protein acetylation through altered SIRT3 activity.29
The initiating stages of ALD are reported to involve sustained oxidative stress and marked lipid accumulation (steatosis).36 The precise role SIRT3 plays in regulating the biochemical consequences of ethanol metabolism has not been elucidated, however, SIRT3 has been shown to regulate antioxidant enzymes such as isocitrate dehydrogenase 2 (IDH2) and superoxide dismutase (SOD2).13,37 Separate reports have shown that SIRT3 ablation and ethanol consumption both lead to increased mitochondrial protein acetylation; therefore we examined the mitochondrial acetylome as a possible mechanism contributing to the pathogenesis of ALD.7, 38 Here, we applied a well-characterized model of early-stage ALD to both WT and SIRT3 KO mice. The resulting analysis of the mitochondrial acetylome in the WT animal group reveals that ethanol-induced protein hyperacetylation targets similar proteins as the SIRT3 KO animals and impacts comparable biochemical pathways. Relevant to the pathologies associated with ALD, we have identified numerous targets for lysine acetylation associated with fatty acid and lipid metabolism, antioxidant response, electron-transport chain and amino acid biosynthesis, suggesting a central role for SIRT3 and altered protein acetylation in the pathogenesis of ALD.
EXPERIMENTAL PROCEDURES
Animal Model
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Colorado and were performed in accordance with published National Institutes of Health guidelines. Male, wild-type or SIRT3 KO C57/BL6J mice were utilized for the analysis and characterization of ethanol-mediated liver damage. SIRT3 KO mice were obtained from Dr. Eric Verdin and genotyped as previously described.6, 31 Briefly, mice were fed a modified Lieber-DeCarli liquid based-diet (Bio-Serv, Frenchtown, NJ) for 6 weeks. The diet consisted of 44% fat-derived calories, 16% protein-derived calories and the remaining balance being comprised of either carbohydrate or ethanol-derived calories. Ethanolfed mice began the study on a diet consisting of 2% (v/v) ethanol, with the ethanol-derived calories increased on a weekly basis until sacrifice; week 6 consisted of 6% ethanol (v/v) or 31.8% ethanol-derived calories. Pair-fed animals were each matched to an ethanol-fed mouse where ethanol-derived calories were replaced with calories from a carbohydrate source (maltosedextrin). Upon completion of the study, animals were anesthetized via intraperitoneal injection of sodium pentobarbital and euthanized via exsanguination. Livers were excised, weighed, and frozen for biochemical characterization, or subjected to differential centrifugation using a sucrose gradient for subcellular fractionation as previously describe.39
2-Dimensional Gel Electrophoresis of Mitochondrial Proteins
Mitochondrial fractions isolated from the livers of the wild-type and SIRT3 knockout control and ethanol-fed mice were subjected to two-dimensional gel electrophoresis on IPG strips (pH 3-11) and separated on 11cm gels (Bio-Rad, Hercules, CA) in duplicate, with each experiment repeated a minimum of 3 times. One set of gels was transferred to a Hybond-P membrane (GE Healthcare, Buckinghamshire, UK) while the others were stained utilizing Imperial Protein Stain (Pierce, Rockfor, IL). Membranes were blocked for 30 minutes with a tris-buffered saline solution containing 1% Tween-20 (TBST) and 5% non-fat dry milk (NFDM) and subsequently probed with primary antibodies directed against acetylated-lysine residues (Acetyl-Lys) (Abcam, Cambridge, MA). A horseradish peroxidase conjugated secondary (Jackson Labs, Bar Harbor, ME) was then applied and membranes were developed using ECL-Plus Reagent from GE Healthcare. Chemiluminescence was visualized using a Storm 860 scanner from Molecular Dynamics (Sunnyvale, CA). Blots were then aligned to the corresponding spots on the Imperial stained companion gels and spots were excised from the gels, digested with sequencing-grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate overnight at 37°C and the resulting peptides prepared for analysis by LC-MS/MS as previously described.40
LC-MS/MS Identification of Mitochondrial Acetylated Proteins
Protein digests from excised 2D-SDS-PAGE spots were introduced using an electrospray (ESI) source into an Agilent SL ion trap mass spectrometer (Palo Alto, CA) with an Agilent 1100 capillary HPLC system and a 1.0 × 150 mm Jupiter Proteo 90 Å column (Phenomenex, Torrance, CA). The flow rate was 5 µL/min of 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Mobile-phase composition was held at 2% solvent B for 5 min and then increased to 40% using a linear gradient over 40 min. The instrument was operated under data-dependent MS/MS conditions with helium as the collision gas. Compound lists of peptide fragment ions were generated using DataAnalysis software package (Bruker) and exported as Mascot Generic Format files and searched against the comprehensive Mass Spectrometry Database (MSDB) obtained from ftp.ncbi.nih.gov/repository/MSDB via the Mascot search engine (v 2.1.04, www.matrixscience.com). Mascot search parameters included trypsin as the digestion enzyme with 2 missed cleavages. Peptide and MS/MS tolerance were 1.2 Da and 0.6 Da, respectively. Variable modifications of cysteine carbamidomethyl, methionine oxidation and lysine acetylation were also included. Searches resulted in the assignment of a UniProt (Universal Protein Resource http://www.uniprot.org/) accession number which was used for subsequent bioinformatics analysis utilizing ExPASy (www.expasy.org).
Immunoprecipitation of acetylated peptides with anti-acetyl lysine agarose was performed to verify the results of the 2D spot picking identifications and to provide insight into the acetylation of specific lysine residues with this model (ImmuneChem Pharmaceuticals Inc., Cat# ICP0388).5 Isolated tryptic peptides were analyzed by multidimensional protein identification technology (MudPIT) by strong cation exchange and reverse-phase biphasic trapping (Integra Frit column, New Objective). Samples were injected and eluted with a series of ammonium acetate salt plugs (25 mM, 100 mM, 500 mM, 1 M) followed by a reverse-phase ramp. Nano liquid chromatography (EASY-nLC, Proxeon) at a flow rate of 300 nL/min with a gradient of 5 to 40% ACN (0.2% formic acid) over 40 min on a C18 EASY-Column analytical column (100 × 0.075 mm). The nLC was coupled to a nano-ESI source and Esquire HCT ion trap mass spectrometer (Bruker Daltonics, Inc., Billerica, MA). The instrument was operated using data-dependent CID MS/MS with a threshold of 10,000 TIC. Data analysis was performed using Mascot (v 2.1.04, www.matrixscience.com) and Esquire v5.2 data analysis package (Bruker Daltonics). Mascot was used to generate a merged file from each MudPIT analysis (1 sample injection and 4 salt plugs). The merged file was then imported into Scaffold 3.1 (Proteome Software, Inc.) in order to analyze, interpret and organize the complex 2D-LC-MS/MS MudPIT data. Only protein IDs with a probability of 80% or greater are reported, while peptide IDs required a 90% cutoff. Lysine acetylation was a required modification. Manual validation of each acetylated peptide ID was performed to discard false positives. Overall, the MudPIT experiment employed 3 levels of acetyl-Lys analysis: 1) an acetyl-Lys IP procedure, 2) Scaffold analysis using the given thresholds and 3) manual validation of the MS/MS spectra to discard false-positives. This 3rd step resulted in the removal of 4 IDs from the search results.
Immunoblotting
Mitochondrial protein was subjected to standard SDS-PAGE and transferred to a Hybond-P membrane (GE Healthcare, Buckinghamshire, UK) (n=3, biological replicates). Membranes were blocked for 30 minutes with TBST and 5% NFDM. Membranes were then probed with primary antibodies directed against Acetyl-Lys (Cell Signaling, Boston, MA), IDH2, SOD2, ALDH2 or VDAC/Porin (Abcam, Cambridge, MA). Following 3 washes with TBST, a horseradish peroxidase conjugated secondary was applied and membranes were developed as described above. In each 1-D Western blot, band intensities were quantified using ImageJ (version 1.46b) and normalized to the mitochondrial loading control VDAC/Porin to ensure equal protein load.
Immunoprecipitation of Acetylated Proteins
Mitochondrial protein (200µg) was incubated with antibodies directed against acetylated-lysine epitopes (5.0µL) overnight at 4°C. 100µL of protein G agarose (Sigma) was then applied to the samples and allowed to rotate overnight at 4°C. Supernatants were removed and the beads were washed 5 times with TBST; beads were denatured in standard loading buffer and proteins were separated via standard SDSPAGE. Immunoblotting was then carried out as outlined above for IDH2, SOD2 and ALDH2.
Statistical Analysis
Statistical analysis and generation of graphs was performed using GraphPad Prism 4.02 (GraphPad Software, San Diego, CA). Differences between the four treatment groups were assessed using a two-way ANOVA analysis with a Bonferroni post-test; statistical significance was determined if P < 0.05.
RESULTS
Mitochondrial Protein Acetylation
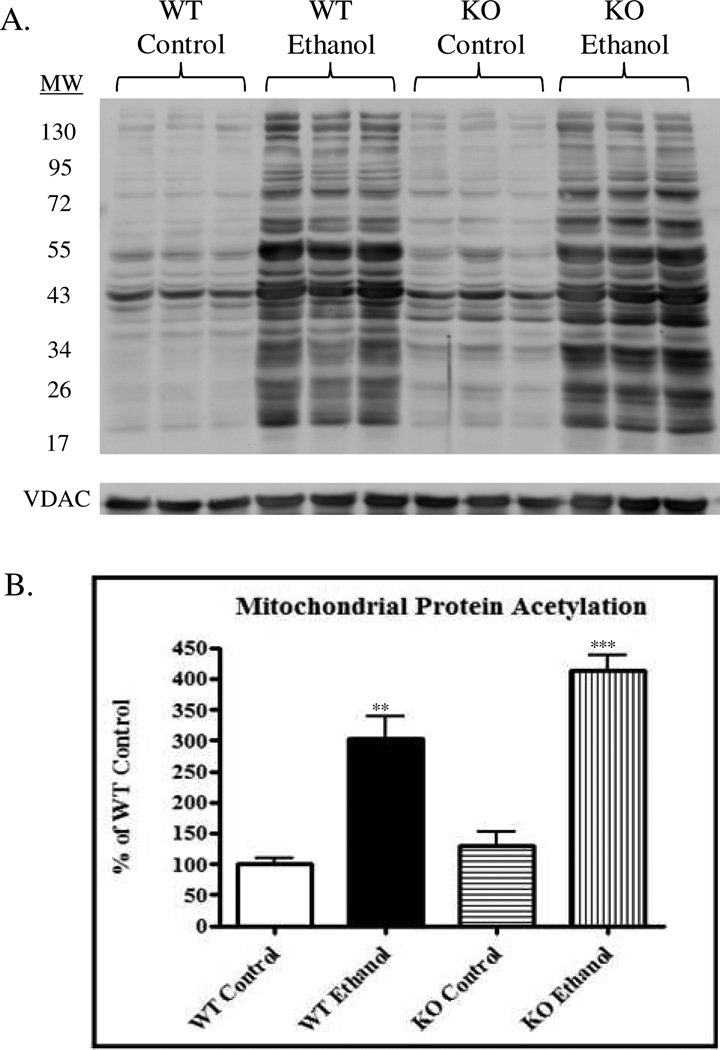
Mitochondrial protein acetylation has become an attractive target for therapeutic intervention in numerous disease states, including ALD.16, 17, 29 A recent report from our laboratory using WT mice demonstrated that increased mitochondrial protein acetylation correlated with increased liver damage in a murine model for ALD 25; the precise targets of acetylation, however, were not identified.26 Additionally, we identified increased protein carbonylation of SIRT3 in our ethanol model and demonstrated the inhibitory effects of this protein modification. To further investigate the effects of mitochondrial protein acetylation in this model, WT and SIRT3 KO mice were utilized. WT mice fed an ethanol-supplemented diet for 6 weeks had a 3-fold increase in mitochondrial protein acetylation (Figure 1) (P < 0.01). The SIRT3 KO ethanol-fed mice also revealed an increase in protein acetylation compared to the KO control-fed mice (a 4-fold increase; P < 0.001). These data further validate previous work outlining increased mitochondrial protein acetylation due to ethanol consumption.29, 30 Further analysis using a two-way ANOVA revealed no significant interaction between the genetic background and ethanol consumption. However, genetic background was found to significantly impact acetylation (P < 0.05) and was attributed to 6.93% of variance. As anticipated, ethanol consumption was found to be the major contributing factor in protein acetylation, accounting for 83.59% of variation (P < 0.001). In summary, these data confirm that mitochondrial protein acetylation is significantly increased as a consequence of ethanol consumption and SIRT3 deficiency and prompted further investigation into the acetylome in our model.
Figure 1.
Immunoblotting reveals a significant increase in mitochondrial protein acetylation following ethanol consumption in WT and SIRT3 KO mice. (A.) Acetylation of hepatic mitochondrial proteins in WT control, WT ethanol, SIRT3 KO control and SIRT3 KO ethanolfed mice. (B.) Statistical analysis determined a significant increase in ethanol-fed groups compared to control groups, n = 3, biological replicates. Two-way ANOVA reveals no significant interaction, while SIRT3 KO accounts for 6.93% (P < 0.05) of the total variance and ethanol-feeding accounts for 83.59% (P < 0.001) of the total variance.
Identification of Acetylated Proteins
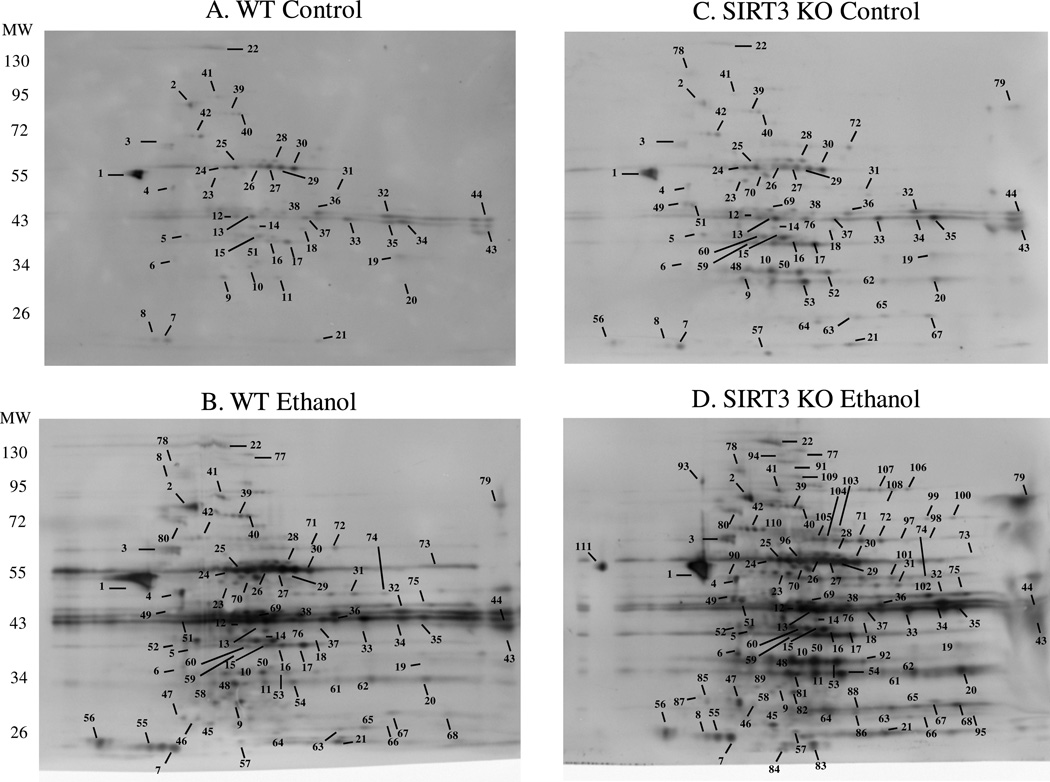
To further understand the effects of mitochondrial protein acetylation in our model, we sought to identify specific targets of acetylation in all 4 treatment groups. As shown in Figure 2, 2-dimensional gel electrophoresis, in conjunction with standard immunoblotting procedures were employed to identify target proteins of acetylation. A total of 44 immuno-positive spots were picked for identification in the WT control-fed mice (Figure 2A). Following LC-MS/MS analysis, a total of 72 proteins were identified from these spots, 46 of which were unique identifications (e.g., 46 proteins were identified in total from the 44 spots picked). The same approach was then applied to the other treatment groups; Figure 2B shows the WT ethanol-fed group, outlining 80 spots picked, yielding 128 identifications, of which 80 were unique proteins. The SIRT3 KO control-fed group is shown in Figure 2C, displaying 65 unique spots, 107 identifications and 71 unique proteins. Lastly, the SIRT3 KO ethanol-fed mice revealed 111 immunopositive spots for 177 identifications, yielding 91 unique protein identifications (Figure 2D). In order to account for a change in protein expression, imperial stained gels were visualized and are shown in Supplemental Figure 1.
Figure 2.
2-dimensional gel electrophoresis reveals numerous targets of acetylation. Numbered proteins were picked for identification utilizing LC-MS/MS in WT control (A.), WT ethanol (B.), KO control (C.), KO ethanol (D.). These proteins are identified in Table 1.
PTMs have the ability to alter the migration of a protein in a 2-dimensional gel by altering both the pI, as well as the molecular weight. We therefore included all proteins identified, which yielded numerous IDs per spot. Presented in Table 1 is the list of identified proteins along with the number of times the protein was identified for each treatment group. Following a query of ExPASy, the molecular function of each identified protein was included and is also presented in Table 1. To fully understand the intricacies of protein acetylation following ethanol consumption and SIRT3 KO, we assessed the molecular processes and biological functions of the identified proteins. Supplemental Tables 1–4 contain the specific protein identifications for each treatment group and are labeled with a “Spot #” corresponding to the protein spot number as noted in Figure 2.
Table 1.
Ethanol consumption and SIRT3 ablation reveals numerous targets of mitochondrial protein acetylation.
| Protein ID | Accession # |
WT Cont |
WT EtOH |
KO Cont |
KO EtOH |
Biological Process |
|---|---|---|---|---|---|---|
| Ornithine aminotransferase | P29758 | 0 | 1 | 1 | 2 | Amino Acid Biosynthesis |
| ATP synthase subunit alpha | Q03265 | 0 | 1 | 0 | 5 | ATP Biosynthesis |
| NADH dehydrogenase (ubiquinone) 1 alpha | Q99LC3 | 0 | 1 | 1 | 1 | Electron Transport Chain |
| Calreticulin | P14211 | 0 | 0 | 0 | 1 | Protein Folding |
| Stress-70 protein | P38647 | 1 | 1 | 1 | 1 | Protein Folding |
| Heat shock cognate 71 kDa protein | P63017 | 1 | 1 | 1 | 1 | Stress Response/Protein Folding |
| Hypoxia up-regulated protein 1 | Q9JKR6 | 0 | 1 | 1 | 1 | Stress Response/Protein Folding |
| Electron transfer flavoprotein subunit alpha | Q99LC5 | 0 | 2 | 1 | 4 | Electron Transport Chain |
| Electron transfer flavoprotein subunit beta | Q9DCW4 | 0 | 3 | 2 | 3 | Electron Transport Chain |
| Beta-lactamase-like protein 2 | Q99KR3 | 0 | 1 | 0 | 1 | Not reported |
| Valacyclovir hydrolase | Q8R164 | 0 | 0 | 0 | 1 | Not reported |
| Peroxisomal coenzyme A diphosphatase | Q99P30 | 0 | 3 | 0 | 3 | Acetyl-CoA Catabolic Process |
| 3-hydroxyisobutyryl-CoA hydrolase | Q8QZS1 | 1 | 1 | 1 | 1 | Amino Acid Catabolism |
| Fumarylacetoacetase | P35505 | 0 | 1 | 1 | 1 | Amino Acid Catabolism |
| Epoxide Hydrolase 2 | P34914 | 1 | 1 | 1 | 1 | Aromatic Hydrocarbon Catabolism |
| ATP synthase subunit beta | P56480 | 1 | 1 | 1 | 3 | ATP Biosynthesis |
| Acyl coenzyme A thioesterase 13 | Q9CQR4 | 0 | 1 | 1 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Bile acid-CoA:amino acid N-acyltransferase | Q91X34 | 0 | 1 | 1 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Fructose-1,6-bisphosphatase 1 | Q9QXD6 | 0 | 1 | 1 | 1 | Gluconeogenesis |
| Cathepsin B | P10605 | 0 | 0 | 0 | 1 | Regulation of Catalytic Activity |
| ATP synthase subunit d | Q9DCX2 | 2 | 2 | 2 | 2 | ATP Biosynthesis |
| Delta (3,5)-Delta (2,4)-dienoyl-CoA isomerase | O35459 | 1 | 1 | 1 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Alpha-methylacyl-CoA racemase | O09174 | 1 | 1 | 1 | 1 | Isoprenoid Catabolic Process |
| Adenylate kinase 2 | Q9WTP6 | 0 | 1 | 1 | 2 | ATP Biosynthesis |
| Acyl-CoA synthetase family member 2 | Q8VCW8 | 0 | 0 | 0 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Propionyl-CoA carboxylase alpha chain | Q91ZA3 | 0 | 0 | 0 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Pyruvate carboxylase | Q05920 | 1 | 2 | 1 | 3 | Gluconeogenesis/Lipid Metabolism |
| Succinyl-CoA ligase [ADP-forming] subunit beta | Q9Z219 | 0 | 1 | 1 | 1 | Proteolysis |
| Succinyl-CoA ligase [GDP-forming] subunit beta | Q9Z218 | 0 | 1 | 1 | 1 | Proteolysis |
| Carbamoyl phosphate synthase | Q8C196 | 1 | 2 | 1 | 3 | Urea Cycle |
| Arginosuccinate sythase | P16460 | 0 | 1 | 1 | 1 | Urea Cycle/Amino Acid Biosynthesis |
| Citrate Lyase subunit beta-like protein | Q8R4N0 | 1 | 1 | 1 | 1 | Cellular Aromatic Compound Metabolic Process |
| 2-hydroxyacyl-CoA lyase 1 | Q9QXE0 | 1 | 1 | 1 | 2 | Fatty Acid Metabolism |
| Enoyl-CoA hydratase | Q8BH95 | 0 | 1 | 0 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Enoyl-CoA hydratase domain-containing protein 2 | Q3TLP5 | 0 | 1 | 1 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Glutamine synthetase | P15105 | 0 | 1 | 1 | 1 | Glutamine Biosynthetic Process |
| Carbonic anhydrase 3 | P16015 | 0 | 2 | 2 | 2 | One-Carbon Metabolic Process |
| Aconitase hydratase | Q99KI0 | 0 | 0 | 0 | 2 | TCA Cycle |
| Cytochrome b-c1 complex subunit 1 | Q9CZ13 | 1 | 2 | 2 | 3 | Electron Transport/Respiratory Chain |
| Myelin basic protein | P04370 | 1 | 2 | 1 | 2 | Myelination |
| 2-oxoisovalerate dehydrogenase subunit beta | Q6P3A8 | 0 | 1 | 0 | 1 | Amino Acid Catabolism |
| 4-hydroxyphenylpyruvate dioxygenase | P49429 | 1 | 1 | 1 | 1 | Amino Acid Catabolism |
| Dimethylglycine dehydrogenase | Q9DBT9 | 2 | 2 | 2 | 2 | Amino Acid Catabolism/Oxidation-Reduction |
| Sarcosine dehydrogenase | Q99LB7 | 1 | 1 | 1 | 2 | Amino Acid Catabolism/Oxidation-Reduction |
| Superoxide dismutase [Mn] | P09671 | 1 | 1 | 1 | 1 | Antioxidant |
| Glutamate dehydrogenase 1 | P26443 | 3 | 3 | 3 | 4 | Cellular Amino Acid Metabolism |
| NADH-cytochrome b5 reductase 3 | Q9DCN2 | 0 | 1 | 1 | 1 | Cholesterol Biosynthesis/Lipid Metabolism |
| Dihydrolipoyl dehydrogenase | O08749 | 1 | 1 | 1 | 1 | Electron Transport |
| Cytochrome b-c1 complex subunit Rieske | Q9CR68 | 0 | 0 | 0 | 1 | Electron Transport Chain |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | Q921G7 | 0 | 0 | 0 | 1 | Electron Transport Chain |
| Succinate dehydrogenase [ubiquinone] flavoprotein | Q8K2B3 | 0 | 1 | 0 | 1 | Electron Transport Chain/TCA Cycle |
| NADH dehydrogenase [ubiquinone] flavoprotein 2 | Q9D6J6 | 0 | 1 | 0 | 1 | Electron Transport/Respiratory Chain |
| Hydroxyacyl-coenzyme A dehydrogenase | Q61425 | 1 | 2 | 1 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Long-chain specific acyl-CoA dehydrogenase | P51174 | 1 | 2 | 2 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Medium-chain specific acyl-CoA dehydrogenase | P45952 | 4 | 4 | 4 | 4 | Fatty Acid Metabolism/Lipid Metabolism |
| Peroxisomal acyl-coenzyme A oxidase | Q9R0H0 | 1 | 3 | 2 | 5 | Fatty Acid Metabolism/Lipid Metabolism |
| Short-chain specific acyl-CoA dehydrogenase | Q07417 | 2 | 2 | 2 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Glycerol-2-phosphate dehydrogenase | P13707 | 0 | 1 | 1 | 1 | Gluconeogenesis |
| Pyruvate dehydrogenase E1 component subunit beta | Q9D051 | 1 | 1 | 1 | 1 | Glycolysis |
| Coproporphyrinogen-III oxidase | P36552 | 1 | 1 | 1 | 1 | Heme Biosynthesis/Porphyrin Biosynthesis |
| Aldehyde dehydrogenase | P47738 | 4 | 5 | 5 | 6 | Oxidation-Reduction Process |
| Alpha-aminoadipic semialdehyde dehydrogenase | Q9DBF1 | 1 | 1 | 1 | 2 | Oxidation-Reduction Process |
| Delta-1-pyrroline-5-carboxylate dehydrogenase | Q8CHT0 | 0 | 1 | 1 | 2 | Oxidation-Reduction Process |
| Hydroxyacid-oxoacid transhydrogenase | Q8R0N6 | 1 | 1 | 1 | 1 | Oxidation-Reduction Process |
| Isovaleryl-CoA dehydrogenase | Q9JHI5 | 1 | 1 | 1 | 1 | Oxidation-Reduction Process |
| Methylmalonate-semialdehyde dehydrogenase | Q9EQ20 | 1 | 1 | 1 | 1 | Oxidation-Reduction Process |
| Uricase | P25688 | 3 | 4 | 4 | 6 | Purine Metabolism |
| Isocitrate dehydrogenase [NAD] subunit apha | Q9D6R2 | 1 | 1 | 1 | 1 | TCA Cycle |
| Malate dehydrogenase | P14152 | 1 | 1 | 1 | 1 | TCA Cycle |
| Isocitrate dehydrogenase [NADP] | P54071 | 2 | 2 | 2 | 2 | TCA Cycle/Stress Response |
| Dihydropteridine reductase | Q8BVI4 | 0 | 1 | 1 | 1 | Tetrahydrobiopterin Biosynthesis |
| Peroxisomal bifunctional enzyme | Q9DBM2 | 0 | 1 | 1 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Peroxisomal multifunctional enzyme type 2 | P51660 | 1 | 1 | 2 | 2 | Fatty Acid Metabolism/Lipid Metabolism |
| Trifunctional enzyme subunit alpha | Q8BMS1 | 0 | 1 | 1 | 1 | Fatty Acid Metabolism/Lipid Metabolism |
| Catalase | P24270 | 0 | 1 | 0 | 5 | Antioxidant |
| Glutathione peroxidase 1 | P11352 | 0 | 1 | 1 | 1 | Antioxidant |
| Peroxiredoxin-1 | P35700 | 0 | 0 | 0 | 2 | Antioxidant |
| Voltage-dependent anion-selective channel protein 1 | Q60932 | 1 | 2 | 2 | 2 | Ion Transport |
| Betaine homocysteine S-methyltransferase | O35490 | 2 | 5 | 2 | 5 | Amino Acid Biosynthesis |
| Ornithine carbamoyltransferase | P11725 | 2 | 2 | 2 | 2 | Amino Acid Biosynthesis |
| Glutathione S-transferase Mu 1 | P10649 | 0 | 1 | 0 | 2 | Antioxidant |
| Hydroxymethylglutaryl-CoA synthase | P54869 | 1 | 3 | 2 | 6 | Cholesterol Biosynthesis/Lipid Metabolism |
| 3-ketoacyl-CoA thiolase | Q8BWT1 | 6 | 6 | 6 | 6 | Fatty Acid Metabolism/Lipid Metabolism |
| Acetyl-CoA acetyltransferase | Q8QZT1 | 6 | 6 | 6 | 6 | Ketone Body Metabolism |
| Aspartate aminotransferase | P05202 | 1 | 1 | 1 | 1 | Lipid Transport |
| non-specific lipid-transfer protein | P32020 | 0 | 2 | 2 | 4 | Lipid Transport |
| 4-aminobutyrate aminotransferase | P61922 | 0 | 0 | 0 | 1 | Neurotransmitter Catabolic Process |
| 3-Mercaptopyruvate sulfurtransferase | Q99J99 | 1 | 1 | 1 | 1 | Sulfate Transport |
| Thiosulfate sulfurtransferase | P52196 | 0 | 0 | 0 | 1 | Sulfate Transport |
| Citrate synthase | Q9CZU6 | 1 | 1 | 1 | 1 | TCA Cycle |
| 72 | 128 | 107 | 177 | |||
Acetylated peptides identified using MudPIT analysis are included in Supplemental Tables 5 and 6. Supplemental Table 5 reports the number of acetylated peptides identified per protein for each treatment group; Supplemental Table 6 shows the acetylated peptide identified for each reported protein. In aggregate, the MudPIT analysis identified 116 proteins and confirmed 56 of the 90 IDs obtained by 2D comparative western analysis. Of the 33 protein IDs not verified by MudPIT, 18 are reported as targets of protein acetylation.5, 41 When compared with the MS IDs by comparative Western blot spot picking, these results provide confirmation that ethanolinduced protein hyperacetylation occurs through both ethanol-dependent and SIRT3 KO-associated mechanisms. It is important to note, however, the global scale by which both ethanol consumption and SIRT3 ablation alter mitochondrial protein acetylation, as Western blot analysis via anti-acetyl Lys antibody clearly demonstrate protein target similarities (Figure 2).
Cellular Effects of Mitochondrial Protein Acetylation
As indicated, this model for early-stage ALD results in a substantial increase in acetylated proteins; these effects were found to be exacerbated following SIRT3 ablation. Our acetylomic approach using comparative western blot analysis and MS identification revealed 178 total proteins, resulting in 91 unique protein identifications. As shown in Table 1, the same proteins were identified on multiple occasions. To account for these redundancies, list exclusion was performed; these results are presented in Table 1 showing both the number of proteins identified, as well as the number of unique protein identifications. This then allowed for the assessment of acetylation on a global scale, focusing on both molecular function, as well as biological processes, while not focusing on specific targets of acetylation. To characterize the effects of chronic ethanol consumption and SIRT3 ablation on mitochondrial protein acetylation in our mouse model for early-stage ALD, Expasy was queried to assess the molecular function and biological processes associated with our list of acetylated proteins. As listed in Table 1, a total of 17 molecular functions were associated with mitochondrial protein acetylation. Not surprisingly, given the nature of the mitochondria, oxidoreductase-like functions represented the greatest occurrence (37 proteins).
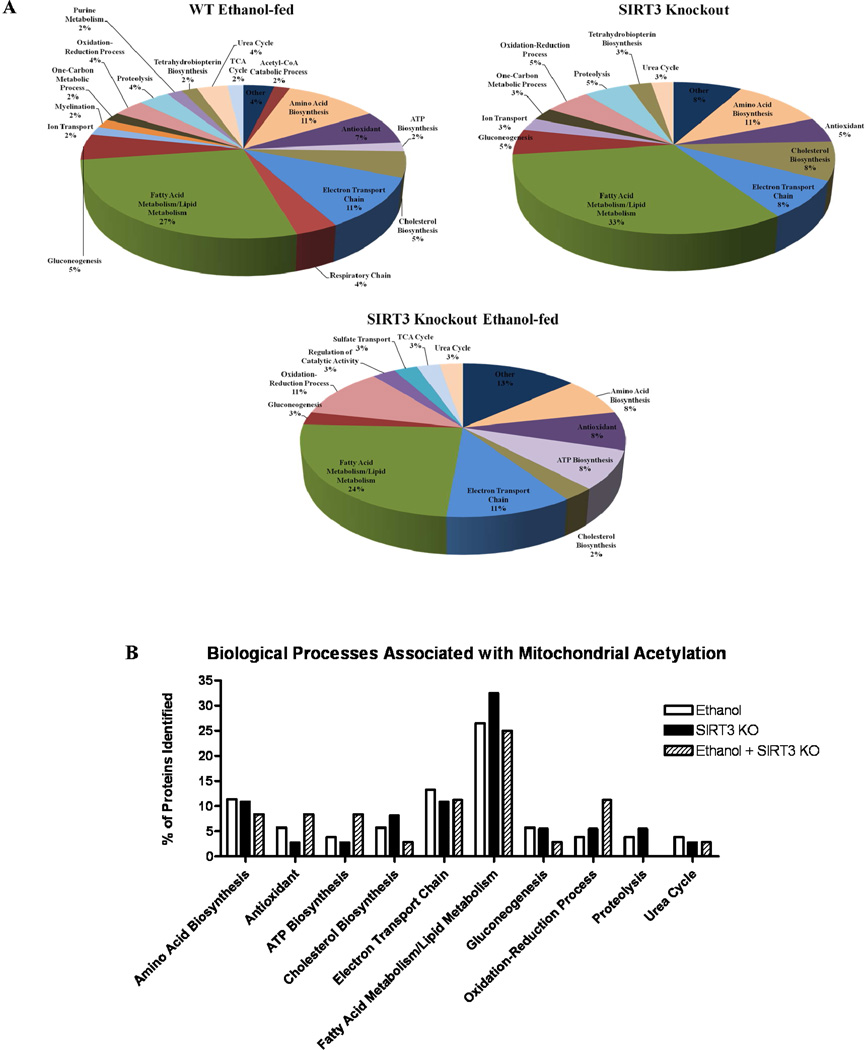
To further understand the mechanisms behind acetylation in the pathogenesis of ALD, we assessed the biological function of the identified proteins; Table 1 provides a composite of these findings. A total of 55 proteins displayed increased acetylation following ethanol consumption in WT mice. In comparison, 36 proteins were hyperacetylated in the control SIRT3 KO mice. Finally, a total of 51 proteins displayed hyperacetylation following ethanol consumption in SIRT3 KO mice. Utilizing list exclusion, we first assessed the impact of acetylation between our WT control and ethanol-fed mice. As illustrated in Figure 3A, sustained ethanol consumption affects a host of biological processes in the mitochondria. Relevant to the pathogenesis of ALD, proteins associated with lipid and fatty acid metabolism (15 proteins; 27% of proteins in this list), antioxidant response (4 proteins; 7%), amino acid biosynthesis (6 proteins; 11%) and the electron-transport chain (6 proteins, 11%) were all found to be affected by ethanol-induced hyperacetylation. When comparing WT and SIRT3 KO control-fed mice, these pathways were found to be acetylated in a similar manner, with lipid and fatty acid metabolism (12 proteins; 33%), antioxidant response (2 proteins; 5%), amino acid biosynthesis (4 proteins; 11%) and the electron-transport chain (3 proteins, 8%) all affected (Figure 3A). Lastly, the WT control and SIRT3 KO ethanol-fed groups were compared, revealing a similar trend, with lipid and fatty acid metabolism (9 proteins; 24%), antioxidant response (3 proteins; 8%), amino acid biosynthesis (3 proteins; 8%) and the electron-transport chain (4 proteins, 11%) (Figure 3A). It should be noted that although the percentage of these identifications was not found to alter drastically between treatment groups, this was largely due to an increase in identifications following SIRT3 KO. A direct comparison of biological processes associated with ALD are presented in Figure 3B. The precise impact of acetylation on these processes remains to be elucidated; however, this proteomic survey provides attractive avenues to assess the impact of acetylation on the regulation of oxidative stress and lipid metabolism following chronic ethanol consumption.
Figure 3.
Ethanol consumption and SIRT3 KO lead to altered protein acetylation in numerous biological processes. (A.) Protein targets identified in Figure 2 outline the effects of sustained ethanol consumption in WT Ethanol-fed and SIRT3 KO mice. (B.) A direct comparison of the percent of proteins identified for each biological process demonstrates an impact on similar pathways (i.e. Electron Transport Chain).
Increased Acetylation of Oxidative Stress Related Proteins
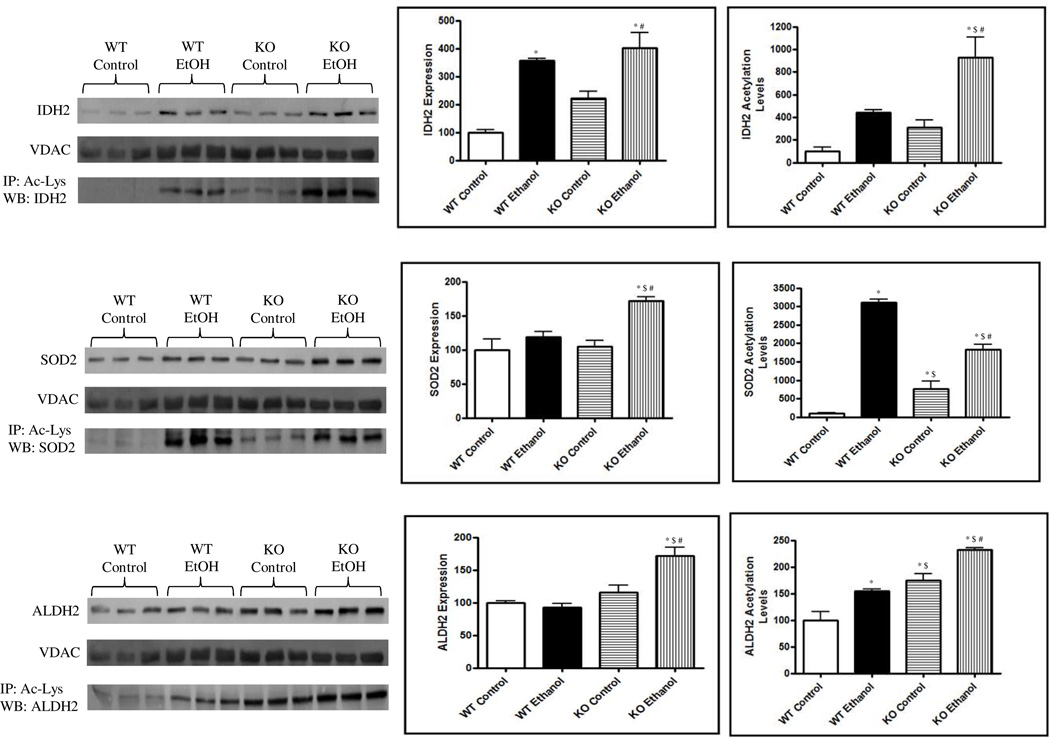
As previously indicated, oxidative stress is a known result of sustained ethanol consumption.42 Critical components of cellular antioxidant capacity were identified, including SOD2, IDH2 and ALDH2. The enzymatic capacity of SOD2 and IDH2 and their role in maintaining basal oxidant levels has recently been demonstrated to be regulated, in part, through protein acetylation.7, 13, 37, 43, 44 Figure 4a and 4b details the relative expression of IDH2, SOD2 and ALDH2 following ethanol feeding and SIRT3 ablation. The expression of IDH2 was found to be significantly induced following ethanol consumption and SIRT3 KO (P < 0.01); these effects were increased when combining SIRT3 KO and ethanol consumption (P < 0.001). SOD2 and ALDH2, however, were only found to be significantly induced in the ethanol consuming SIRT3KO mice (P < 0.01). To assess the acetylation status of these proteins, immunoprecipitation was performed utilizing acetyl-lys antibodies followed by standard Western blotting for IDH2, SOD2 and ALDH2. While IDH2 and ALDH2 acetylation appears consistent when normalized to protein expression, SOD2 acetylation is significantly increased when compared to expression levels. This acetylomic survey of ALD and SIRT3 ablation provides an important mechanistic link by which redox balance in the hepatic mitochondria is affected through ethanol consumption and altered protein acetylation, directly implicating a mechanistic role for SIRT3 in the development of early-stage ALD.
Figure 4.
Ethanol consumption and/or SIRT3 KO lead to a significant increase in the acetylation of IDH2, SOD2 and ALDH2. Standard immunoblotting reveals a significant increase in IDH2 expression in ethanol-fed groups. SOD2 and ALDH2 expression was only significantly increased in the SIRT3KO, ethanol-consuming mice. Bands were normalized to VDAC and are expressed as a % of WT control. Immunoprecipitation with anti-acetyl-lys antibodies reveals varying altered acetylation patterns of IDH2, SOD2 and ALDH2. Bands were quantified, adjusted for protein expression and are presented as a % of WT control. * P < 0.01 from WT control; $ P < 0.01 from KO control; # P < 0.01 from WT ethanol.
DISCUSSION
The regulation of protein function through acetylation is now widely recognized as a major factor in cellular responses mediated through pathological situations.14 Altered protein acetylation as a consequence of chronic ethanol consumption is well documented; however, the underlying mechanisms resulting in this hyperacetylation and resulting pathophysiological impact relating to ALD remain unknown.25, 29, 30, 45, 46 Our acetylome analysis reveals that both ethanol consumption and SIRT3 ablation contribute to the observed increases in acetylation; however, ethanol accounts for the majority of this increase. We also demonstrate that numerous metabolic and antioxidant pathways are impacted through acetylation in both ethanol-fed and SIRT3-KO mice. Furthermore, this effect was found to be visibly increased, as ethanol-fed SIRT3-KO mice displayed the highest level of protein acetylation, which suggests ethanolinduced hyperacetylation may include both SIRT3-targetted and non-SIRT3-targetted pathways. Other factors such as increased acetyl-CoA, acetate and acetyltransferase activity provide a likely hypothesis for the observed non-SIRT3 related hyperacetylation.29, 30, 47–49
Alcohol-induced oxidative stress is a direct result of ethanol metabolism through a number of pathways regulated by microsomal and mitochondrial systems.50, 51 The generation of reactive oxygen species (ROS) and depletion of cellular glutathione creates a well-documented prooxidant environment that results in damage to both protein and lipid membranes. Our findings reveal that ethanol-induced protein acetylation may play a crucial role in a number of biological processes associated with antioxidant, fatty acid and lipid metabolism, electron transport and stress responses. Specifically, IDH2, SOD2 and ALDH2, identified in this report, are known to play important roles in the clearance of ROS and may provide a mechanistic link between altered acetylation and the progression of ALD. A review by Bell et al. highlights a protective role for SIRT3 against mitochondrially derived ROS.52 Ethanol-derived superoxides (O2−) are metabolized through SIRT3 via SOD2 detoxification to hydrogen peroxide (H2O2), which is then converted to water by glutathione peroxidase (GPX).52 GPX activity requires reduced glutathione which is supplied by glutathione reductase (GSR). The enzymatic activity of GSR requires NADPH which is generated via IDH2 activity.52 Indeed, IDH2 is implicated in the regulation of ethanol-induced toxicity through the generation of NADPH.38, 53 SIRT3 has been shown to directly regulate IDH2 and SOD2 activities through deacetylation, as hyperacetylation of each is known to be inhibitory.7, 12, 13, 37, 44, 54 As a key enzyme involved in the oxidative metabolism of ethanol-derived acetaldehyde, altered ALDH2 acetylation may play a functional role in the initiation of ALD. The acetylation of ALDH2 was recently described in a model of acetaminophen induced liver damage, however, the precise role that acetylation plays on ALDH2 activity remains to be characterized.55 These pathways are highlighted in Figure 5, and represent a proposed role for protein acetylation in both antioxidant responses as well as lipid accumulation. Future studies should aim to examine the impact of ethanol-induced hyperacetylation on IDH2, SOD2 and ALDH2 activities and confirm which specific residues are responsible for altered activity.
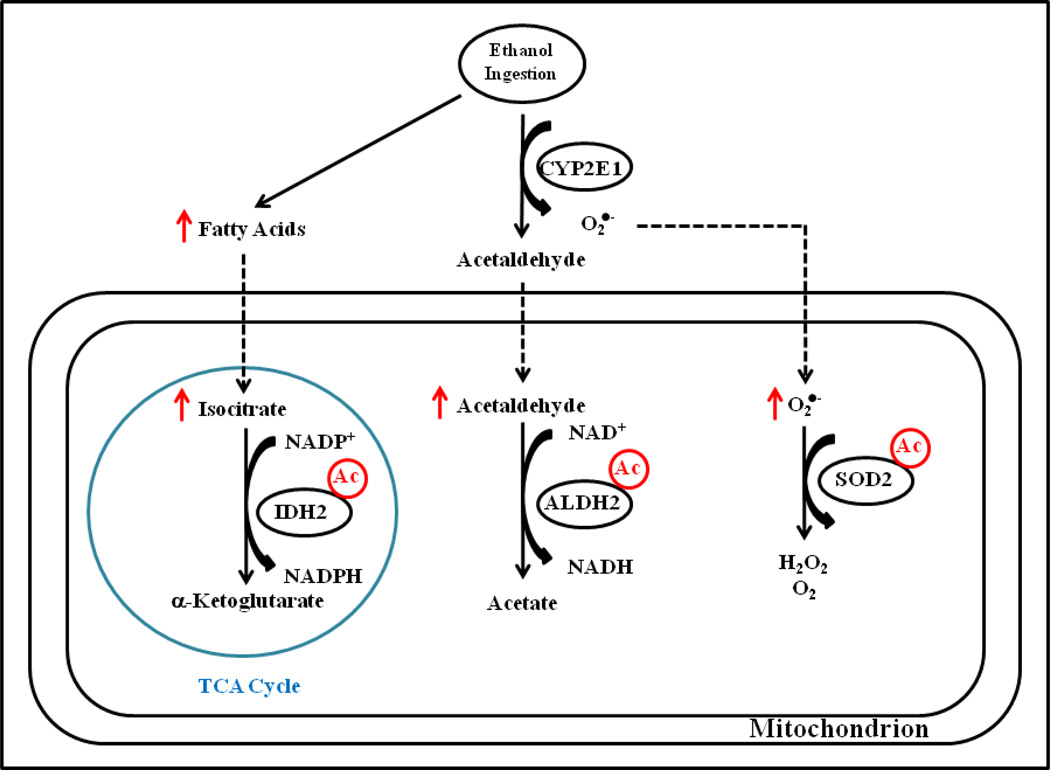
Figure 5.
Ethanol consumption and SIRT3 KO results in the hyperacetylation of key antioxidant proteins. Figure 5 presents a proposed mechanism for consequences of IDH2, ALDH2 and SOD2 hyperacetylation. Inhibition of SIRT3 leads to hyperacetylation of IDH2, leading to altered NADP+/NADPH ratio as well as increased fatty acid production due to a dysregulation in the TCA cycle. Increased acetylation of ALDH2 likely leads to a decrease in the detoxification of acetaldehyde, leading to increased aldehyde-associated ROS generation. Acetylation of SOD2 also leads to a decrease in activity, resulting in increased superoxide and further oxidative stress.
Among the biological processes we found to contain proteins with altered acetylation status, cholesterol biosynthesis, lipid metabolism, fatty acid metabolism and lipid transport are of significant interest to the pathologies associated with ALD. The first pathological indication of ALD is the accumulation of lipid droplets within the liver, termed steatosis. SIRT3 activity has been implicated in reducing lipid accumulation through adenosine monophosphate kinase (AMPK) and acetyl-CoA carboxylase (ACC).56 Furthermore, a recent study reports that SIRT3 regulates the activity of a central mitochondrial fatty acid oxidation enzyme, long chain acyl-CoA dehydrogenase (LCAD).6 Here, our findings identify the acetylation of LCAD as well as other family members, medium chain acyl-CoA dehydrogenase (MCAD) and short-chain acyl-CoA dehydrogenase (SCAD). Additionally, our results support the recent findings by Hallows et al. that SIRT3 may potentially modulate β-oxidation through short-chain L-3-hydroxyacyl-CoA dehydrogenase (SCHAD), short/branched chain acyl-CoA dehydrogenase (ACADSB) and very-long-chain acyl-CoA dehydrogenase (VLCAD).57 A host of proteins central to lipid metabolism are identified in Table 1 and provide numerous mechanistic targets for the regulation of fatty acid oxidation by SIRT3. Further investigation is required to elucidate mechanisms of ethanolinduced hyperacetylation through mechanisms independent and dependent of SIRT3 action.
A number of proteomics studies have identified non-nuclear protein targets of lysine acetylation and reveal that this modification is highly conserved and more abundant than previously recognized. The continued characterization of protein acetylation is likely to provide numerous target pathways for investigation; leading to the identification of biochemical implications of these acetylation events. Utilizing models for hyperacetylation such as sustained ethanol-feeding and SIRT3 KO mice, it remains possible to identify novel targets for this PTM. Interestingly, a recent report highlights what little understanding exists regarding the role of SIRT3 in various pathologies, leading the authors to state, “SIRT3 might act as a double-edged sword, a word of caution regarding therapeutic strategies aimed at potentiating SIRT3 activity”.58 Future studies will reveal a crucial role for SIRT3 in numerous disease pathologies, including ALD. Using our 2D spot-picking analysis, our findings have revealed 91 mitochondrial targets for protein acetylation in a model of early-stage ALD. These proteins have been shown to play major roles in multiple biological processes and highlight a number of pathologies associated with the initiation and progression of the ALD.
Supplementary Material
ACKNOWLEDGEMENTS
Studies were supported by the National Institutes of Health/National Institutes of Alcoholism and Alcohol Abuse under grant numbers R37AA09300 (DRP), R01DK074487-01 (DRP) and F31AA018606-01 (JJG).
Footnotes
Supporting Information Available: Supplemental material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90(1):9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atadja PW. HDAC inhibitors and cancer therapy. Prog Drug Res. 2011;67:175–195. doi: 10.1007/978-3-7643-8989-5_9. [DOI] [PubMed] [Google Scholar]
- 3.Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- 4.Ghizzoni M, Haisma HJ, Maarsingh H, Dekker FJ. Histone acetyltransferases are crucial regulators in NF-kappaB mediated inflammation. Drug Discov Today. 2011;16(11–12):504–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallows WC, Smith BC, Lee S, Denu JM. Ure(k)a! Sirtuins Regulate Mitochondria. Cell. 2009;137(3):404–406. doi: 10.1016/j.cell.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105(38):14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382(3):790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 13.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Close P, Creppe C, Gillard M, Ladang A, Chapelle JP, Nguyen L, Chariot A. The emerging role of lysine acetylation of non-nuclear proteins. Cell Mol Life Sci. 2010;67(8):1255–1264. doi: 10.1007/s00018-009-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7(10):841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 16.Alcain FJ, Villalba JM. Sirtuin activators. Expert Opin Ther Pat. 2009;19(4):403–414. doi: 10.1517/13543770902762893. [DOI] [PubMed] [Google Scholar]
- 17.Alcain FJ, Villalba JM. Sirtuin inhibitors. Expert Opin Ther Pat. 2009;19(3):283–294. doi: 10.1517/13543770902755111. [DOI] [PubMed] [Google Scholar]
- 18.Sauve AA. Sirtuin chemical mechanisms. Biochim Biophys Acta. 2010;8:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35(12):669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286(28):24694–24701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20(8):615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7(2):104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804(8):1645–1651. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376(3):615–619. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 26.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem Res Toxicol. 2011;24(5):651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32(1):11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 28.Mantena SK, King AL, Andringa KK, Eccleston HB, Bailey SM. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic Biol Med. 2008;44(7):1259–1272. doi: 10.1016/j.freeradbiomed.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepard BD, Tuma PL. Alcohol-induced protein hyperacetylation: mechanisms and consequences. World J Gastroenterol. 2009;15(10):1219–1230. doi: 10.3748/wjg.15.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Exp Res. 2010;34(2):280–291. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber CS. Metabolism of alcohol. Clin Liver Dis. 2005;9(1):1–35. doi: 10.1016/j.cld.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Sastre J, Serviddio G, Pereda J, Minana JB, Arduini A, Vendemiale G, Poli G, Pallardo FV, Vina J. Mitochondrial function in liver disease. Front Biosci. 2007;12:1200–1209. doi: 10.2741/2138. [DOI] [PubMed] [Google Scholar]
- 34.Lieber CS. Clinical biochemistry of alcohol and its metabolic and hepatic effects. Journ Annu Diabetol Hotel Dieu. 1992:183–210. [PubMed] [Google Scholar]
- 35.Veech RL, Guynn R, Veloso D. The time-course of the effects of ethanol on the redox and phosphorylation states of rat liver. Biochem J. 1972;127(2):387–397. doi: 10.1042/bj1270387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramaiah S, Rivera C, Arteel G. Early-phase alcoholic liver disease: an update on animal models, pathology, and pathogenesis. Int J Toxicol. 2004;23(4):217–231. doi: 10.1080/10915810490502069. [DOI] [PubMed] [Google Scholar]
- 37.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3(2):102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang ES, Park JW. Regulation of ethanol-induced toxicity by mitochondrial NADP(+)-dependent isocitrate dehydrogenase. Biochimie. 2009;91(8):1020–1028. doi: 10.1016/j.biochi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Roede JR, Stewart BJ, Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45(11):1551–1558. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 40.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In vitro and in silico characterization of peroxiredoxin 6 modified by 4-hydroxynonenal and 4-oxononenal. Chem Res Toxicol. 2008;21(12):2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 41.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 42.Vidali M, Stewart SF, Albano E. Interplay between oxidative stress and immunity in the progression of alcohol-mediated liver injury. Trends Mol Med. 2008;14(2):63–71. doi: 10.1016/j.molmed.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Yang ES, Lee JH, Park JW. Ethanol induces peroxynitrite-mediated toxicity through inactivation of NADP+-dependent isocitrate dehydrogenase and superoxide dismutase. Biochimie. 2008;90(9):1316–1324. doi: 10.1016/j.biochi.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieber CS, Leo MA, Wang X, Decarli LM. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373(2):246–252. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370(1):44–48. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Niemela O. Aldehyde-protein adducts in the liver as a result of ethanol-induced oxidative stress. Front Biosci. 1999;4:506–513. doi: 10.2741/niemela. [DOI] [PubMed] [Google Scholar]
- 48.Kendrick SF, O'Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, Day CP. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51(6):1988–1997. doi: 10.1002/hep.23572. [DOI] [PubMed] [Google Scholar]
- 49.Dormeyer W, Ott M, Schnolzer M. Probing lysine acetylation in proteins: strategies, limitations, and pitfalls of in vitro acetyltransferase assays. Mol Cell Proteomics. 2005;4(9):1226–1239. doi: 10.1074/mcp.M500047-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Albano E. Alcohol, oxidative stress and free radical damage. Proc Nutr Soc. 2006;65(3):278–290. doi: 10.1079/pns2006496. [DOI] [PubMed] [Google Scholar]
- 51.Das SK, Vasudevan DM. Alcohol-induced oxidative stress. Life Sci. 2007;81(3):177–187. doi: 10.1016/j.lfs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Bell EL, Guarente L. The SirT3 Divining Rod Points to Oxidative Stress. Mol Cell. 2011;42(5):561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang ES, Lee SM, Park JW. Silencing of cytosolic NADP+-dependent isocitrate dehydrogenase gene enhances ethanol-induced toxicity in HepG2 cells. Arch Pharm Res. 2010;33(7):1065–1071. doi: 10.1007/s12272-010-0713-4. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler MD, Nakagami M, Bradford BU, Uesugi T, Mason RP, Connor HD, Dikalova A, Kadiiska M, Thurman RG. Overexpression of manganese superoxide dismutase prevents alcohol-induced liver injury in the rat. J Biol Chem. 2001;276(39):36664–36672. doi: 10.1074/jbc.M105352200. [DOI] [PubMed] [Google Scholar]
- 55.Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011 doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11(1):55–62. doi: 10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- 57.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41(2):139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silberman DM, Mostoslavsky R. SIRT3 deacetylase: the Jekyll and Hyde sirtuin. EMBO Rep. 2011;12(8):746–747. doi: 10.1038/embor.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.