Abstract
In this issue, Pathania et al (2011) report the involvement of BRCA1 in UV damage response at stalled replication forks which extends the function of BRCA1 beyond its established role in the repair of DNA double-strand breaks, raising the complexity of how this tumor suppressor maintains genomic stability.
BRCA1 (Breast Cancer gene 1) is one of the major tumor suppressors underlining familial breast cancer. To date the most prevailing understanding is that BRCA1 is recruited to and acts on DNA double strand breaks (DSBs) during late S and G2 phases. BRCA1 is critical for the initiation of end resection and for the assembly of recombinogenic DNA structure, two essential steps in the repair of DSBs via homologous recombination (Ciccia and Elledge 2010; Polo and Jackson, 2011). Whilst the focus of BRCA1 investigation has been on DSB processing, its potential role in dealing with other types of DNA damage is relatively under-explored. In this issue of Molecular Cell, Pathania et al (Pathania et al., 2011) present an intriguing discovery that BRCA1 is important in dealing with UV-induced DNA lesions, revealing a novel role of BRCA1 distinct from its widely-held axiom in DSB repair.
DNA bulky adducts are a major class of DNA helix-distorting lesions that occur very frequently. They are formed either by covalent attachment of alkylating molecules to nitrogenous bases or by ultraviolet (UV) light-induced covalent bonds between adjacent pyrimidines. Bulky adducts are efficiently removed by the nucleotide excision repair (NER) pathway (de Laat et al., 1999). Defects in NER lead to the profoundly cancer-prone genetic syndrome xeroderma pigmentosum. Bulky adduct lesions differ fundamentally from DSBs in that the former are repaired primarily in an error-free manner. Excision of the lesion-containing ssDNA patch allows the damaged site to be restored faithfully using the undamaged complementary strand, which retains the original sequence. However, when a replication fork encounters a bulky adduct lesion, the stalled fork structure presents an onerous scenario. Findings by Pathania et al (Pathania et al., 2011) shed the first light on how BRCA1 deficiency influences repair of UV-induced photo lesions in the context of a stalled replication fork.
Although BRCA1 mutant cells are well-characterized for their ionizing radiation and interstrand crosslink sensitivity, Pathania et al uncovered clues hinting at a role for BRCA1 in UV damage response. Cell lines derived from BRCA1 patients were more susceptible to UV killing. BRCA1 was recruited to UV-microirradiated subnuclear spots. BRCA1-depletion reduced the elimination of UV lesions. Interestingly, these impacts were found to be specific to S phase cells, manifesting during active DNA synthesis and indicative of lesion-stalled replication forks. One would expect that BRCA1 is attracted to and acts on DSBs resulting from UV damage because high doses of shortwave UV readily generate DSBs; alternatively, excessive blockage of replication forks presumably would lead to their collapse. However, the authors used a moderate UV dose range within which DSBs were undetectable both by the comet assay and by 53BP1 staining. More compellingly, in UV-irradiated S phase cells, the number of DSBs increased markedly upon BRCA1-depletion at late time points. This observation implicates BRCA1 in UV-damage response prior to any fork collapsing and before the possible onset of DSBs.
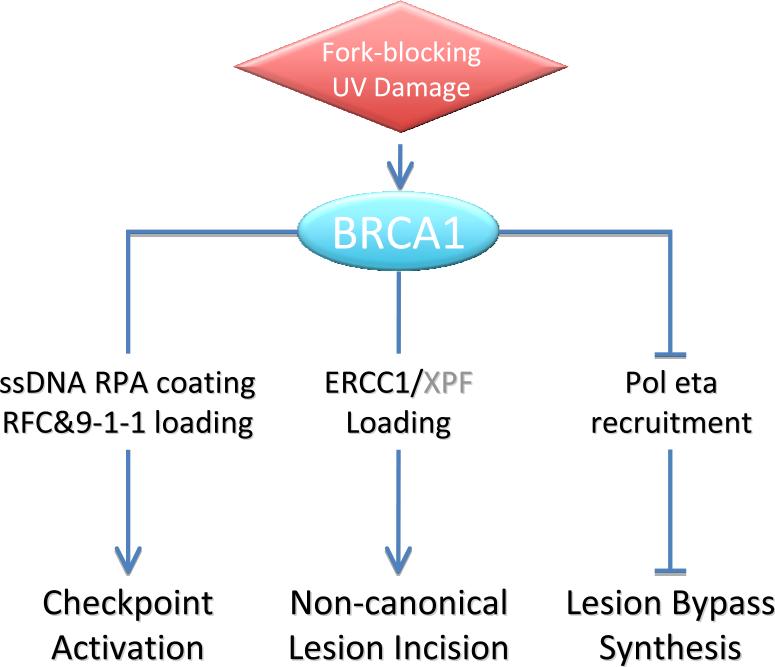
Cellular sensitivity to UV damage is caused by deficiencies in one or more of the three mechanisms, namely cell cycle checkpoint control, repair of UV-induced photo dimers, or lesion bypass synthesis. Remarkably, in deciphering how BRCA1 deficiency renders UV sensitivity, Pathania et al. showed that all three mechanisms are affected by BRCA1 deficit directly or otherwise (Figure 1).
Figure 1.
BRCA1 affects three distinct aspects of UV-damage response when an elongating replication fork encounters a UV-induced photo lesion.
Pathania et al noted that generation of RPA-coated ssDNA region upon UV damage was much reduced in the absence of BRCA1. In UV-damaged S phase cells, an excessive stretch of ssDNA is created at a stalled replication fork when the Mcm2-7 helicase is uncoupled and strays from the lesion-halted replicative polymerase (Byun et al., 2005; Cortez, 2005). Because RPA-coated ssDNA at stalled forks serves as a recruitment anchor for checkpoint sensing factors and for the signal initiation kinase Atr/Atrip (Zou and Elledge, 2003), failure to form RPA-coated ssDNA impairs the proper activation of the intra S phase checkpoint. Indeed, this prediction was validated by a full set of experiments, from weakened 9-1-1 and Atrip recruitments to a detectable defect in the intra-S-phase checkpoint in BRCA1-deprived cells upon UV exposure. These corroborating observations prompted the question of how BRCA1 is attracted to a stalled replication fork if no DSBs are present. Equally important is how BRCA1 regulates the generation of ssDNA at stalled forks.
In addressing this question, Pathania et al probed replication factors immediately present upon lesion encountering. They showed that damage-mediated BRCA1 recruitment was unaffected by Mcm2-7 depletion and that BRCA1 and the RFC complex exhibited mutual dependence on their damage-mediated enrichment at stalled replication sites. These results provide a general placement of BRCA1 at the immediate early stage of fork stress. An intact BRCT motif was found to be necessary for BRCA1 loading to UV-stalled replication forks. Whether a BRCT binding target(s) directs BRCA1 to the stalled replication fork remains a mechanistically worthy question.
In testing BRCA1's involvement in UV-induced chromatin enrichment of repair proteins, Pathania et al clearly established that BRCA1 recruitment did not rely on the NER lesion-sensing factors and vice versa. However, they found that recruitment of Ercc1, a subunit of the Ercc1/Xpf excinuclease, was significantly reduced by BRCA1-depletion. This finding is both intriguing and provocative. In a stalled replication fork, the blocking lesion is situated either in ssDNA or at a single-double strand junction (cf Fig. 7F). The NER damage recognition factor XPC-HHR23B recognizes a lesion only in the context of dsDNA by binding to the undamaged strand within the helix distortion, rather than binding to the lesion-containing strand (Min and Pavletich, 2007). As such, XPC is unlikely to be able to recognize a fork-blocking lesion to initiate the canonical NER process. The BRCA1-dependent loading of Ercc1/Xpf, with yet-to-be determined significance, may provide a plausible measure to eliminate the lesion in avoidance of an otherwise inevitable translesion synthesis (TLS). This is to the cells’ benefit because TLS is known for its low fidelity. However, it remains unclear as to whether and how Ercc1/Xpf could apply its endonuclease activity without generating a DSB at the stalled fork, given that Ercc1/Xpf has defined substrate and polarity. It is possible that the flap-clipping single-strand nuclease is needed at a later stage to resolve intermediate structures generated during recombination-mediated fork re-establishment. Moreover, how BRCA1 facilitates Ercc1/Xpf recruitment poses another interesting question.
An S-phase-specific function for BRCA1 in UV lesion repair is congruent with elevated TLS activity, as Pathania et al observed significant increases in PCNA monoubiquitination and in Pol eta recruitment to UV-damaged sites in the absence of BRCA1. This is also functionally reflected by an increase in UV-induced mutagenesis in BRCA1-deficient cells. On the one hand, this may implicate a regulatory role for BRCA1 in suppressing TLS. The up-regulation of TLS may compensate for the UV repair defect of BRCA1-mutant cells, partially masking their UV sensitivity. On the other hand, elevated TLS could be a passive response to a surge in unrepaired fork-blocking lesions caused by BRCA1 deficiency.
The BRCA1-mutants measured by Pathania et al displayed a moderate UV sensitivity of 3-fold over normal breast epithelial controls. Putting this finding in perspective, one should realize that BRCA1 function affords an evident yet relatively minor protection against acute UV-exposure, because classical NER-deficient or Pol eta-deficient XPV cells are an order of magnitude more sensitive than the tested BRCA1 mutants. However, BRCA1's multiple roles in bulky adduct processing may have an indispensible influence on the long-term maintenance of genomic stability, particularly in dealing with intrinsic sources of damage. As Pathania et al postulated, certain estradiol metabolites can form DNA bulky adducts and might be a contributing element to genomic instability that leads to breast cancer. Lending additional support to this notion are clinical studies showing that BRCA1-deficient breast and ovarian cancer patients have a higher rate than BRCA1-wt patients in their response to cisplatin (Boyd et al., 2000; Silver et al., 2010), which damages DNA predominantly in the form of intrastrand diadduct. Findings from Pathania et al may provide mechanistic underlying for these clinical observations.
The involvement of BRCA1 in post-replication UV damage response adds to the complexity of how its tumor suppressor function is defined. To extend the observation to bulky adducts at large, alkylating agents that forms various monoadduct can be tested on isogenic cell strains to see if BRCA1 has lesion-specific functions and to gauge BRCA1's role in non-UV bulky adduct response. At present, the model proposed by Pathania et al suggests that the UV sensitivity of BRCA1-deficient cells maybe a compounded phenotype from perturbed intra-S-phase and G2/M checkpoints, lesion processing, and TLS. It will be interesting to determine which of these defects is the prevailing contributor to the UV sensitivity.
When tending to DSBs, BRCA1 is brought in via an elaborate signaling and recruitment cascade to load downstream components and to assemble specialized complexes. For its newly discovered roles in UV damage response, it is imperative to shed light on how BRCA1 is recruited to a fork-stalling lesion and to identify its downstream targets/effectors. Connections at the molecular level, particularly with regard to defined or novel biochemical properties of BRCA1, will help plowing deeper into this new territory of BRCA1 function beyond the boundary of DNA double-strand breaks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, et al. JAMA. 2000;283:2260–2265. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NGJ, Hoeijmakers JHJ. Genes & Development. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- Min JH, Pavletich NP. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- Pathania S, Nguyen J, Hill SJ, Scully R, Feunteun J, Livingston DM. Molecular Cell. 2011;43 doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]