Abstract
Specific human leukocyte antigens (HLAs), notably HLA-B*27 and HLA-B*57 allele groups, have long been associated with control of HIV-1. Although the majority of HIV-specific CD8+ T cells lose proliferative capacity during chronic infection, T cells restricted by HLA-B*27 or HLAB*57 allele groups do not. Here we show that CD8+ T cells restricted by 'protective' HLA allele groups are not suppressed by Tref cells, whereas, within the same individual, T cells restricted by 'nonprotective' alleles are highly suppressed ex vivo. This differential sensitivity of HIV-specific CD8+ T cells to Tref cell–mediated suppression correlates with their expression of the inhibitory receptor T cell immunoglobulin domain and mucin domain 3 (Tim-3) after stimulation with their cognate epitopes. Furthermore, we show that HLA-B*27– and HLA-B*57–restricted effectors also evade Tref cell–mediated suppression by directly killing Tref cells they encounter in a granzyme B (GzmB)-dependent manner. This study uncovers a previously unknown explanation for why HLA-B*27 and HLA-B*57 allele groups are associated with delayed HIV-1 disease progression.
INTRODUCTION
Cytotoxic T lymphocytes (CTLs) are crucial in controlling HIV-1 infection (reviewed in ref. 1), which is emphasized by the influence of human leukocyte antigen (HLA) class I alleles on the rate of progression to AIDS2, 3. Long-term nonprogressors (LTNPs) are a rare, heterogeneous population of chronically infected individuals with low viral loads and sustained CD4+ T cell counts who are relatively spared from disease progression even without antiretroviral therapy. The mechanisms allowing for such control of HIV-1 are not known, but a number of specific HLA allele groups, notably HLA-B*27 and HLA-B*57, are highly enriched in LTNP populations4. Possession of HIV-1–specific CD8+ CTLs restricted by HLA-B*27 and HLA-B*57 during early HIV-1 infection correlates with longer AIDS-free survival5 and better defines disease progression compared to HLA genotype alone, suggesting that HIV-specific CD8+ CTLs, and not the presence of a particular HLA allele, determines disease progression5.
LTNPs possess HIV-specific CD8+ CTLs restricted by HLA-B*27 or HLA-B*57 that can continue to proliferate throughout chronic infection, whereas the majority of HIV-specific CD8+ CTLs restricted by other HLA alleles lose their proliferative capacity6, 7, 8. Proliferative ability is linked to upregulation of perforin and is, therefore, associated with the cytotoxic capabilities of virus-specific CD8+ CTLs6. We show here that HLA-B*27– and HLA-B*57–restricted HIV-specific CD8+ CTLs possess an additional feature in that they evade suppression mediated by Tref cells. Tref cells, CD4+ (or CD8+) T cells with suppressive activity, have a key role in maintaining peripheral tolerance, preventing autoimmune diseases and limiting chronic inflammatory responses (reviewed in refs. 9,10). However, they may also have detrimental effects by suppressing effective antigen-specific immune responses. Several mechanisms are used by Tref cells to exert immunosuppressive function; however, our observation of differential suppression of CD8+ CTLs restricted by distinct HLA alleles has not been previously described to our knowledge.
Tim-3 negatively regulates T helper type 1 responses upon interaction with its ligand, galectin-9 (Gal-9, encoded by LGALS9) expressed on lymphocytes and other cell types11. In humans, defects in Tim-3 expression contribute to multiple sclerosis pathology12, suggesting that expression of Tim-3 on effector T cells is involved in inducing and maintaining peripheral tolerance of these T cells. Tref cells constitutively express Gal-9 (ref. 13) and, thus, could be providing the ligand for inducing tolerance in Tim-3–expressing effectors. Furthermore, a recent study demonstrated that Tim-3 expression defines a distinct population of dysfunctional effector T cells in progressing HIV-1–infected individuals14.
Here we show that Tref cells suppress proliferation of nonprotective HIV-specific CD8+ CTLs through Tim-3–Gal-9 interactions during chronic infection, but they do not suppress proliferation of HIV-specific CD8+ CTLs restricted by HLA-B*27 or HLA-B*57. These HLA-B*27– or HLA-B*57–restricted CTLs can continue to proliferate and kill infected targets during chronic infection, which may account for delayed disease progression in persons with HLA-B*27 and HLA-B*57 allele groups. In addition, this finding may explain why the HLA-B*27 allele group is also associated with resolution of other chronic infections (for example, hepatitis C virus) and why both HLA-B*27 and HLA-B*57 allele groups are associated with autoimmunity15, 16, 17.
RESULTS
Tref cells disparately suppress CTLs based on HLA allele
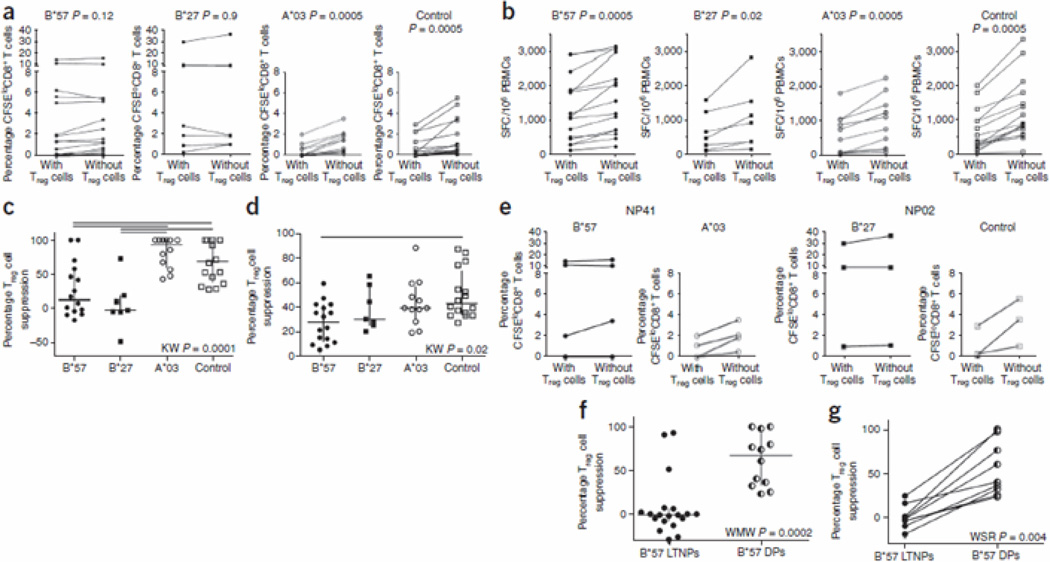
To assess the susceptibility of HIV-1–specific CTLs to Tref cell–mediated suppression, we measured proliferation of numerous HIV-1 epitope–specific CD8+ CTLs from LTNPs (individual clinical data are listed in Supplementary Table 1; epitope responses measured are listed in Supplementary Table 2) by measuring carboxyfluorescein succinimidyl ester (CFSE) dilution in the presence or absence of Tref cells8, 18 in vitro. Tref cells differentially suppressed proliferation of CD8+ CTLs restricted by different HLA alleles (Fig. 1). Epitope-specific CD8+ CTLs restricted by the protective HLA allele groups (HLA-B*27 and HLA-B*57) were not suppressed by Tref cells (P = 0.9 and P = 0.12, respectively; median percentage Tref cell suppression −1.0 (indicating that effector T cells proliferated more in the presence of Tref cells than in their absence) and 13.3, respectively), whereas proliferation of epitope-specific CD8+ CTLs restricted by other HLA-A and HLA-B alleles (control alleles) was significantly (P = 0.0005) suppressed (Fig. 1a,c). The control allele groups were HLA-A*02, HLA-A*24 and HLAB*08, allele groups with at least three epitope responses found in at least two HIV-1 infected individuals. Of note, responses were differentially susceptible to suppression, with CTLs restricted by HLA-A*03 being particularly susceptible (P = 0.0005; Fig. 1a; median percentage Tref cell suppression 92.8). Of note, Tref cells suppressed interferon-γ (IFN-γ) secretion from all CD8+ CTLs (Fig. 1b,d; P = 0.02 for HLA-B*27–restricted responses and P = 0.005 for HLAB*57, HLA-A*03 and control allele group). This suggests that only the proliferative ability of effector CD8+ CTLs is differentially suppressed by Tref cells, whereas cytokine secretion is always suppressed, regardless of the HLA restriction.
Figure 1.
Treg cell suppression of in vitro proliferative ability or cytokine secretion of CD8+ T cells restricted by HLA-B*57, HLA-B*27, HLA-A*03 and control HLAs (HLA-A*02, HLA-A*24 and HLA-B*08). (a) Background-subtracted percentage CFSEloCD8+ T cells in PBMCs with or without Treg cells when cultured in the presence of HIV-1 epitopes recognized by CD8+ T cells restricted by various HLA alleles. (b) Background-subtracted HIV-specific IFN-γ ELISPOT responses in the presence and absence of Treg cells. HLA-B*27 or HLA-B*57–restricted and non–HLA-B*27– and HLA-B*57–restricted responses are shown after stimulation with their cognate epitopes. SFC, spot-forming cell. In a and b, Wilcoxon signed-rank (WSR) test was used. (c) Percentage suppression of proliferation grouped according to HLA restricting allele. (d) Percentage suppression of cytokine secretion grouped according to HLA restricting allele. In c and d, Kruskal-Wallis (KW) test was used for grouped comparisons with a post hoc Dunn’s test showing significant subgroup comparisons with horizontal lines. (e) Differential suppression of proliferation of HLA-B*27– and HLA-B*57– versus HLA-A*03– and control HLA–restricted HIV-specific CD8+ CTLs within the same person. NP02 and NP41 are two LTNPs. (f) Percentage suppression of proliferation by Treg cells of HLA-B*57–restricted CD8+ CTLs in HLAB*57+ LTNP versus HLA-B*57+ delayed progressors (DP). (g) Longitudinal analyses of percentage suppression of proliferation by Treg cells of HLA-B*57–restricted CD8+ CTLs before and after progression in HLA-B*57+ individuals.
We also observed differential suppression of HIV-specific CTLs within individuals (for example, subjects NP02 and NP41, Fig. 1e). Thus HIV-specific CTLs restricted by HLA-B*27 or HLAB*57 were not suppressed by Tref cells, whereas HIV-specific CTLs restricted by other alleles from the same individual were suppressed, suggesting that the suppression was due to a difference in the CTLs (for example, in the strength of T cell receptor (TCR) signaling) rather than the Tref cells.
Although many individuals with HLA-B*57 allele group have longer AIDS-free survival than individuals with other HLA alleles, the majority will eventually progress to disease. Indeed, it has been shown that HLA-B*57+ LTNPs who eventually progress to disease have HLA-B*57–restricted T cells with lower proliferative capacity when compared to HLA-B*57+ LTNPs who have not progressed8. This reduced proliferation correlated with an increased susceptibility to Tref cell–mediated suppression when we compared HIV-specific, HLA-B*57–restricted CD8+ CTLs in HLA-B*57+ LTNPs versus HLA-B*57+ delayed progressors (P = 0.0002; Fig. 1f). Longitudinal analyses before and after progression in three individuals confirmed that when clinical progression to disease occurred, previously Tref cell–resistant HLA-B*57–restricted CTLs became susceptible to Tref cell-mediated suppression (P = 0.004; Fig. 1g).
Differential suppression is independent of CTL frequency
HLA-B*27– and HLA-B*57–restricted CD8+ CTLs proliferate more than HIV-specific CD8+ CTLs restricted by other HLAs8, 19. Thus, cells capable of high proliferation may escape Tref cell–mediated suppression regardless of their allele restriction. We examined whether percentage suppression by Tref cells (defined in Methods) correlated with either the initial precursor frequency of epitope-specific CD8+ CTLs (measured by IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assay) or with magnitude of proliferation. Only about 10% of the data could be explained by either initial precursor frequency (r2 = 0.1, P = 0.01; Supplementary Fig. 1a) or magnitude of proliferation (r2 = 0.13, P = 0.003; Supplementary Fig. 1b) suggesting that neither initial precursor frequency nor magnitude of proliferation has a substantial role in determining the susceptibility of the CTLs to Tref cell–mediated suppression.
Because all of the HLA-A*03–restricted responses were of low magnitude (below 3% CFSEloCD8+ CTLs) (Fig. 1a), we extended these analyses to look at only low-frequency responses below 3%. Even then, only 21% of the data variations were explained by the magnitude of proliferation (r2 = 0.21, P = 0.0004; Supplementary Fig. 1c), and this weak correlation seemed to be mainly driven by HLA-A*03–restricted responses (n = 11; r2 = 0.82, P < 0.0005), as responses restricted by HLA-B*57 were not correlated with their ability to be suppressed (n = 12; P = 0.2). These data suggested that for some low-frequency responses (for example, restricted by HLA-A*03), the magnitude of proliferation might explain susceptibility to Tref cell–mediated suppression, with lower frequency responses being more susceptible. However, this does not explain why HLA-B*27– and HLA-B*57–restricted T cells are resistant to Tref cell–mediated suppression.
Tref cells mediate differential suppression via Gal-9–Tim-3
Upon activation with their cognate epitopes, effector CD8+ CTLs upregulate many inhibitory receptors (for example, programmed death-1 (PD-1) and Tim-3) as a means of self-limiting the inflammatory response. Tref cells constitutively express the ligands for Tim-3 (Gal-9) and PD-120, 21. We hypothesized that the differential sensitivity to Tref cell–mediated suppression could be due to differential upregulation of inhibitory receptors on CD8+ CTLs restricted by HLAB*27 and HLA-B*57 versus CD8+ CTLs restricted by other HLA alleles. Tim-3 was of particular interest, as Tim-3–knockout mice are resistant to peripheral tolerance22, and elevated frequencies of dysfunctional Tim-3+CD8+ CTLs are found in HIV-infected individuals with progressive disease14. Furthermore, although in recent years PD-1 has been the primary marker for 'exhausted' T cells23, new data have shown that Tim-3 expression correlates better with dysfunctional CD8+ CTLs than expression of PD-1 (ref. 24).
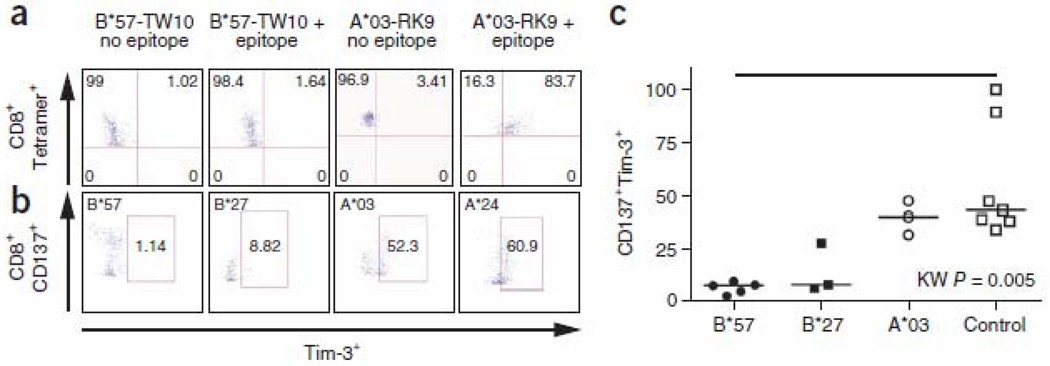
Peripheral blood mononuclear cells (PBMCs) from LTNPs are not exhausted and do not constitutively express PD-1 or Tim-3. However, they do upregulate these inhibitory receptors after antigenic stimulation. We stimulated PBMCs with HIV-1 epitopes bound by various HLAs and determined the frequency of Tim-3 expression on antigen-specific CD8+ CTLs (as measured by either tetramer staining (Fig. 2a) or CD137 upregulation25 (Fig. 2b)). As predicted, HIV-specific CD8+ CTLs restricted by HLA-B*27 or HLA-B*57 allele groups upregulated significantly less Tim-3 after stimulation than HIV-specific CD8+ CTLs restricted by other HLA alleles (Fig. 2c, P = 0.005), suggesting that Tref cells may mediate differential suppression of effector CD8+ CTLs via Gal-9–Tim-3 interactions. To test this, we masked Gal-9 on Tref cells by adding lactose (Fig. 3), which is a ligand of the galectin family of lectins, to which Gal-9 belongs. Addition of lactose during in vitro proliferation assays26 prevented Tref cell–mediated suppression of proliferation of HIV-specific CD8+ CTLs restricted by any HLA alleles other than the HLA-B*27 or HLA-B*57 allele groups but had no effect on proliferation of HIV-specific CD8+ CTLs restricted by HLA-B*27 or HLA-B*57 (Fig. 3a,c). Because lactose is not specific for Gal-9, but rather binds all lectins, we also isolated Tref cells from PBMCs, silenced LGALS9 using siRNA (Supplementary Fig. 2) and then added the cells back to Tref cell–depleted PBMCs, followed by stimulation with their cognate HIV-1 epitopes. In agreement with the lactose studies, knockdown of LGALS9 in Tref cells prevented their suppression of proliferation of HIV-specific CD8+ CTLs restricted by any non–HLA-B*27 or non–HLA-B*57 allele groups but had no effect on their suppression of proliferation of HIV-specific CD8+ CTLs restricted by HLAB*27 or HLA-B*57 (Fig. 3b,d). Therefore, differential suppression of proliferation of HIV-specific CD8+ CTLs by Tref cells is mediated through Gal-9–Tim-3 interactions; HLA-B*27– or HLA-B*57–restricted HIV-specific CD8+ CTLs upregulate less Tim-3 when they encounter their cognate epitopes than HIV-specific CTLs restricted by other HLAs, and, thus, they are less susceptible to Tref cell–mediated suppression.
Figure 2.
Frequency of CD8+Tim-3+ T cells following stimulation with their cognate epitopes. (a) Percentage of Tim-3+ CD8+ T cells using allophycocyanin-labeled HLA-A*03–RLRPGGKKK tetramer or phycoerythrin-labeled HLA-B*57-TSTLQEQIGW tetramer staining of PBMCs before and after stimulation with their cognate epitopes. Top right quadrant shows percentage of Tim-3+tetramer+ CD8+ CTLs. (b) Percentage of Tim-3+ on CD8+ T cells using CD137 to identify antigen-specific T cells after stimulation with their cognate epitopes. (c) Percentage of CD137+Tim-3+ T cells after stimulation of PBMCs from different individuals with their corresponding epitopes.
Figure 3.
CFSE dilution data showing inhibition of Gal-9–Tim-3 interactions by lactose and siRNA. (a) Examples of proliferation of PBMCs stimulated with their corresponding epitopes, showing percentage CFSEloCD8+ T cells in the absence or presence of lactose. (b) Examples of proliferation of CFSE-labeled, Treg cell–depleted PBMCs stimulated with their cognate epitopes in the presence of Treg cells treated with either LGALS9 siRNA or siControl (at 1:0.25 ratio). The measures of coculture suppression by Treg cells in the presence or absence of lactose or siRNA are shown for a representative experiment from three repeat experiments for each approach. (c) Percentage of Treg cell suppression calculated after stimulation of CD8+ T cells with their corresponding epitopes in the presence of lactose. (d) Percentage of Treg cell suppression calculated after stimulation of CD8+ T cells with their cognate epitopes in the presence of LGALS9 siRNA–treated Treg cells (at 1:0.25 ratio).
Protective CTLs kill Tref cells via granzyme B
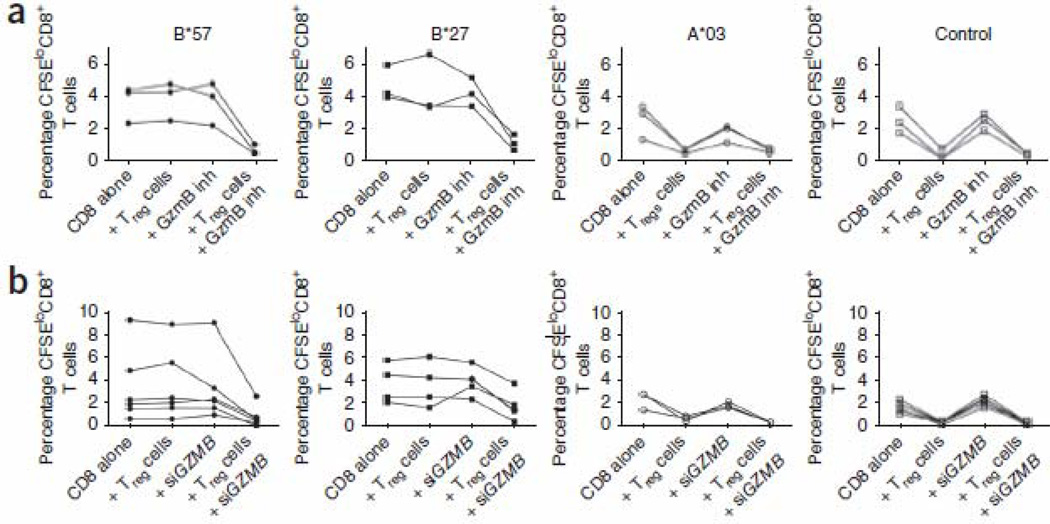
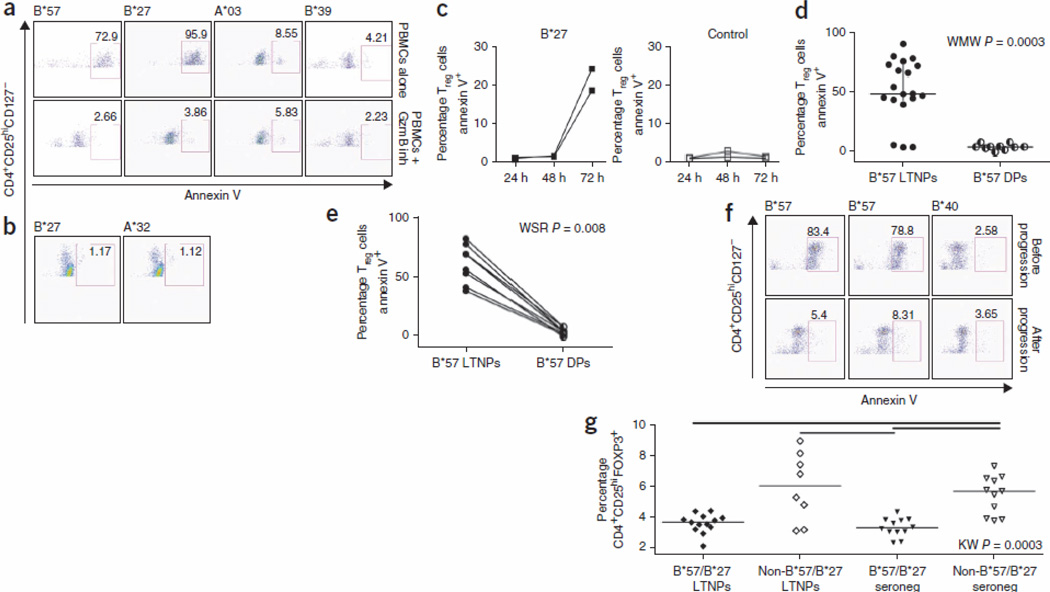
A recent study showed that HIV-specific HLA-B*27– and HLA-B*57–restricted CD8+ CTLs upregulate more granzyme B (GzmB, encoded by GZMB) on a per-cell basis than HIV-specific CD8+ CTLs restricted by other HLAs19. This upregulation is independent of their enhanced proliferative ability. Moreover, it has also been shown that CD4+ effectors that express high amounts of GzmB escape Tref cell–mediated suppression by killing Tref cells that they encounter27. This mechanism of Tref cell evasion has not been observed for human CD8+ effector cells. To determine whether this mechanism could also be involved in the observed allele-specific differential suppression by Tref cells, we performed epitope-specific CFSE dilution assays in the presence of z-AAD-CMK (z-Ala-Ala-Asp(OMe)-chloromethyl ketone), a potent and selective GzmB peptide inhibitor. When we preincubated CD8+ T cells with z-AAD-CMK, CD8+ CTLs restricted by HLA-B*27 and HLA-B*57 became susceptible to Tref cell suppression (Fig. 4a and Supplementary Fig. 3). Similarly, if we silenced GZMB in CD8+ CTLs with GZMB siRNA (Supplementary Fig. 2b) before stimulation with cognate epitope in the presence or absence of Tref cells, the HLA-B*27– and HLA-B*57–restricted CD8+ CTLs were rendered susceptible to Tref cell–mediated suppression, whereas CD8+ CTLs restricted by HLA-B*27 and HLA-B*57 treated with an irrelevant siRNA were not susceptible to Tref cell suppression (Fig. 4b and Supplementary Fig. 3b). This suggests that either expression of Tim-3 or lack of expression of GzmB in CD8+ effector CTLs may increase their susceptibility to Tref cell–mediated suppression. Furthermore, the resistance of HLA-B*27– and HLA-B*57–restricted CD8+ CTLs to Tref cells resulted from their directly killing Tref cells when they were stimulated with their cognate epitopes, whereas we observed low killing of Tref cells in cultures stimulated with non–HLA-B*27 and non–HLA-B*57 epitopes (Fig. 5a). When we blocked GzmB activity with z-AAD-CMK, the frequency of apoptotic Tref cells was reduced greatly in cultures stimulated with HLA-B*27– and HLA-B*57–restricted epitopes compared with the absence of GzmB inhibitor (Fig. 5a). In contrast, blockade of GzmB activity in cultures stimulated with non–HLA-B*27 and non–HLA-B*57 epitopes had minimal effect on the frequency of apoptotic Tref cells. Thus, HLA-B*27– and HLA-B*57–restricted effector CD8+ CTLs killed the Tref cells they encountered in a GzmB-dependent manner. Killing mediated by HLA-B*27– and HLAB*57–restricted HIV-specific CD8+ CTLs was not limited to Tref cells, as they also killed non-Tref CD4+ cells (Supplementary Fig. 4). HLA-B*27 and HLA-B*57–restricted CTLs killed considerably more Tref cells than non–HLA-B*27 and HLA-B*57–restricted CTLs (Fig. 5a). Notably, both HLA-B*27 or HLA-B*57–restricted effectors and non–HLA-B*27 or HLAB*57–restricted effectors induced more apoptosis of Tref cells than of non-Tref cells (P < 0.0001 for HLA-B*27 or HLA-B*57–restricted effectors and P = 0.001 for non–HLA-B*27 or HLAB*57–restricted effectors; Supplementary Fig. 4). Thus, Tref cells are not seen as 'Tref cells' by protective CD8+ CTLs but rather as any other CD4+ T cell presenting their epitope.
Figure 4.
CFSE dilution data showing CD8+ T cells restricted by HLA-B*57 and HLA-B*27 resist Treg cell-mediated suppression in a GzmB dependent manner. (a) Percentage CFSEloCD8+ T cells after CFSE-labeled isolated CD8+ T cells were stimulated with their corresponding epitopes alone or together with Treg cells (at 1:0.25 ratio), and also in the presence or absence of a GzmB peptide inhibitor (z-AAD-CMK). Examples of flow data are shown in Supplementary Figure 3a. (b) Percentage CFSEloCD8+ T cells after electroporation with GZMB siRNA or nonhybridizing negative control (siControl) siRNA oligonucleotides and stimulation with their cognate epitopes alone or with Treg cells (at 1:0.25 ratio). Examples of flow data are shown in Supplementary Figure 3b.
Figure 5.
CD8+ T cells restricted by HLA-B*27 and HLA-B*57 induce Treg apoptosis in a GzmB-dependent manner. (a–c) Percentage annexin V+ Treg cells (CD3+CD4+CD25hiCD127lo) in PBMCs stimulated for 4 d (a), 24 h (b) or 24–72 h (c) with HLA-B*27–, HLA-B*57–, HLA-B*39– or HLA-A*03–restricted epitopes in the presence or absence of GzmB peptide inhibitor. These data are representative of three separate experiments from different LTNPs. (d) Percentage annexin V+ Treg cells in PBMCs stimulated with HLA-B*57–restricted epitopes from HLA-B*57+ LTNP versus HLA-B*57+ DPs. (e,f) Percentage annexin V+ Treg cells in PBMCs stimulated with HLA-B*57–restricted epitopes before and after progression to disease. (g) Treg cell frequencies in HIV-1–seronegative individuals versus HIV-1 infected HLA-B*27+ or HLA-B*57+ and HLA-B*27− or HLA-B*57− LTNPs. Percentages of CD4+CD25hiFOXP3+ Treg cells are shown in PBMCs from 12 HIV-seronegative HLA-B*27+ or HLA-B*57+ individuals, 12 HLA-B*27– or HLA-B*-57– individuals, 13 HLA-B*27+ or HLA-B*57+ LTNPs and 8 HLA-B*27− or HLA-B*57− LTNPs.
Maximal expression of lytic effector molecules (perforin and GzmB) requires many days of proliferation6, 19. This explains why Tref cells were able to suppress the IFN-γ secretion capabilities of HLA-B*27– and HLA-B*57–restricted T cells, as there would be insufficient GzmB upregulation to kill Tref cells during this short assay (24 h). In agreement with this, we did not observe killing of Tref cells in cultures stimulated for 24 h with HLA-B*27 and HLA-B*57 epitopes (Fig. 5b). However, after 72 h, we observed a substantial increase in the killing of Tref cells by HLA-B*27–restricted CD8+ CTLs (Fig. 5c).
In agreement with the data showing that HLA-B*57– restricted CD8+ CTLs became susceptible to Tref cell–mediated suppression of proliferation upon disease progression, HIV-specific HLAB*57–restricted CD8+ CTLs from delayed progressors failed to kill Tref cells (P = 0.0003; Fig. 5d). Analyses before and after progression in HLA-B*57+ individuals confirmed that when clinical progression to disease occurred, HLA-B*57–restricted HIV-specific CTLs lost their ability to kill Tref cells (P = 0.008; Fig. 5e,f).
The fact that stimulated HLA-B*27– and HLA-B*57–restricted CD8+ CTLs kill Tref cells suggested that there should be fewer Tref cells in individuals who express these HLA alleles. Indeed, we found a significant decrease (P = 0.0003) in Tref cell frequency in HLA-B*27+ or HLA-B*57+ LTNPs compared with non–HLA-B*27+ and non–HLA-B*57+ LTNPs and seronegative individuals (Fig. 5g), suggesting that deletion of Tref cells occurs in vivo in HLAB* 27+ and HLA-B*57+ individuals despite their maintenance of normal CD4+ T cell counts.
Our data show that HLA-B*27– and HLA-B*57–restricted CTLs are resistant to Tref cell–mediated suppression irrespective of the epitopes recognized by the T cells (Fig. 1a and Supplementary Fig. 5), suggesting that resistance to Tref cell–mediated peripheral tolerance is related to allele restriction rather than epitope specificity. To determine whether this occurs only during HIV infection, we assessed Tref cell susceptibility of HLA-B*27–restricted versus HLA-A*02– or HLA-B*07–restricted herpes simplex virus (HSV-2) or Epstein Barr virus (EBV) epitopes in HIV-seronegative individuals. HSV- and EBV-specific HLA-B*27–restricted CD8+ CTLs were also not susceptible to Tref cell–mediated suppression, whereas HSV-2– and EBV-specific CD8+ CTLs restricted by other alleles were susceptible (Supplementary Fig. 6), proving that ability to evade Tref cells is due to the allele restriction of the CTL and not to the specific infection.
DISCUSSION
Although the role of human Tref cells in preservation of self-tolerance is well documented, less is known about their influence on chronic viral infection. Several studies have shown that Tref cell frequencies are either increased28, 29 or decreased in HIV infection30, 31. Despite the conflicting data, some recent in vivo studies suggest that high frequencies of Tref cells during HIV infection are detrimental. Specifically, a recent in vivo phase 3 trial showed that IL-2 therapy preferentially expands Tref cells in infected individuals and that individuals with the greatest expansion are more likely to progress to disease32, 33.
We show that there are two divergent outcomes for HIV-specific CD8+ CTLs during chronic infection: the majority of HIV-specific CTLs upregulate Tim-3 when they encounter their cognate epitopes and are subsequently suppressed by Tref cells; however, CD8+ CTLs restricted by protective HLA allele groups upregulate less Tim-3 but more GzmB upon recognition of their cognate epitopes. They are subsequently less susceptible to Tref cell–mediated suppression, instead killing Tref cells they encounter. Thus, our data suggest a previously unknown model of how HLA-B*27– and HLA-B*57–restricted, HIV-specific CD8+ CTLs may evade Tref cells and subsequently control HIV replication (Fig. 6). Moreover, we provide data showing that HIV-specific, HLA-B*57–restricted CD8+ CTLs can be suppressed by Tref cells once progression to disease occurs. However, we cannot at present determine whether disease progression in these individuals is a cause or effect of loss of the ability to evade Tref cells.
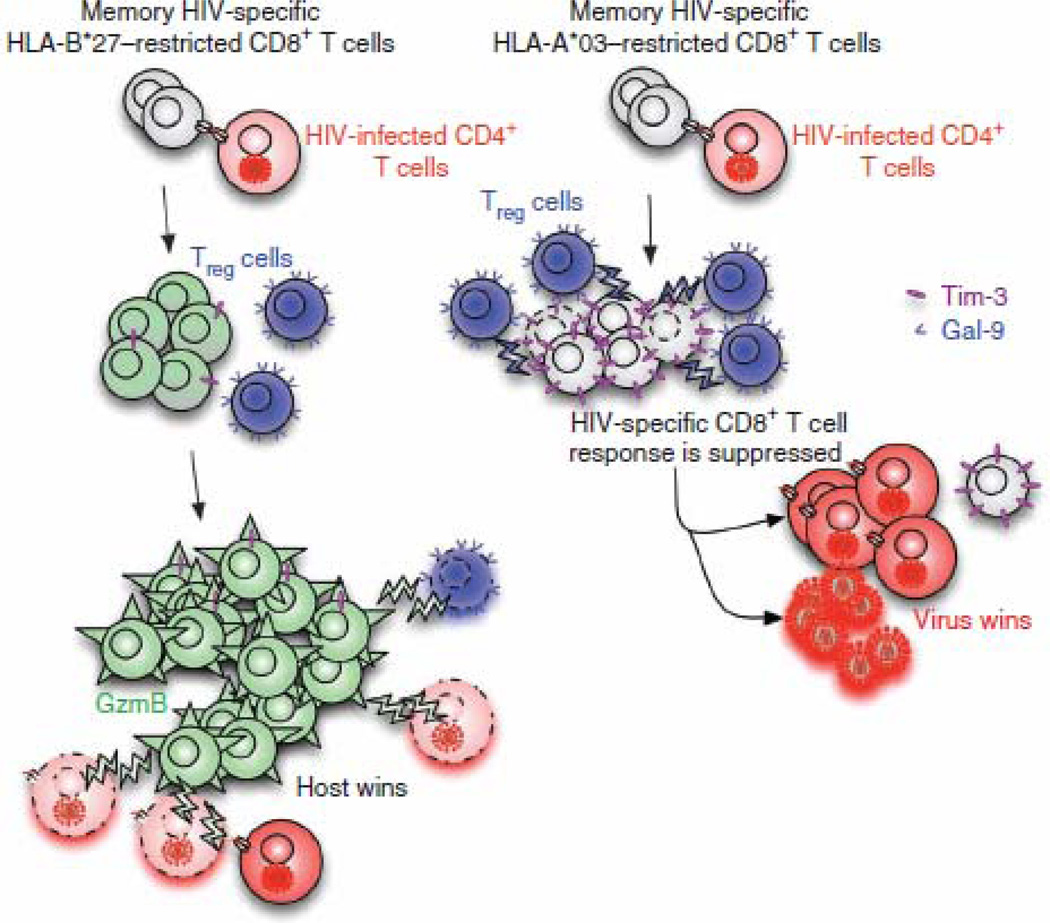
Figure 6.
Model depicting how HLA-B*27– or HLA-B*57–restricted HIV-specific CD8+ T cells evade Treg cell suppression and subsequently control HIV replication. HIV-specific, HLA-B*27–restricted CD8+ T cells do not upregulate surface expression of Tim-3 upon recognition of their cognate epitopes on HIV-infected CD4+ T cells, whereas HIV-specific, HLA-A*03–restricted CD8+ T cells upregulate high surface expression of Tim-3. Treg cells suppress HLA-A*03–restricted CD8+ T cells owing to their high expression of Tim-3 but cannot suppress proliferation of HLA-B*27–restricted CD8+ T cells. Highly proliferating HLA-B*27–restricted CD8+ T cells upregulate high levels of GzmB and kill not only infected CD4+ T cells but also infected Treg cells that they encounter. Thus, HLA-B*27–restricted CD8+ T cells can control HIV replication during chronic infection, whereas HLA-A*03–restricted CD8+ T cells cannot.
The mechanisms accounting for why HLA-B*27– and HLA-B*57–restricted T cells upregulate less Tim-3 and more GzmB than other HIV-specific T cells upon recognition of their cognate epitopes are not known. Understanding these differences in TCR signaling will be crucial for determining potential therapeutic interventions that 'switch on' virus-specific CTLs that are restricted by other HLAs. Such mechanisms may include a higher-avidity TCR on CD8+ CTLs restricted by HLA-B*27 and HLA-B*57 (ref. 34). In addition, HLA-B*27 is an unusual allele group in that it can form heavy-chain homodimers and trimers, which may lead to alternative TCR interactions35.
The observation that Tref cells are killed more efficiently than non-Tref CD4+ T cells suggests that, during HIV infection, Tref cells are more susceptible to effector-induced apoptosis. This is consistent with studies demonstrating that Tref cells have increased turnover and higher expression of caspase-3 during chronic infection36. Increased intracellular levels of caspase-3 have been shown to increase susceptibility of T cells to apoptosis37. Peripheral Tref cell frequencies are similar between HLA-B*27+ or HLA-B*57+ LTNPs and HLA-B*27+ or HLAB*57+ uninfected subjects. Most of the LTNPs studied here have low or undetectable viral load in their peripheral blood, so there is minimal antigen to stimulate killing of Tref cells by HLA-B*27 or HLA-B*57–restricted CTLs. We presume that HLA-B*27 or HLA-B*57–restricted CTLs will kill Tref cells where antigen is present, so it is not surprising that we did not observe significant differences in Tref cell frequencies in the periphery between uninfected and infected individuals. However, in lymph nodes and gut we would expect to see fewer Tref cells in infected individuals with HLA-B*27 or HLA-B*57. This is supported by data from another group showing fewer Tref cells in lymph nodes from LTNPs than in those from infected individuals with progressive infection29. Therefore, it is possible that loss of effector cell evasion of Tref cells results in accumulation of Tref cells in lymph nodes and disease progression.
Our study indicates that CD8+ CTLs are differentially susceptible to Tref cell–mediated suppression of proliferation dependent on HLA restriction but independent of the epitope specificity of the suppressed cell. Thus, HIV-specific CTLs restricted by HLA-B*27 and HLAB*57 are resistant to Tref cells, allowing them to continue to function maximally during chronic infection. This may explain why these HLAs are associated with delayed progression to disease. Resistance of HLA-B*27–restricted CTLs to Tref cell–mediated suppression may also explain why this allele group is associated with better clinical outcomes of other chronic infections, such as hepatitis C virus15. Both HLA-B*27 and HLA-B*57 allele groups are highly associated with autoimmunity: HLA-B*27 allele groups with ankylosing spondylitis16 and HLA-B*57 allele groups with psoriasis38. Thus, possession of these allele groups seems to be a double-edged sword. Because T cells restricted by them cannot be tolerized, these allele groups are beneficial in chronic infection but detrimental in autoimmunity. Therefore, we feel this work has far-reaching implications for both control of chronic infection and autoimmunity.
METHODS
Study group
LTNPs were enrolled through the HIV Vaccine Trial Unit (M.J.M.). They were defined as HIV infected for more than 11 years, with repeated CD4+ T cell counts over 500 cells per µl or CD4% over 28% and viral load <10,000 copies per ml in the absence of antiretroviral therapy. Clinical data and HLA genotypes are shown in Supplementary Table 1. An additional subject was recruited through the University of Washington Primary Infection Clinic (A.C.C.), meeting early criteria of an elite controller (viral load 58 copies per ml), infected for about ten years but recently progressed. We studied 17 LTNPs and 7 LTNPs who eventually progressed (delayed progressors), defined as HIV infected with viral load >10,000 RNA (copies per ml) or declining CD4+ T cell count in the absence of antiretroviral therapy. All subjects were males of European descent except subject NP14 who reported African-American ethnicity. HIV-1 seronegative–individuals were recruited at Seattle BioMed.
Fourteen HIV-seronegative individuals (seven males and seven females) who were seropositive for HSV-2 were recruited and HLA-typed as described39 (Supplementary Table 1). They were assumed to be EBV seropositive if they had an IFN-γ response to known EBV CD8+ epitopes because most adults are EBV seropositive.
The appropriate Institutional Review Boards at the University of Washington, Fred Hutchinson Cancer Research Center and Seattle BioMed approved the studies, and all individuals provided written informed consent.
Epitope mapping
HIV epitope specificities were mapped in each HIV+ individual as previously described8. Some LTNPs were mapped using potential T cell epitopes provided by The National Institutes of Health AIDS Research and Reference Reagent Program. The epitope-specific responses assessed for each individual are shown in Supplementary Table 2. HSV-2+ individuals were mapped as previously described39. HSV and EBV epitope-specific responses assessed for each individual are shown in Supplementary Table 2.
Cell separation and flow cytometry
We purified Tref cells by magnetic separation (STEMCELL Technology) and assessed purity by flow cytometry using a LSRII flow cytometer (BD Biosciences). We analyzed flow data with FlowJo software v7.2.2 (Tree Star). To determine levels of Tref cell apoptosis, we labeled epitope-stimulated PBMCs with annexin V (BD Bioscience).
In vitro cytokine measurements
We measured cytokine responses by performing ELISPOT or intracellular cytokine secretion (ICS) assays40, 41. In brief, we cultured 2 × 105 (ELISPOT) and 1 × 106 (ICS) cells per well and stimulated them with 2 µg ml−1 of their cognate epitopes. For ELISPOT we called a response positive when the number of spot-forming cells was twice the background level and there were at least 50 spot-forming cells per 1 × 106 PBMCs. For ICS, we called a response positive when the percentage of bright cytokine+CD8+ T cells was twice that of the negative control.
Proliferation assays
We performed CFSE dilution assays as previously described8. Where indicated, we added Tref cells (1:4) to PBMCs or CD8+ T cell cultures, although we obtained similar data when we used a ratio as low as 1:16 Tref cells:PBMCs (that is, an approximate Tref cell frequency of 6%, which we have shown is in the physiological range of Tref cell frequencies in vivo; Supplementary Fig. 5). We performed some assays in the presence of lactose (Sigma-Aldrich; 30 mM) or the GzmB inhibitor z-AAD-CMK (Calbiochem; 10 µg ml−1).
Small interfering RNA transduction
For silencing of LGALS9, we performed RNA interference experiments on isolated Tref cells according to the manufacturer's protocol (Santa Cruz Biotechnology). Knockdown of LGALS9 in Tref cells is shown in Supplementary Figure 2a. Control Tref cells received a scrambled I duplex RNA. Seven hours after transfection, we replaced the transfection medium with complete medium and cultured Tref cells for another 24 h before using them in CFSE proliferation assays. For silencing of GZMB, we electroporated PBMCs or CD8+ T cells with GZMB siRNA (Applied Biosystems/Ambion) following the manufacturer's guidelines. We rested the cells for 36 h before using them in CFSE proliferation assays. Expression of intracellular GzmB after electroporation is shown in Supplementary Figure 2b.
Statistical analyses
We calculated percentage suppression of proliferation by the formula (percentage CFSEloCD8+ without Tref cells − percentage CFSEloCD8+ with Tref cells) / percentage CFSEloCD8+ without Tref cells × 100, grouped according to HLA restricting allele. Two responses with CFSEloCD8+ cells without Tref cells were zero, and the lowest measured response (0.012) was substituted to prevent a division by zero error. We similarly calculated percentage suppression by lactose and LGALS9 siRNA by substituting the differential suppression via these compounds. We calculated percentage suppression of cytokine secretion by the formula (spot-forming cells per million PBMCs without Tref cells–spot-forming cells per million PBMCs with Tref cells) / spot-forming cells per million PBMCs without Tref cells × 100. We used signed Wilcoxon tests to compare paired data with and without the suppressing agent (Tref cells, lactose, siRNA). For comparisons of grouped data, we used Kruskal-Wallis tests with a post hoc Dunn's test for subgroup comparisons. We used Spearman rank tests for correlation analyses. All tests were two-tailed, with a P value of less than 0.05 considered statistically significant. We conducted statistical analyses and graphing with GraphPad Prism 5.02 (GraphPad) and Adobe Illustrator Creative Suite 3, 13.0.2 (Adobe Systems).
Supplementary Material
ACKNOWLEDGMENTS
We thank our study volunteers for providing samples and supporting this work, as well as the clinical staff for their dedication to this research. This work was supported by US National Institutes of Health (NIH) grants R01 AI65328, R21 AI089373, U01 AI4674, U01 AI 46725, P01 AI57005, P30 AI27757, a New Investigator Award (for S.E.) from the University of Washington/Fred Hutchinson Cancer Research Center Center for AIDS Research (CFAR), AI30731, AI 081060, R37 AI042528 and M01-RR-00037 (the University of Washington General Clinical Research Center). This research was supported by the University of Washington Center for AIDS Research (CFAR), an NIH funded program (P30 AI027757) which is supported by the NIH National Institute of Allergy and Infectious Diseases, National Cancer Institute, National Institute of Mental Health, National Institute on Drug Abuse, National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute and National Center for Complementary and Alternative Medicine). We also acknowledge the support of the James. B. Pendleton Charitable Trust.
Footnotes
AUTHOR CONTRIBUTIONS
S.E. designed and performed all the experiments and wrote part of the manuscript. N.L. assisted S.E. to perform some of the experiments. W.L.D. performed statistical analysis and graphing design. K.J.L. performed epitope mapping for individuals infected with HSV. D.M.K. advised on the HSV experiment. D.M.K., K.J.L., M.J.M. and A.C.C. supplied samples from subjects. H.H. designed and supervised all of the research and wrote the manuscript. All authors revised and edited the manuscript.
References
- 1.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 2.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 3.Carrington M, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 4.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu. Rev. Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 5.Dinges WL, et al. Virus-specific CD8+ T cell responses better define HIV disease progression than HLA genotype. J. Virol. 2010;84:4461–4468. doi: 10.1128/JVI.02438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 7.Lichterfeld M, et al. Loss of HIV-1–specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1–specific CD4+ T cells. J. Exp. Med. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton H, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J. Immunol. 2006;177:7406–7415. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- 9.Vila J, Isaacs JD, Anderson AE. Regulatory T cells and autoimmunity. Curr. Opin. Hematol. 2009;16:274–279. doi: 10.1097/MOH.0b013e32832a9a01. [DOI] [PubMed] [Google Scholar]
- 10.Galgani M, Di Giacomo A, Matarese G, La Cava A. The Yin and Yang of CD4+ regulatory T cells in autoimmunity and cancer. Curr. Med. Chem. 2009;16:4626–4631. doi: 10.2174/092986709789878201. [DOI] [PubMed] [Google Scholar]
- 11.Zhu C, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 12.Koguchi K, et al. Dysregulated T cell expression of TIM3 in multiple sclerosis. J. Exp. Med. 2006;203:1413–1418. doi: 10.1084/jem.20060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XC, Turka LA. An update on regulatory T cells in transplant tolerance and rejection. Nat. Rev. Nephrol. 2010;6:577–583. doi: 10.1038/nrneph.2010.101. [DOI] [PubMed] [Google Scholar]
- 14.Jones RB, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J. Exp. Med. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann-Haefelin C, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 16.Mathieu A, et al. The interplay between the geographic distribution of HLA-B27 alleles and their role in infectious and autoimmune diseases: a unifying hypothesis. Autoimmun. Rev. 2009;8:420–425. doi: 10.1016/j.autrev.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Henseler T. Genetics of psoriasis. Arch. Dermatol. Res. 1998;290:463–476. doi: 10.1007/s004030050338. [DOI] [PubMed] [Google Scholar]
- 18.Migueles SA, Connors M. Frequency and function of HIV-specific CD8(+) T cells. Immunol. Lett. 2001;79:141–150. doi: 10.1016/s0165-2478(01)00276-0. [DOI] [PubMed] [Google Scholar]
- 19.Migueles SA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. Tim-3-Galectin-9 pathway involves the suppression induced by CD4+CD25+ regulatory T cells. Immunobiology. 2009;214:342–349. doi: 10.1016/j.imbio.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabatos CA, et al. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat. Immunol. 2003;4:1102–1110. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 23.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 24.Sakuishi K, et al. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehler TC, et al. Rapid identification and sorting of viable virus-reactive CD4+ and CD8+ T cells based on antigen-triggered CD137 expression. J. Immunol. Methods. 2008;339:23–37. doi: 10.1016/j.jim.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Chabot S, et al. Regulation of galectin-9 expression and release in Jurkat T cell line cells. Glycobiology. 2002;12:111–118. doi: 10.1093/glycob/12.2.111. [DOI] [PubMed] [Google Scholar]
- 27.Ashley CW, Baecher-Allan C. Cutting Edge: Responder T cells regulate human DR+ effector regulatory T cell activity via granzyme B. J. Immunol. 2009;183:4843–4847. doi: 10.4049/jimmunol.0900845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson J, et al. The prevalence of regulatory T cells in lymphoid tissue is correlated with viral load in HIV-infected patients. J. Immunol. 2005;174:3143–3147. doi: 10.4049/jimmunol.174.6.3143. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson J, et al. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. 2006;108:3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsunemi S, et al. Relationship of CD4+CD25+ regulatory T cells to immune status in HIV-infected patients. AIDS. 2005;19:879–886. doi: 10.1097/01.aids.0000171401.23243.56. [DOI] [PubMed] [Google Scholar]
- 31.Eggena MP, et al. Depletion of regulatory T cells in HIV infection is associated with immune activation. J. Immunol. 2005;174:4407–4414. doi: 10.4049/jimmunol.174.7.4407. [DOI] [PubMed] [Google Scholar]
- 32.Abrams D, et al. Interleukin-2 therapy in patients with HIV infection. N. Engl. J. Med. 2009;361:1548–1559. doi: 10.1056/NEJMoa0903175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss L, et al. In vivo expansion of naive and activated CD4+CD25+FOXP3+ regulatory T cell populations in interleukin-2–treated HIV patients. Proc. Natl. Acad. Sci. USA. 2010;107:10632–10637. doi: 10.1073/pnas.1000027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoniou AN, Ford S, Taurog JD, Butcher GW, Powis SJ. Formation of HLA-B27 homodimers and their relationship to assembly kinetics. J. Biol. Chem. 2004;279:8895–8902. doi: 10.1074/jbc.M311757200. [DOI] [PubMed] [Google Scholar]
- 36.Xing S, et al. Increased turnover of FoxP3high regulatory T cells is associated with hyperactivation and disease progression of chronic HIV-1 infection. J. Acquir. Immune Defic. Syndr. 2010;54:455–462. doi: 10.1097/QAI.0b013e3181e453b9. [DOI] [PubMed] [Google Scholar]
- 37.Sabbagh L, et al. The selective increase in caspase-3 expression in effector but not memory T cells allows susceptibility to apoptosis. J. Immunol. 2004;173:5425–5433. doi: 10.4049/jimmunol.173.9.5425. [DOI] [PubMed] [Google Scholar]
- 38.Feng BJ, et al. Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5:e1000606. doi: 10.1371/journal.pgen.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laing KJ, et al. Diversity in CD8+ T cell function and epitope breadth among persons with genital herpes. J. Clin. Immunol. 2010:703–722. doi: 10.1007/s10875-010-9441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horton H, et al. Induction of human immunodeficiency virus type 1 (HIV-1)–specific T-cell responses in HIV vaccine trial participants who subsequently acquire HIV-1 infection. J. Virol. 2006;80:9779–9788. doi: 10.1128/JVI.00794-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horton H, et al. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J. Immunol. Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.