Abstract
Six bacterial genera containing species commonly used as probiotics for human consumption or starter cultures for food fermentation were compared and contrasted, based on publicly available complete genome sequences. The analysis included 19 Bifidobacterium genomes, 21 Lactobacillus genomes, 4 Lactococcus and 3 Leuconostoc genomes, as well as a selection of Enterococcus (11) and Streptococcus (23) genomes. The latter two genera included genomes from probiotic or commensal as well as pathogenic organisms to investigate if their non-pathogenic members shared more genes with the other probiotic genomes than their pathogenic members. The pan- and core genome of each genus was defined. Pairwise BLASTP genome comparison was performed within and between genera. It turned out that pathogenic Streptococcus and Enterococcus shared more gene families than did the non-pathogenic genomes. In silico multilocus sequence typing was carried out for all genomes per genus, and the variable gene content of genomes was compared within the genera. Informative BLAST Atlases were constructed to visualize genomic variation within genera. The clusters of orthologous groups (COG) classes of all genes in the pan- and core genome of each genus were compared. In addition, it was investigated whether pathogenic genomes contain different COG classes compared to the probiotic or fermentative organisms, again comparing their pan- and core genomes. The obtained results were compared with published data from the literature. This study illustrates how over 80 genomes can be broadly compared using simple bioinformatic tools, leading to both confirmation of known information as well as novel observations.
Electronic supplementary material
The online version of this article (doi:10.1007/s00248-011-9948-y) contains supplementary material, which is available to authorized users.
Introduction
The first bacterial genome sequences were published in 1995, and within 15 years, over a thousand fully sequenced bacterial genomes have become publicly available [16]. A number of these genome sequences are derived from bacteria used as probiotics or starter cultures in food fermentation, or both. Reid and co-workers [21] defined probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host”. A number of bacterial species from various genera are in use as probiotics, including members of Lactobacillus, Lactococcus and, less commonly, Leuconostoc. These Firmicutes are sometimes collectively described as lactic acid bacteria (LAB). Other commonly used probiotic species belong to Bifidobacterium, a genus within the phylum Actinobacteria. These genera exclusively contain species that are unlikely to cause disease while colonizing the intestine, and although some species (e.g. Bifidobacterium dentium) have been associated with dental disease, these are more commonly members of a normal oral flora. The distinction between normal gut flora (commensals) and probiotic bacteria having a beneficial effect on their host’s health cannot always be made, for which reason we collectively describe them here as ‘non-pathogens’. Species belonging to LAB or Bifidobacterium are also frequently used in food fermentation, another application where the bacterial load of food is desirably increased. Besides LAB and Bifidobacterium, fermentation starter cultures can typically comprise of Streptococcus thermophilus, a non-pathogenic member of this genus that mostly contains pathogenic species. Some strains of Enterococcus are also in use as starter cultures or probiotics, whereby the used species also contain pathogenic strains. These two genera are therefore of interest, and their species that are used as starter cultures are included in our general description of ‘non-pathogens’. Other types of bacteria (particular strains of Escherichia coli, Pediococcus species and others) or yeasts used as starter cultures or probiotics are not treated here.
For all six genera of interest, multiple genome sequences are publicly available. In many cases, several genomes per species have been sequenced, so that the variation between and even within species can be assessed. One obvious question that could be addressed by comparison of these genomes is: what genes (if any) are common to all genomes of non-pathogens and distinct from genes found in (related) pathogens? Such a comparison requires including multiple species and genera of multiple bacterial phyla (in this case, the phylum of Firmicutes and Actinobacteria). As a general rule, genetic diversity increases with evolutionary distance, so that the genetic variation in such a collection of genomes will be enormous. One way of extracting information from such complex data is by grouping genes into functional groups or families, so that gene families rather than individual genes are compared. Such grouping is based on protein sequence similarity, as this approximately predicts conservation of gene function, ignoring the exceptions resulting from parallel evolution where function similarity does not coincide with sequence conservation. Slight differences in function, resulting from minor differences in sequences, are usually ignored in these groupings, so that fewer but broader groups can be achieved.
In this contribution, 2 approaches were used to compare over 80 genomes from 6 bacterial genera of interest. First, all protein-coding genes from these genomes were grouped into gene families based on sequence identity using a defined similarity cut-off, after which comparisons between and across genera could be performed. Genomes were then compared within their genus for both conserved and variable genes. Second, clusters of orthologous groups (COG) of genes were used to produce functional groups of genes. An attempt was made to identify differences in functional gene distribution between pathogenic and non-pathogenic members of the six genera of interest.
Materials and Methods
Selection of Genomes Used in This Study
Publicly available genomes of the six bacterial genera analyzed here were identified from the NCBI web pages. All completely sequenced genomes (as of July 2010) of 4 Lactococcus lactis strains, 3 Leuconostoc species and 21 Lactobacillus strains from 14 species were included. For Bifidobacterium, 11 completely sequenced and 8 incomplete genomes were selected; the latter were chosen when fewer than 70 contigs resulting in 19 genomes from 9 species. Since only 1 complete Enterococcus genome was available at the time of analysis, this genome was combined with 10 incomplete sequences, provided they were represented in fewer than 80 contigs, whereby animal isolates were excluded. This allowed inclusion of 2 strains obtained from normal gut flora to give 11 genomes from 4 species. For Streptococcus, all S. thermophilus genomes were included. All other species of this genus for which genome sequences were available are pathogens, and a selection of these was made of three genomes per species. These were chosen based on their strain characteristics to cover common but diverse serotypes. Animal isolates were excluded, although Streptococcus suis (a typical pig pathogen) was included as it has been responsible for a large human outbreak in China. This resulted in 23 genomes from 12 species. All genomes are listed in Table 1, which also provides characteristics such as their size, GC content and the strain description. The latter was extracted from the Genome Project pages at NCBI but checked in the corresponding genome publication when available. This resulted in a few small differences from descriptions listed on the Genome Project Description pages at NCBI. The derived proteomes (protein-coding sequences translated from the DNA sequence) were extracted from GenBank for completed sequences or produced with Prodigal [14] for incomplete sequences.
Table 1.
Genomes selected for analysis
| GPID | Strain namea | Size, bp or Mb | % CG | Contigs | Number of genes | Strain characteristics |
|---|---|---|---|---|---|---|
| 82 | Lactobacillus acidophilus NCFM | 1,993,560 | 34.7 | 1 | 1,862 | Commercial strain for yogurt, fluid milk production |
| 404 | Lactobacillus brevis ATCC 367 | 2,340,228 | 46.1 | 3 | 2,218 | Starter culture for beer, sourdough, and silage |
| 402 | Lactobacillus casei ATCC 334 | 2,924,325 | 46.6 | 2 | 2,771 | Starter culture for milk fermentation and flavour development of cheese |
| 30359 | Lactobacillus casei BL23 | 3,079,196 | 46.3 | 1 | 3,044 | Probiotic strain |
| 46813 | Lactobacillus crispatus ST1 | 2,043,161 | 36.9 | 1 | 2,024 | Normal oral/vaginal flora, chicken isolate |
| 16871 | Lactobacillus delbrueckii bulgaricus ATCC 11842 | 1,864,998 | 49.7 | 1 | 2,096 | Yogurt |
| 403 | Lactobacillus delbrueckii bulgaricus ATCC BAA-365 | 1,856,951 | 49.7 | 1 | 1,721 | Thermophilic starter culture for yogurt, Swiss and Italian-type cheeses |
| 18979 | Lactobacillus fermentum IFO 3956 | 2,098,685 | 51.5 | 1 | 1,843 | Not specified |
| 84 | Lactobacillus gasseri ATCC 33323 | 1,894,360 | 35.3 | 1 | 1,755 | Human isolate, type strain |
| 17811 | Lactobacillus helveticus DPC 4571 | 2,080,931 | 37.1 | 1 | 1,610 | Cheese culture |
| 36575 | Lactobacillus johnsonii FI9785 | 1,785,116 | 34.4 | 1 | 1,737 | Competitive exclusion strain in chicken |
| 9638 | Lactobacillus johnsonii NCC 533 | 1,992,676 | 34.6 | 1 | 1,821 | Probiotic strain |
| 32969 | Lactobacillus plantarum JDM1 | 3,197,759 | 44.7 | 1 | 2,948 | Probiotic strain |
| 356 | Lactobacillus plantarum WCFS1 | 3,348,625 | 44.4 | 4 | 3,101 | Human saliva |
| 15766 | Lactobacillus reuteri DSM 20016 | 1,999,618 | 38.9 | 1 | 1,900 | Type strain, human isolate |
| 19011 | Lactobacillus reuteri JCM 1112 | 2,039,414 | 38.9 | 1 | 1,820 | Human isolate |
| 32195 | Lactobacillus rhamnosus GG | 3,010,111 | 46.7 | 1 | 2,944 | Probiotic strain |
| 40637 | Lactobacillus rhamnosus GG ATCC53103 | 3,005,051 | 46.7 | 1 | 2,834 | Human isolate |
| 32197 | Lactobacillus rhamnosus Lc 705 | 3,033,106 | 46.7 | 2 | 2,992 | Probiotic strain |
| 13435 | Lactobacillus sakei sakei 23K | 1,884,661 | 41.3 | 1 | 1,885 | Fermenting |
| 13280 | Lactobacillus salivarius UCC118 | 2,133,977 | 33.0 | 4 | 2,014 | Probiotic strain |
| 18797 | Lactococcus lactis cremoris MG1363 | 2,529,478 | 35.7 | 1 | 2,516 | Plasmid-cured NCDO712, lab strain |
| 401 | Lactococcus lactis cremoris SK11 | 2,598,348 | 35.8 | 6 | 2,504 | Cheese production |
| 72 | Lactococcus lactis lactis Il1403 | 2,365,589 | 35.3 | 1 | 2,266 | Laboratory strain |
| 41115 | Lactococcus lactis lactis KF147 | 2,635,654 | 34.9 | 1 | 2,575 | Fermenting, non-dairy |
| 16062 | Leuconostoc citreum KM20 | 1,896,614 | 38.9 | 5 | 1,823 | Kimchi (food, Korea) |
| 40837 | Leuconostoc kimchii IMSNU11154 | 2,101,787 | 37.0 | 1 | 2,130 | Kimchi? not specified |
| 315 | Leuconostoc mesenteroides mesenteroides ATCC 8293 | 2,075,763 | 37.7 | 2 | 2,005 | Food fermentation, not specified |
| 70 | Enterococcus faecalis V583 | 3,359,974 | 37.4 | 4 | 3,265 | Clinical, blood isolate, vancomycin resistant |
| 32949 | Enterococcus faecalis T11 | 2,729,089 | 37.7 | 49 | 2,522 | Urine isolate |
| 32941 | Enterococcus faecalis E1Sol | 2,853,151 | 37.5 | 75 | 2,737 | Faecal isolate, antibiotic-naïve, normal flora |
| 20843 | Enterococcus faecalis OG1RF | 2,739,625 | 37.7 | 1 | 2,515 | No info - lab strain? |
| 32919 | Enterococcus faecalis T3 | 2,821,089 | 37.6 | 40 | 2,603 | Urine isolate |
| 32927 | Enterococcus gallinarum EG2 | 3,134,429 | 40.6 | 49 | 2,985 | No info |
| 32931 | Enterococcus casseliflavus EC10 | 3,423,270 | 42.5 | 54 | 3,243 | No info |
| 32935 | Enterococcus casseliflavus EC20 | 3,392,502 | 42.8 | 57 | 3,121 | No info |
| 46979 | Enterococcus faecium PC4.1 | 2,811,160 | 37.9 | 78 | 2,705 | Human microbiome, normal flora |
| 32965 | Enterococcus faecium Com12 | 2,685,402 | 38.1 | 67 | 2,573 | No info |
| 32967 | Enterococcus faecium Com15 | 2,771,455 | 38.3 | 70 | 2,698 | No info |
| 330 | Streptococcus agalactiae 2603V/R | 2,160,267 | 35.6 | 1 | 2,124 | Clinical isolate, common in adults |
| 326 | Streptococcus agalactiae A909 | 2,127,839 | 35.6 | 1 | 1,996 | No info |
| 334 | Streptococcus agalactiae NEM316 | 2,211,485 | 35.6 | 1 | 2,134 | Blood isolate |
| 27849 | Streptococcus dysgalactiae equisimilis GGS 124 | 2,106,340 | 39.6 | 1 | 2,100 | No info |
| 34729 | Streptococcus gallolyticus UCN34 | 2,350,911 | 37.6 | 1 | 2,261 | Normally rumen flora, this is a clinical human isolate from endocarditis |
| 66 | Streptococcus gordonii str. Challis CH1 | 2,196,662 | 40.5 | 1 | 2,051 | Causes caries and periodontal diseases |
| 20527 | Streptococcus infantarius infantarius ATCC BAA-102 | 1,925,087 | 37.6 | 22 | 1,962 | Human microbiome project, normal flora |
| 16302 | Streptococcus mitis B6 | 2,146,611 | 40.0 | 1 | 2,018 | Clinical isolate |
| 28997 | Streptococcus mutans NN2025 | 2,013,587 | 36.8 | 1 | 1,895 | Normally oral flora, can cause caries, endocarditis. Clinical isolate |
| 333 | Streptococcus mutans UA159 | 2,030,921 | 36.8 | 1 | 1,960 | Oral flora, can cause caries, caries isolate |
| 31233 | Streptococcus pneumoniae ATCC 700669 | 2,221,315 | 39.5 | 1 | 2,135 | Alternative name Spain 23FST81. Pandemic, high prevalence, invasive |
| 29047 | Streptococcus pneumoniae G54 | 2,078,953 | 39.7 | 1 | 2,115 | Resistant clinical isolate |
| 277 | Streptococcus pneumoniae TIGR4 | 2,160,842 | 39.7 | 1 | 2,125 | Virulent clinical isolate |
| 269 | Streptococcus pyogenes M1 GAS SF370 | 1,852,441 | 38.5 | 1 | 1,696 | Group A |
| 16364 | Streptococcus pyogenes MGAS10270 | 1,928,252 | 38.4 | 1 | 1,987 | Sequenced for comparative genome analysis |
| 286 | Streptococcus pyogenes MGAS8232 | 1,895,017 | 38.5 | 1 | 1,845 | Serotype M18 |
| 13942 | Streptococcus sanguinis SK36 | 2,388,435 | 43.4 | 1 | 2,270 | Indigenous oral bacteria, causes dental decay, oral plaque isolate |
| 17153 | Streptococcus suis 05ZYH33 | 2,096,309 | 41.1 | 1 | 2,186 | Causes disease in pigs and occasionally humans |
| 32237 | Streptococcus suis BM407 | 2,170,808 | 41.0 | 2 | 2,058 | Human clinical isolate |
| 18737 | Streptococcus suis GZ1 | 2,038,034 | 41.4 | 1 | 1,978 | Causes meningitis, arthritis, pneumonia in pigs human epidemic in China |
| 13163 | Streptococcus thermophilus CNRZ1066 | 1,796,226 | 39.1 | 1 | 1,915 | Isolated from yogurt for industrial dairy fermentations |
| 13773 | Streptococcus thermophilus LMD-9 | 1,864,178 | 39.1 | 3 | 1,716 | Used in the manufacture of fermented dairy foods |
| 13162 | Streptococcus thermophilus LMG 18311 | 1,796,846 | 39.1 | 1 | 1,889 | Isolated from yogurt for industrial dairy fermentations |
| 16321 | Bifidobacterium adolescentis ATCC 15703 | 2,089,645 | 59.2 | 1 | 1,631 | Normal gut flora |
| 19423 | Bifidobacterium animalis lactis AD011 | 1,933,695 | 60.5 | 1 | 1,528 | Normal gut flora |
| 42883 | Bifidobacterium animalis lactis BB-12 | 1,942,198 | 60.5 | 1 | 1,642 | Normal gut flora |
| 32897 | Bifidobacterium animalis lactis Bl-04 | 1,938,709 | 60.5 | 1 | 1,567 | Normal gut flora |
| 32893 | Bifidobacterium animalis lactis DSM 10140 | 1,938,483 | 60.5 | 1 | 1,566 | Normal gut flora |
| 32515 | Bifidobacterium animalis lactis V9 | 1,944,050 | 60.4 | 1 | 1,572 | Normal gut flora |
| 28807 | Bifidobacterium animalis lactis HN019 | 1,915,892 | 60.4 | 28 | 1,578 | Normal gut flora |
| 17583 | Bifidobacterium dentium Bd1 | 2,636,367 | 58.5 | 1 | 2,129 | Normal oral and gut flora, can cause caries, caries isolate |
| 20555 | Bifidobacterium dentium ATCC 27678 | 2,642,081 | 58.5 | 2 | 2,151 | Human microbiome, faeces isolate |
| 18773 | Bifidobacterium longum DJO10A | 2,389,526 | 60.2 | 3 | 2,003 | Normal gut flora, probiotic |
| 328 | Bifidobacterium longum NCC2705 | 2,260,266 | 60.1 | 2 | 1,729 | Normal gut flora, probiotic |
| 17189 | Bifidobacterium longum infantis ATCC 15697 | 2,832,748 | 59.9 | 1 | 2,416 | Normal gut flora, probiotic |
| 30065 | Bifidobacterium longum infantis CCUG 52486 | 2,453,376 | 60.2 | 55 | 2,085 | Normal gut flora, human microbiome project |
| 47579 | Bifidobacterium longum longum JDM301 | 2,477,838 | 59.8 | 1 | 1,959 | Normal gut flora, probiotic |
| 29261 | Bifidobacterium angulatum DSM 20098 | 2,007,108 | 59.4 | 17 | 1,586 | Normal gut flora, type strain |
| 30055 | Bifidobacterium bifidum NCIMB 41171 | 2,186,140 | 62.8 | 33 | 1,810 | Normal gut flora, probiotic |
| 30749 | Bifidobacterium catenulatum DSM 16992 | 2,058,429 | 56.1 | 31 | 1,720 | Normal gut flora |
| 30751 | Bifidobacterium gallicum DSM 20093 | 2,019,802 | 57.5 | 27 | 1,580 | Human microbiome project |
| 30373 | Bifidobacterium pseudocatenulatum DSM 20438 | 2,304,808 | 56.3 | 36 | 1,870 | Human microbiome project |
aThe official abbreviation ‘subsp.’ between species and subspecies name has been deleted throughout this contribution
GPID genome project identification number (NCBI: see http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi), NA not available
Definition of Gene Families and Pan- and Core Genome
The pan-genome of a collection of genomes represents all genes encountered in these genomes [27]. In order to define a pan-genome, the criteria to score a gene as ‘conserved’ or ‘novel’ were used as previously described [12]. Simply put, two genes are considered to belong to the same gene family and thus ‘conserved’ when their amino acid sequence is at least 50% identical over at least 50% of the length of the longest gene. All genes of a genome are thus grouped into gene families. Multiple genes per genome can belong to a single gene family, resulting in a lower number of gene families per genome than the reported number of genes. A gene not finding a match with the given criteria is put in its own gene family as a singleton.
An accumulative pan-genome was constructed according to Friis et al. [11], who built on work by Tettelin and co-workers [27]. A resulting pan-genome curve increases in size as more genomes are analyzed, and its shape is order-dependent, though the accumulative pan-genome is not influenced by the order of analysis. Similarly, a core genome is defined as all gene families conserved in all analyzed genomes, and this decreases in size as more genomes are analyzed.
Pairwise pan- and core genomes were calculated for all genome combinations as above, and for each combination, the obtained core genome was expressed as the fraction of the pan-genome. These percentages were visualized in a BLAST Matrix [11].
Core Genome Consensus Tree
Phylogenetic trees were constructed of all core genes that were conserved within the analyzed Firmicute genomes. Multiple alignments of all core sequences were performed with MUSCLE software [7]. PAUP was used to construct a set of core trees [10]. Later, these trees were compared and a best-fit consensus tree was constructed as described by Retief [22].
In Silico MLST Analysis
In silico multilocus sequence typing (MLST) analysis was performed with gene fragments extracted from the genome sequences. For Bifidobacterium, gene fragments from clpC, fusA, gyrB, IleS, purF, rplB and rpoB were extracted, according to the method proposed for Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium longum [6]. For Enterococcus, the gene set of gdh, gyd, pstS, gki, aroE, xpt and yqlI, which is advised for use in Enterococcus faecalis (http://www.mlst.net), was compared with that designed for Enterococcus faecium, which is based on atpA, ddl, gdh, purK, gyd, pstS and adk. For Lactobacillis, de Las Rivas and co-workers [4] described an MLST gene set specified for Lactobacillis plantarum based on the target genes pgm, ddl, gyrB, purK1, gdh, mutS and tkt4. Two alternative combinations of genes have been proposed for Lactobacillis casei: ftsZ, polA, mutL, metRS, nrdD and pgm [1] or fusA, ileS, lepA, leuS, pyrG, recA and recG (http://www.pasteur.fr). A fourth gene set (gdh, gyrA, mapA, nox, pgmA and pta) has recently been described for Lactobacillis sanfranciscensis [20], but since this species is not represented in our dataset, this scheme was not used. For each genus, after concatenation of the gene fragments, a maximum likelihood phylogenetic tree was constructed.
Analysis of Variable Gene Content
The variable gene content of the analyzed genomes was compared using the method by Snipen and Ussery [24]. This method calculates Manhattan distances based on a matrix in which the presence or absence for each gene in each genome is scored with the binary score of 0 (absent) or 1 (present). Core genes and singletons are ignored. BLAST Atlases were produced according to Hallin and co-workers [12].
COG Analysis
COG is a database of proteins where each sequence is assigned to some group. All proteins within a group are believed to have a common ancestor and are likely to share a common function. The various groups are again clustered into some super-groups called functional groups [26]. In this analysis, each found protein was compared to the COG database using BLASTP to identify the functional groups to which they belong. An R-script was used to analyze the protein composition in pan- and core genomes, and the results were visualized in a pie chart. This was done using standard operating procedures [19].
Results
Comparison of Pan-Genomes
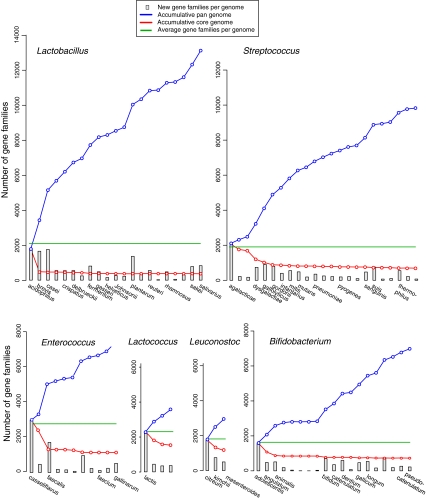
After the selection of genome sequences as described in the “Materials and Methods” section, 81 genome sequences were obtained from organisms listed in Table 1. These represented 43 different species and coded for 147,074 protein genes in total. Table 2 summarizes some average findings for each of the analyzed genera. Enterococcus has the largest average genome size and Leuconostoc the smallest, a difference that is reflected in their average number of genes, since gene density is generally conserved in these bacteria. Bifidobacterium has a significantly higher CG content, which was one of the reasons to place this genus in the Actinobacteria [9]. The CG content varied most within the genus of Lactobacillus, with a CG content below 37.2% for Lactobacillus acidophilus, Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus helveticus, Lactobacillus johnsonii and Lactobacillus salivarius; genomes of the other members of this genus contain at least 38.9% CG. The average number of gene families (as defined in the “Materials and Methods” section) is also shown in Table 2. Since multiple genes per genome can belong to a single gene family, there are fewer gene families than genes per genome, but the difference is small for Bifidobacterium. This indicates that there is little gene redundancy in that genus. Lastly, the pan- and core genomes of these genera (based on the analyzed genomes) are quantified in Table 2. The plots resulting in these running totals are shown in Fig. 1, where the average number of gene families present per genome is given as a green line. In all graphs, the pan-genome and core genome curves strongly diverge, indicative of a large variation in gene content between the analyzed genomes within each genus. The largest difference between the pan- and core genome, as a measure for the variance within the analyzed genera, is seen with Lactobacillus (21 genomes of 14 species) and Streptococcus (23 genomes of 12 species). The variance is larger in four genomes of Lc. lactis than in three different Leuconostoc species. Thus, intra-species variation in gene content of Lc. lactis exceeds inter-species variation of Leuconostoc, at least for these analyzed genomes.
Table 2.
Average findings per genus and their pan- and core genome
| Genus | Number of genomes included | Number of species | Average genome size (kbp) | Average % CG | Average number of genes (min–max values) | Average number of gene families (min–max values) | Pan-genomea | Core genomea |
|---|---|---|---|---|---|---|---|---|
| Lactobacillus | 21 | 14 | 2,369 | 42.4 | 2,235 (1,562–3,059) | 2,071 (1,437–2,873) | 13,069 | 363 |
| Lactococcus | 4 | 1 | 2,532 | 35.4 | 2,465 (2,266–2,504) | 2,238 (2,118–2,341) | 3,389 | 1,522 |
| Leuconostoc | 3 | 3 | 2,025 | 37.9 | 1,986 (1,820–2,130) | 1,896 (1,724–2,050) | 2,927 | 1,164 |
| Enterococcus | 11 | 4 | 3,041 | 36.6 | 3,078 (2,573–2,515) | 2,707 (2,439–3,114) | 7,519 | 1,092 |
| Streptococcus | 23 | 12 | 1,981 | 38.9 | 2,018 (1,696–2,270) | 1,923 (1,643–2,180) | 9,785 | 638 |
| Bifidobacterium | 19 | 9 | 2,209 | 59.5 | 1,796 (1,528–2,416) | 1,746 (1,497–2,287) | 6,980 | 724 |
aNumber of gene families is given
Figure 1.
Pan- and core genome plots of the six analyzed genera. The genomes were analyzed in alphabetical order of species names
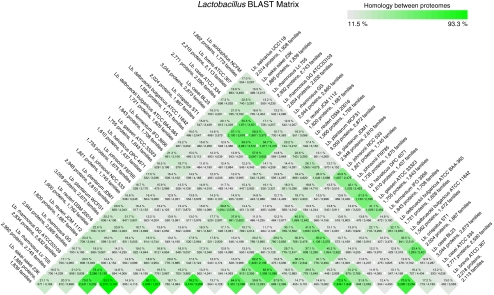
The pan- and core genomes of pairwise genome comparisons were also determined to establish the percentage identity for each combination. This identity was expressed as the pairwise core genome divided by its pan-genome and was visualized by colour intensity in a BLAST Matrix. Figure 2 shows the BLAST Matrix for the Lactobacillus genomes. The strongest green, indicative of the highest fraction of genes found similar between two genomes, are reported for comparisons within a species, shown at the bottom of the figure. Some species also share a large fraction of genes between them. For instance, the two Lb. casei genomes share between 55.5% and 59.3% of their genes with those of the three Lactobacillus rhamnosus genomes (represented in the six darker green cells in the upper part of the matrix). An even higher similarity (62.2–62.8%) is found between Lb. gasseri and Lb. johnsonii. The highest similarity recorded is 93.3%, between two Lb. rhamnosus strains, and the lowest is 11.5%, between Lb. casei BL23 and Lactobacillus delbrueckii bulgaricus ATCCBAA-365.
Figure 2.
BLAST Matrix for the Lactobacillus genomes. To the side, the total number of protein genes and gene families are listed for each genome. In the matrix cells, the shared protein genes are given as a percentage, based on the ratio of the core genome and pan-genome of each pair, as indicated
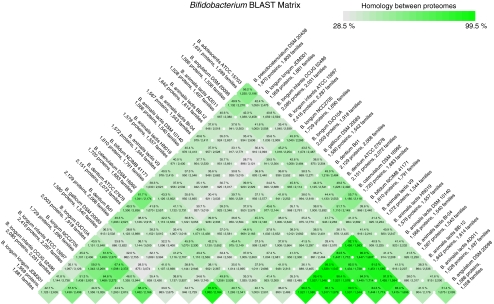
A similar matrix is shown for Bifidobacterium in Fig. 3. In this case, the similarity between the six Bifidobacterium animalis genomes is obvious (visible as 15 strongly coloured cells at the bottom right). Two of these genomes reach a similarity of 95.5%. The lowest degree of similarity is seen between Bifidobacterium gallicum and B. longum infantis strain ATCC 15697 (28.5%).
Figure 3.
BLAST Matrix for Bifidobacterium genomes. Note that the colour scale is different to that of Fig. 2
When a BLAST Matrix was constructed with all genomes included in the analysis, the similarity between Bifidobacterium genomes and those of the other genera remained below 3%, illustrative of the difference of Bifidobacterium compared to the Firmicutes (results not shown). Thus, despite their sharing of an ecological niche, these bacteria share relatively few genes. A comparison of all Firmicute genomes is provided as Supplementary Fig. S1. As expected, the found percentage identity within any of these genera is much higher than that between genera. For instance, the three Leuconostoc genomes produced a similarity of 49.5–52.3% between them, but around 8% to 10% to genomes of other genera. The four Lc. lactis genomes gave slightly higher similarities of 16.1–18.4% to all other Firmicute genomes whilst sharing 59.5–66.1% between themselves. An Enterococcus and a Streptococcus genome typically share 10% to 15% of their genes, and two genomes of Enterococcus and Lactococcus 14% to 16%. Different Enterococcus species share around 30% of their genes, but multiple genomes within one species of this genus have around 70% of their genes being similar.
Comparison of Core Genomes and Conserved Genes
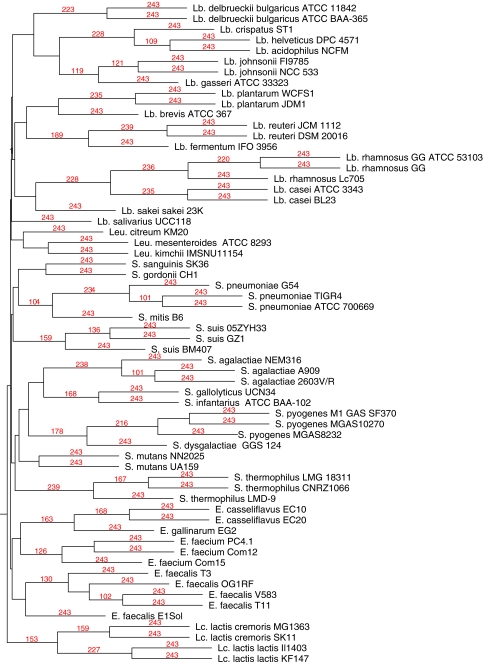
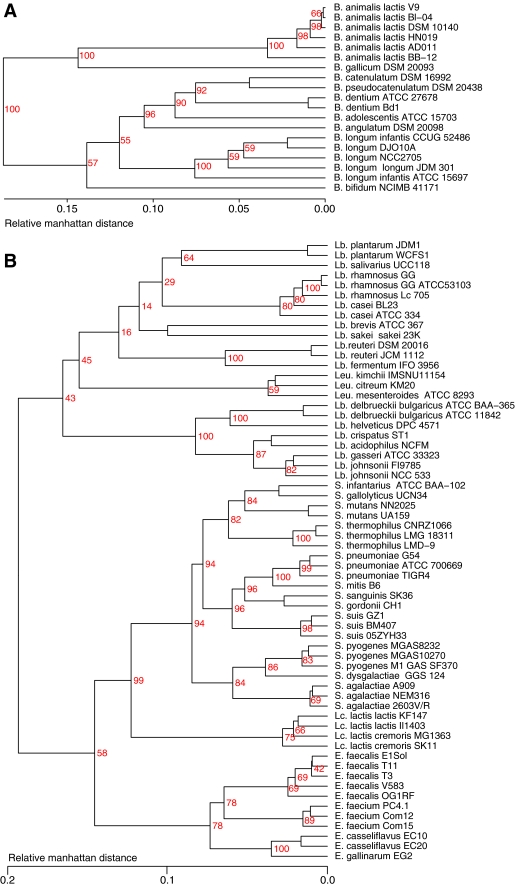
The pan-genomes of all six genera were combined to calculate the core genome shared by all genera. This resulted in only 63 core gene families out of a pan-genome of 37,053 gene families, using the criteria of gene similarity as described in the “Materials and Methods” section. These are listed in Supplementary Table S1. Exclusion of the distinct Bifidobacterium genus retained 243 core gene families for the Firmicute genomes that together produced a pan-genome of 30,615 gene families. Since these core genes are conserved in all Firmicute genomes analyzed here, phylogenetic trees could be generated and a consensus tree was generated, as shown in Fig. 4. The consensus core gene tree split all Lactobacillus genomes into three main clusters, though Lb. salivarius is excluded from these groups. The cluster shown at the top of the figure contains most Lactobacillus species with lower CG content, though it also includes L. delbrueckii, whose CG content is quite a bit higher. This clustering, based on these core genes, corroborates the inter-strain similarities already reported for their complete genomes, as shown in Fig. 2. The Streptococcus genus is separated into two large clusters in Fig. 4. Two clusters are also observed for the Enterococcus species, while Lactococcus is placed outside all other genera.
Figure 4.
Consensus tree of 243 core genes conserved in all analyzed Firmicutes. The number of genes supporting the branches is shown in red. Values for fewer than 100 genes are not shown
A more commonly used procedure is to compare only a small subset of core genes. In population biology, MLST of six or seven core gene fragments is frequently used to assess evolutionary distances between isolates within a species. MLST analysis is based on DNA sequences. We adapted this approach to perform in silico MLST for all isolates within a genus, as a measure for evolutionary distance of core genes, and used this for analysis of three genera. Unfortunately, despite the reputation of MLST as being generally applicable and despite a considerable number of gene families being conserved even between Firmicutes and Bifidobacteria (63 gene families), different MLST target gene sets have been proposed for various species, and most of these are not conserved between all species (Supplementary Table S1). In order to compare our findings with published data, we have used fragments of various genes depending on the genus, as suggested in the literature.
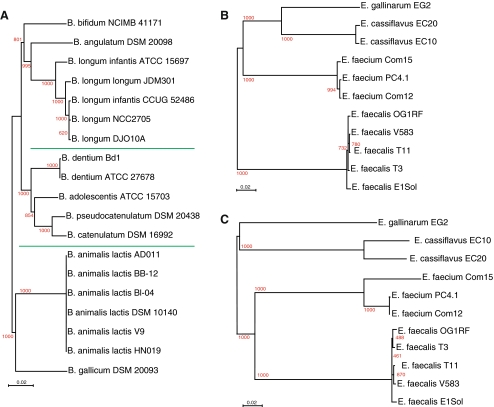
For in silico MLST analysis of Bifidobacterium, 7 gene fragments were extracted according to Delétoile and co-workers [6], as these happened to be conserved in all 19 Bifidobacterium genomes analyzed here. A phylogenetic tree of the extracted and concatenated MLST fragments is shown in Fig. 5. Although MLST was not designed for this purpose, the results show that this approach can reveal phylogenetic relationship of these core genes between species within a genus. All multiple isolates per species are correctly clustered, although subspecies are not correctly grouped (see the position of B. longum longum and B. longum infantis). Three major clusters can be recognized, separated in the figure by green lines. These findings are in accordance to the three groups within this genus recognized by Lee and O’Sullivan [17], based on an extensive 16S ribosomal RNA gene analysis.
Figure 5.
In silico MLST of gene fragments extracted from the genomes of Bifidobacterium (a) and Enterococcus (b, c). b The genes selected for MLST of E. faecium; c the genes selected for E. faecalis were used
The MLST website (http://www.mlst.net) lists two different gene sets to be used for Enterococci. Figure 5 (right side) shows the results obtained with each. Both trees produce little resolution within the species, especially when compared with the consensus tree based on 243 core genes in the previous figure.
For Lactobacilli, four MLST schemes are available: one for L. plantarum [4], two for Lb. casei ([1], http://www.pasteur.fr) and one for L. sanfranciscensis [20], which is not represented in our dataset. The first three MLST schemes were tested, which produced different trees (Supplementary Fig. S2). All three trees clustered multiple strains per species, but the branch positions of these species varied according to the gene set used. It cannot be stated which MLST tree is ‘correct’ as they all display the evolutionary relationship of the genes analyzed in question—but obviously, the phylogeny of core genes is not always conserved within a genome, as it is affected by recombination. This is also visible from the numbers of core genes producing consensus branches in Fig. 4. With this variation in mind, an MLST tree should be interpreted with caution, as it represents only a tiny fraction of the complete core genome of a strain.
Comparison of Variable Gene Content
The pan-genome of a species or genus comprises both conserved core and variable genes. The latter can also be used to establish inter-genome relationships, although not by phylogeny. Instead, clustering of presence or absence of variable genes can be performed [24]. This method calculates Manhattan distances for genes variably present. Obviously, core genes and genes found present in only one genome were excluded from this analysis, as they cannot identify any correlation between genomes. Thus, only genes whose presence varies, found at least in two genomes but absent in at least one genome, are assessed. The resulting clustering is not a phylogenetic tree, since it is not based on phylogeny of individual genes. Instead, it shows which genomes share more of their variable genes than others.
Figure 6 shows the hierarchical clustering of the Bifidobacterium genomes based on their variable gene content. As can be seen, genomes of identical species cluster together and are separated from different species, but the subspecies of B. longum are not correctly separated. Since their variable gene content seems to be mixed, this suggests that these two subspecies share the same gene pool for horizontal gene transfer events. The similarity, in terms of variable gene content, between the two species B. catenulatum and B. pseudocatenulatum is not more than that between various B. longum subspecies. A deep division splits B. animalis combined with B. gallicum from the others, which correlates with the MLST tree shown in Fig. 5.
Figure 6.
Hierarchical clustering of the Bifidobacterium genomes (a) and the Firmicute genomes (b) based on their variable gene content. The scale at the bottom applies to both trees
The analysis of variable gene content can simultaneously be performed with genomes of varying similarity, so that Fig. 6b combines all Firmicute genomes. The 21 Lactobacillus genomes are split into two major groups, which match a deep branch in the phylogenetic tree of 16S rRNA genes of this genus [2]. However, the clustering based on variable gene content produces a different picture to the consensus tree based on core genes (compare Figs. 4 and 6b). This probably reflects different evolutionary forces at play. Genes whose presence is variable may be located on mobile elements or may be more frequently subjected to DNA recombination than core genes. The three Leuconostoc genomes are placed within the Lactobacillus genus; apparently, these share a considerable number of variable genes.
The three major clusters within the Streptococcus genus visible in Fig. 6 largely match their taxonomic relationship as defined by 16S rRNA [8], although the distance between S. thermophilus and Streptococcus infantarius, which are both part of the ‘Salivarius group Streptococci’, is better captured by variable gene content than by 16S rRNA phylogeny. The discrepancy between this clustering and the consensus core gene tree is even more extensive for this genus.
The four Lc. lactis genomes are placed between Streptococcus and Enterococcus, which reminds of their inclusion, prior to the 1980s, into the single genus Streptococcus [25]. Within the genus Enterococcus, the clustering in Fig. 6 separates each of the analyzed species and confirms that Enterococcus casseliflavus and Enterococcus gallinarum are more related to E. faecium than to E. faecalis.
Visualization of Conserved and Variable Gene Content
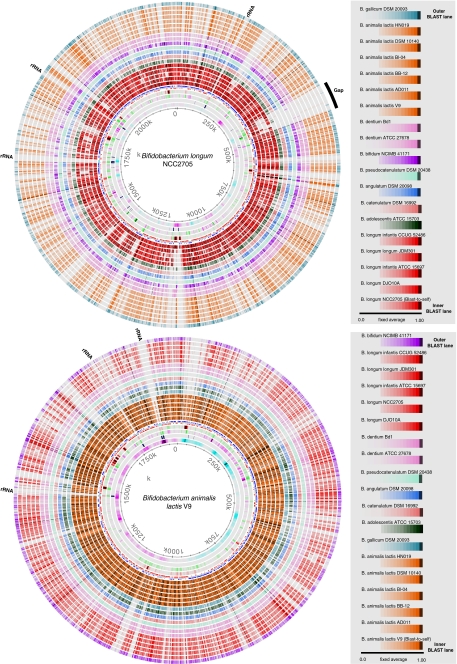
Conservation and variation in gene content between genomes can also be visualized by a BLAST Atlas [12], which contains information on gene location as well as on gene presence, at least for the reference genome on which a BLAST Atlas is based. Two different Bifidobacterium reference genomes were used in the two BLAST Atlases shown in Fig. 7 to which all other Bifidobacterium genomes were compared. Only genes present in the reference genome are captured in these atlases as these are used as query, for which the hits in the other genomes are recorded as colour in the BLAST lanes. The more strongly a protein gene is conserved, the more intense the colour is. Different colours are used to separate the different species, and these colours have been kept constant between the two panels, so that it is obvious that genes are mostly conserved within a species. The most inner BLAST lane included in Fig. 7 is that of the reference genome against itself. This shows the maximum colour that can be obtained for each location. Gaps in this ‘Blast-to-self’ lane where BLAST hits are absent, for instance around 1,700 kb, are due to non-translated genes such as ribosomal RNA copies. In Fig. 7a, a large region around 350–400 kb appears to produce a gap of non-conserved genes in most Bifidobacterium genomes, with the exception of B. longum infantis CCUG 52486 and B. longum DJO10A. This represents a region with variable genes within the B. longum genomes (the red lanes in the atlas), which are completely absent in the other Bifidobacterium genomes. Other than that, there appears to be relatively little variation between the B. longum genomes. Strong conservation within the species is also observed for B. animalis when used as the reference, as shown in Fig. 7b. In that lower panel, the B. animalis lanes are far more darkly coloured than in the top panel, whereas the B. longum lanes are lighter in colour, illustrating that stronger homology is identified within a species than across species. Note that the large gap of the top atlas is no longer visible now, as the genes that were found in B. longum are absent in B. animalis and thus are no longer captured when the latter is used as a reference. Taken together, these data suggest that there is relatively strong conservation within a species of Bifidobacterium, an observation that has been made by others as well [30].
Figure 7.
Blast Atlas of Bifidobacterium genomes with B. longum strain NCC2705 (top) and B. animalis lactis strain V8 (bottom) as the reference. To the right, the BLAST lanes for each atlas are listed. The four circles inwards of the annotation lane of the reference genome represent stacking energy, position preference, global direct repeats and GC skew (from out to in)
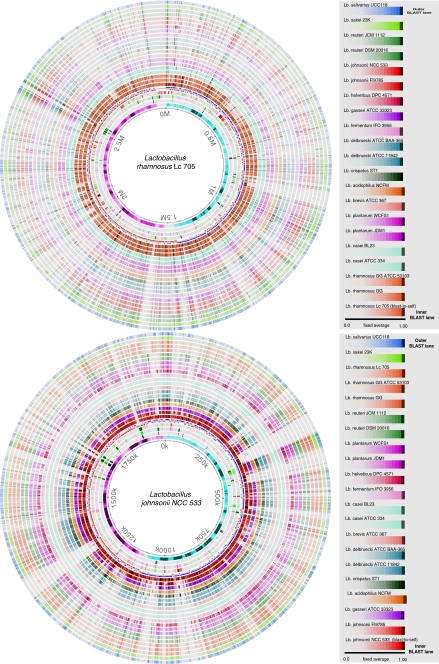
Figure 8 shows two BLAST Atlases of the Lactobacillus genomes. There appears to be considerably less conservation between species of this genus compared to Bifidobacterium. Even within the species of the two reference genomes of both panels, there are multiple gaps. This reflects the higher genetic diversity of the Lactobacillus genus compared to Bifidobacterium.
Figure 8.
BLAST Atlas of Lactobacillus with L. rhamnosus strain Lc705 (top) and Lb. johnsonii strain NCC533 (bottom) as the reference
A BLAST Atlas of Streptococcus genomes with S. thermophilus LMD-9 as the reference is provided as Supplementary Fig. S3. Two non-pathogenic E. faecalis genomes were included as well, since these are normal human flora strains and could be considered to share a similar niche to S. thermophilus, at least when colonizing the human gut. There is quite a bit of variation in protein-coding genes between the three S. thermophilus genomes, and as expected, there is even fewer conservation in other species of Streptococcus or in the two E. faecalis genomes. Apparently, similarity in bacterial lifestyle is not necessarily represented by a significant homology in gene content.
COG Comparison of Pan- and Core Genomes
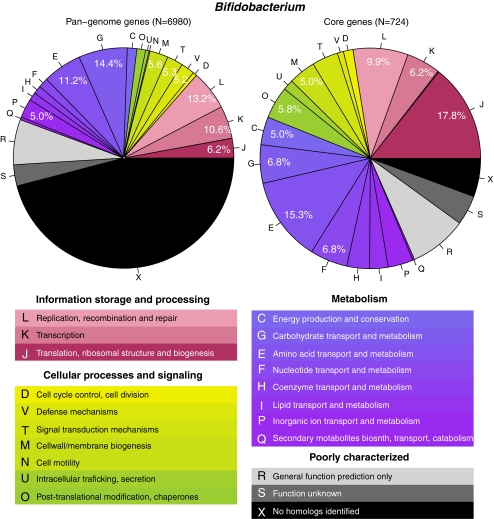
So far, conservation of genes was assessed and reported irrespective of their function, but that information is essential for a biological interpretation. The function of genes is not always known, but a large number of proteins have been assigned to a functional category of orthologous group, based on inference of sequence similarity to functionally characterized proteins. We have extracted the top-level COG groups for the genomes of interest and, in a first step, compared their core and pan-genomes genes. An example of such a statistical analysis for Bifidobacterium is shown in Fig. 9. At the bottom, the legend specifies the 3 top-level COG categories: ‘information storage and processing’, ‘cellular processes and signaling’ and ‘metabolism’, which are divided into 18 groups. The pie charts show what the fraction of the complete pan-genome genes of Bifidobacterium (left) or of the conserved core genes (right) belongs to each COG group. As expected, genes for which a function is not precise or not at all predicted build a significant fraction in the pan-genome, but these are mostly removed from the core genes, as their presence varies. More surprisingly, the three top categories are more or less similarly distributed in the two pie charts (thereby ignoring the contribution of the grey and black fractions), with a slight overrepresentation only of the information storage genes in the core genome compared to the pan-genome. Within these three broad categories, however, differences are visible when comparing the pan-genome or the core genome of these Bifidobacterium genomes. For instance, within ‘information storage and processing’, class J (translation, ribosomal structure and biogenesis) is enriched in the core genome, at the expense of K and L (transcription and replication, respectively). This means that the gene content related to these latter information storage processes is more variable and is hence captured in the pan-genome but less so in the core genome than the genes related to translation and ribosome biogenesis. Of interest is also the shift within the group ‘metabolism’ between classes E and G (for amino acid and carbohydrate transport/metabolism, respectively). The results indicate that the gene content for metabolism of amino acids is more conserved than that for carbohydrates, at least between these Bifidobacterium genomes. Lastly, enrichment in the core genome of class O, for post-translational modification and chaperones, is apparent within the group ‘cellular processes and signaling’.
Figure 9.
COG statistics for the genes found in the pan-genome (left) and core genome (right) of Bifidobacterium genomes. The key for the COG classes is explained below the pie charts. Percentages given in the pie chart are calculated by exclusion of classes R, S and X. Only values ≥5% are shown
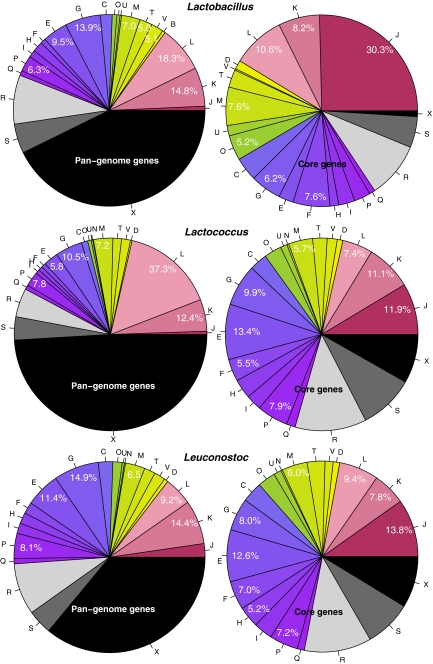
The Bifidobacterium findings can be compared to those of Lactobacillus, shown at the top of Fig. 10. The distribution of the three top-level COG categories in the pan-genome of Lactobacillus is different to that of Bifidobacterium, with more information storage and fewer metabolism genes. This is more obvious from Table 3, which lists the relative fractions of these COG classes when the grey and black fractions are ignored. For the core genes of Lactobacillus, the relative increase (compared to its pan-genome) in the fraction of information, storage and processing genes, at the expense of metabolism genes, is far more pronounced than for Bifidobacterium. Within the information and storage group, the enrichment of class J genes in the core genome of Lactobacillus is also stronger than reported for Bifidobacterium.
Figure 10.
COG distribution of pan-genome genes (left) and core genes (right) for Lactobacillus (top), Lactococcus (middle) and Leuconostoc (bottom)
Table 3.
Relative fractions of COG groups within the functionally annotated genes for the six genera
| COG groups | Bifidobacterium | Lactobacillus | Lactococcus | Leuconostoc | Enterococcus | Streptococcus | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pan (%) | Core (%) | Pan (%) | Core (%) | Pan (%) | Core (%) | Pan (%) | Core (%) | Pan (%) | Core (%) | Pan (%) | Core (%) | |
| Information storage | 30.0 | 33.9 | 34.0 | 49.1 ↑↑ | 50.5 | 30.4 ↓↓ | 28.1 | 31.0 | 26.6 | 33.8 ↑ | 34.7 | 42.6 ↑↑ |
| Cellular process, signalling | 21.9 | 20.2 | 22.7 | 20.3 | 17.1 | 19.1 | 19.1 | 20.0 | 24.4 | 18.9 ↓ | 26.3 | 20.3 ↓ |
| Metabolism | 48.1 | 45.9 | 44.3 | 30.6 ↓↓ | 32.2 | 50.6 ↑↑ | 52.7 | 49.1 | 50.0 | 47.8 | 39.2 | 36.9 |
All percentages are expressed as the fraction of all COG classes C to V. The arrows indicate significant shifts between the pan-genome genes and core genes for a given genus. Percentages do not always add up to 100% due to rounding effects
Figure 10 also shows the plots for Lactococcus (middle) and Leuconostoc (bottom). Although these last two genera are represented by four and three genomes only, all pan-genomes look surprisingly similar. However, when concentrating on the functionally annotated genes only (Table 3), some differences become apparent. The core genes of Lactococcus and Leuconostoc display a similar distribution of the three major COG classes as Bifidobacterium (which is taxonomically removed) that is different to the core genome of Lactobacillus, to which they are much closer related. Note that, in their pan-genomes, these three COG groups are similarly divided in Bifidobacterium and Lactobacillus. The shifts observed between pan-genome and core genome within a genus are contrasting between Lactobacillus and Lactococcus, whereas there is hardly a shift for Leuconostoc. From Fig. 10, it can be seen that, in the pan-genome of Lactococcus, class L genes make up a relatively large proportion. Within the metabolic gene classes, for Lactobacillus, a strong enrichment of nucleotide metabolism genes (class F) is observed in the core genes, whereas genes related to amino acid metabolism (class E) are more favoured in the core genome of Lactococcus. A significant increase in the core genes of COG class O (post-translational modification and chaperones) is observed for all analyzed genera. This could be an indication of the importance for such genes in the natural habitat of these gut bacteria.
The COG distribution plots for the pan-genome genes and the core genes of Enterococcus and Streptococcus is provided as Supplementary Fig. S4; the percentages of the three functionally classified COG top levels are included in Table 3. In contrast to the above examples, these two genera contain both pathogenic and non-pathogenic isolates. As in the previous examples, the large fraction of genes with unknown function is minimized in the core genome, but for both genera. Metabolism genes are neither over- nor underrepresented in the core genome. As before, a strong conservation of genes of COG class J (translation, ribosomal structure and biogenesis) was observed. Carbohydrate transport and metabolism genes (class G) were more frequently found in the Enterococcus pan-genome than in the Streptococcus pan-genome, though this was less pronounced for their core genomes.
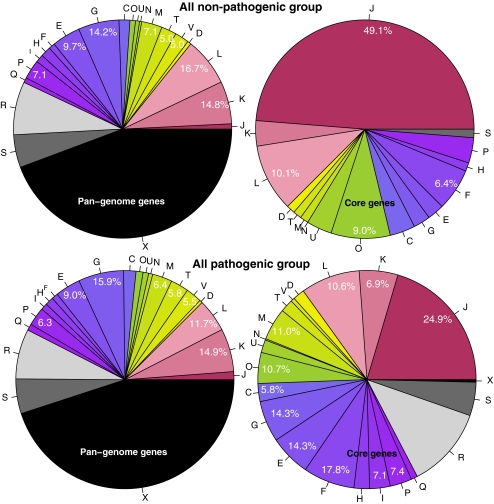
In an attempt to correlate findings with the presence or absence of pathogenicity, all genomes of pathogenic isolates (irrespective of their genus) were combined to collectively compare these with the non-pathogens (probiotic, fermentative and normal gut flora organisms) combined. The pathogenic group consisted of Enterococcus and Streptococcus genomes only, whilst the non-pathogenic group contained genomes of all genera analyzed. The COG analysis was then repeated for these two phenotypic collections, whereby the pan- and core genomes obviously were recalculated. The pathogenic collection had a pan-genome of 14,209 gene families and a core genome of 508. The pan-genome of the non-pathogenic collection was significantly larger (21,087), and this group produced a core genome of only 278 gene families. The results of the COG analysis are shown in Fig. 11. Surprisingly, the two pan-genome statistics look nearly identical, despite the obvious phenotypic differences between these two groups that both consist of diverse organisms, with a skewed genus distribution. However, the COG distribution between the two core genomes differs dramatically. The fraction of genes for which no homologue could be identified has (nearly) disappeared from the core genome of the non-pathogenic group, but a significant fraction of these genes was retained in the core genome of pathogens. The top level of metabolism genes has decreased in both core genomes, but more so in the group of the non-pathogens. Thus, the core genes of the non-pathogenic isolates are more frequently information storage genes and less likely metabolism genes than the core genes of pathogens (Table 4). Zooming in on shifts in single categories between pan- and core genomes, the enrichment of core genes belonging to class J, already observed for all single genus plots shown above, is even more extensive and far more extreme with the collection of non-pathogenic organisms. An enrichment for class O (post-translational modification and chaperones) within the top-level ‘metabolism’ is pronounced in the core genome of both groups, but the pathogens also show enrichment of class M genes (cell wall/membrane biogenesis) which is actually reduced in the core genome of non-pathogens.
Figure 11.
COG statistics for the genes found in the pan-genome (left) and core genome (right) of the collection of genomes from all included organisms, divided into non-pathogenic isolates (probiotic, fermentative and normal human gut flora) at the top and pathogenic isolates at the bottom
Table 4.
Relative fractions of COG groups within the functionally annotated genes for non-pathogens/pathogens. The arrows indicate how the reported percentages increase or decrease in the core genome compared to the pan genome.
| COG groups | Non-pathogens | Pathogens | ||
|---|---|---|---|---|
| Pan (%) | Core (%) | Pan (%) | Core (%) | |
| Information storage | 33.5 | 64.4 ↑↑ | 29.3 | 42.4 ↑↑ |
| Cell. process, signalling | 22.0 | 16.6 ↓ | 25.7 | 18.9 ↓ |
| Metabolism | 44.5 | 20.2 ↓↓ | 44.9 | 38.7 ↓ |
Discussion
The comparative analysis presented here of 81 bacterial genomes, covering 6 genera and 43 different species, could be performed by grouping their genes into gene families and comparing core and pan-genomes of various subsets of genomes. The findings frequently confirmed taxonomic relationships but could not identify common signatures, in terms of gene content, for all non-pathogenic bacteria included in the analysis. This finding is surprising, as all these species occupy a similar niche. Conserved genes were compared by means of a consensus tree, while genes variably present were analyzed by cluster analysis. The latter indicated that Leuconostoc genomes share a considerable number of variable genes with Lactobacillus. Functional analysis of the proteins coded by the genes comprising a genus’ core genome identified the relative strong conservation of information storage genes; this was observed for all genera analyzed. When all genomes were divided into a pathogenic and a non-pathogenic group, the pan-genome of both groups showed a surprisingly similar COG distribution; however, their core genome differed considerably. It was observed that, in the core genome of non-pathogenic genomes, genes for post-translational modification and chaperones were enriched.
A simultaneous comparison of the pan- and core genomes of publicly available genomes of Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, Streptococcus and Bifidobacterium, as was performed here, has not been published before, but similar analyses have been published for smaller selections of organisms. Canchaya and co-workers [2] performed comparative genomics of the then five available Lactobacillus genomes from different species and commented on the high variability within this genus. Schleifer and Ludwig [23] stated that “It is widely recognized that the taxonomy of this genus is unsatisfactory due to the highly heterogeneous nature of its members”. Indeed, data presented here illustrate the diversity within Lactobacillus. However, the heterogeneity of this genus is not larger than that of other bacteria. Using the same comparison criteria as applied here, the pan-genome of 53 E. coli genomes was found to comprise 13,000 gene families, even within this single species [18]. Similarly, an analysis of 27 genomes from 7 Vibrio species produced a pan-genome of nearly 15,000 gene families for this genus [31], and 38 genomes of 5 Burkholderia species contained as much as 26,000 gene families [28]. Thus, the diversity in gene content within the genus Lactobacillus, based on the genome sequences currently available, is not exceptional in the bacterial world.
Our analyses are mainly based on core genomes, an approach that others followed as well [2]. Those authors had defined a core genome for Lactobacillus whose size is similar to our findings. However, the fraction of identified orthologous genes in the pairwise comparisons performed by those authors range from 52.3% to 68.9%, which is much higher than our findings of between 12% and 42%, shown in the BLAST Matrix of Fig. 2. The difference may be due to the way these percentages were calculated. Whereas we express these as the fraction of gene families found in the reciprocal pan-genome of the pair of analyzed genomes, their calculations are different, and they do not state the cut-off used to recognize orthologous genes as such. In view of their limited reported range, we believe our way of expressing pairwise homology is more useful, as it gives a more sensitive measure. In a subsequent publication, comparative genomics was performed with a larger set of 12 Lactobacillus genomes [3]. Inclusion of 7 more genomes reduced their core genome to 141 genes which indicates they used more strict criteria of inclusion than the 50–50 rule we applied. Similar to our analysis, these authors compared the COG classes of the core genes they had identified, and their findings also reported the largest class represented to be genes involved in translation, followed by replication.
Comparative genomics of both Lactobacillus and Bifidobacterium was presented in a review [30], which mentioned the ability of Bifidobacterium to “synthesize at least 19 amino acids and (…) all of the enzymes that are needed for the biosynthesis of pyrimidine and purine nucleotides”. These authors further emphasized the importance of carbohydrate metabolism for Bifidobacterium with its capability to degrade complex sugars. Indeed, top-level metabolism genes form a major part of the Bifidobacterium core genome (Fig. 9) with class E (amino acid metabolism) as the largest fraction within that category. When we compare this core genome with that of Lactobacillus (Fig. 10), our analysis shows that class F genes (nucleotide metabolism) comprise the largest metabolism gene fraction in the Lactobacillus core genome. Ventura and co-workers [30] used a known physiological characteristic (Bifidobacterium species are known for their prototrophy) and looked for evidence of this in the genomes. In contrast, we have done a neutral analysis of pan- and core genome COG class representation and then compared this between genera. Our approach reveals novel insights that would remain unnoticed when known phenotypes are taken as a start, for instance the conservation of COG class O genes, involved in post-translational modification and chaperones, in both of these genera.
The authors of a recent review on Bifidobacterium genomics [17] pointed out that most Bifidobacterium genomes have been sequenced from organisms that have a long history of culture outside their natural habitat, the gut, with the exception of B. longum DJO10A. There is good evidence that the genome of Bifidobacterium is subject to gene reduction to adapt to prolonged culture conditions. This could potentially bias our comparative analysis of Bifidobacterium genomes with that of the other probiotic organisms.
The term ‘lactic acid bacteria’ is commonly used to describe bacteria used as starter cultures and fermentation of foodstuffs. LAB can refer to species from the genera Lactobacillus, Lactococcus, Leuconostoc, Streptococcus, Enterococcus, Pediococcus or all of the Lactobacillales, and sometimes includes Bifidobacterium as well. However, there are good reasons why these bacteria have been placed into different genera and phyla. The analyses presented here support their current taxonomic positions and stress their differences in gene content. The term LAB incorrectly suggests all these organisms are somehow related; a view that is still being presented in the literature [15]. The use of the term LAB is a bit misleading, as the genetic content from these various genera differ significantly. Moreover, some of the genera within LAB comprise only non-pathogenic species (Leuconostoc, Bifidobacterium, Lactobacillus), whereas other genera are a mixture of pathogenic and non-pathogenic species and strains (Streptococcus, Enterococcus). It would be better to refrain from the term LAB as there is no common denominator, other than the production of lactic acid (which is not restricted to these organisms) to collectively describe all species and strains supposedly included in this diverse group of organisms.
An extensive comparative study of Enterococcus genomes could not be identified from the literature. Most studies concentrate on pathogenicity of E. faecalis. Vebø and co-workers [29] compared probiotic and (uro-)pathogenic E. faecalis genomes; however, those comparisons were not based on sequence data. The Enterococcus genomes we have included were mostly from pathogenic organisms (only two non-pathogenic E. faecalis strains whose sequences were nearing completion were publicly available at the time of analysis), which limits the strength of this analysis, as it cannot be used to compare and contrast multiple non-pathogenic with pathogenic Enterococcus genomes. The 11 genomes included represent only 4 species, giving a pan-genome of nearly 8,000 gene families. The first four species of Lactobacillus or Streptococcus genomes in the pan-genome plots of Fig. 1 produce smaller pan-genomes, which could suggest that the diversity of Enterococcus could be at least as extensive as that of Lactobacillus. The pairwise BLAST comparison within this genus resulted in homologues varying from 24% to 84%, again indicating extensive intra-genus diversity.
Streptococcus and Enterococcus are frequently considered as closely related, but the BLAST Matrix comparing all included genomes (Supplementary Fig. S1) does not support this. Instead, somewhat surprisingly, the observed homology between Leuconostoc and Streptococcus genomes is slightly higher than that between Streptococcus and Enterococcus. On the other hand, Lc. lactis was positioned in between these two genera in the tree based on variable gene content. A shared gene pool between these genera can be hypothesized. Based on the conserved core genes, however, Enteroccus is more related to Streptococcus, while Lactococcus is more distinct.
A small comparative study of Streptococcus genomes combined with MLST suggested that S. thermophilus is a relatively young clone, evolved by genome reduction which removed or inactivated Streptococcus virulence genes [13]. It is possible, however, that the reduced genomes observed are the result of prolonged use as starter cultures, as no fresh human isolates have been sequenced to date. In a short review, Delorme [5] states that “S. thermophilus is related to Lactococcus lactis…”. Indeed, from the all-against-all BLAST Matrix, a similarity between 17.3% and 20.2% is recorded between genomes of these two species, which is higher than that between S. thermophilus and any other non-streptococcal genome. However, Lc. lactis also shares 16.0% to 18.0% of reciprocal genes with S. suis, so these overlapping percentages of gene similarity are no indicator of similarity in (probiotic) phenotype. Within the Streptococcus genus, the stated similarity of S. thermophilus with Streptococcus sanguinis (the only member of the viridans group for which a genome sequence is available) is confirmed in our Matrix, but an even higher similarity is found with Streptococcu gordonii.
The COG analysis of the core genomes of separate genera identified both similarities and differences. The three top-level functional COG groups are relatively equally divided over the functionally annotated pan-genes of all species, but their core genomes differ. Notably, Lactobacillus and Leuconostoc both have a smaller fraction of metabolism core genes than the other four genera and a larger information storage gene fraction. Information storage genes are essential, but redundancy allows so much variation between organisms that they are not all maintained in a core genome of diverse species. In the approach presented here, we first identified the core genomes of groups of bacteria and then sorted the genes in these core genomes for top-level COG categories. As a consequence, genes that were insufficiently conserved based on sequence similarity to be maintained in the core genome are removed despite their possible functional conservation. Using this approach, we found no correlation between the diversity within a genus (using the difference of their pan- and core genome as a measure) and the fraction of their information/storage COG genes. This lack of correlation is illustrated by the core genome of Bifidobacterium (724, or 10% of its pan-genome) and Leuconostoc (1,164, or 40% of its pan-genome). These two core genomes contain 34% and 31% information/storage genes, respectively, despite a huge difference in the degree of variation in these two genera.
Of particular interest is the COG analysis where all genomes were divided into a pathogenic and a non-pathogenic group. Virulence genes are not a separate COG category, but from the comparison of the core genomes of the pathogenic group with that of the non-pathogenic group, we can hypothesize that genes belonging to COG categories M (cell wall/membrane biosynthesis) and O (post-translational modification, chaperones) would mostly contribute to virulence. Conversely, it could be assumed that genes highly overrepresented in the core genome of the non-pathogenic group (compared to the core genome of the pathogenic group) most likely contribute to their probiotic or fermentative lifestyle. We observe enrichment for genes belonging to COG class J (translation, ribosomal structure and biogenesis) and again O (post-translational modification and chaperones). The finding that core genes of the non-pathogenic isolates are more frequently information storage genes and less likely metabolic genes than the core genes of pathogens is counter-intuitive. It is generally accepted that commensals and probiotic strains are most adequately equipped to live in the intestine, which would assume they share a large number of (conserved) metabolic genes to do so. Instead, the reduced metabolism gene fraction in their core genome suggests that there is a large variation within these genes, which reflects the diversity of the various commensals, fermentative and probiotic isolates. The vast enrichment for information/storage genes in the core genome of the non-pathogenic organisms is possibly a reflection of the relative poor conservation of all other functional classes in this group, an effect that appears to be less pronounced in the (ecologically more diverse) pathogenic group. The fact that Bifidobacterium are not present in the pathogenic group may have skewed these results slightly. A more accurate prediction for conserved genes with an important role in bacteria with a non-pathogenic lifestyle may become possible in the future, when more non-pathogenic Enterococcus genomes become available, which allows comparison of gene content within a genus or even species.
Conclusions
This study illustrates the value of comparative genomics of multiple genomes within and between related species and genera. The applied tools are relatively simple to analyze a vast number of genes, and the results can support or contradict existing hypotheses and taxonomic divisions, as well as generate novel hypotheses. We believe the data presented here can assist in understanding the commensal and probiotic relationship of bacteria with their human host. The work presented here demonstrates that the used analyses can be applied to large numbers of genomes, when searching for general mechanisms to predict trends even across genera. The presented analyses can be taken as a test case for comparison of multiple genomes from a largely variable dataset.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Functional description of 63 core genes conserved in all analysed genomes. (DOC 35 kb)
(PDF 2570 kb)
(PDF 274 kb)
(PDF 4228 kb)
(PDF 191 kb)
Acknowledgements
The authors are grateful to all research groups that have submitted their genome sequences to public databases, without which this analysis would not have been possible. TMW acknowledges the support provided by the Safety and Environmental Assurance Centre at Unilever for part of this work. OL and DWU received supported by the Center for Genomic Epidemiology at the Technical University of Denmark; part of this work was funded by grant 09-067103/DSF from the Danish Council for Strategic Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Cai H, Rodríguez BT, Zhang W, Broadbent JR, Steele JL. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiol. 2007;153:2655–2665. doi: 10.1099/mic.0.2007/006452-0. [DOI] [PubMed] [Google Scholar]
- 2.Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O’Toole PW. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiol. 2006;152:3185–3196. doi: 10.1099/mic.0.29140-0. [DOI] [PubMed] [Google Scholar]
- 3.Claesson MJ, van Sinderen D, O’Toole PW. Lactobacillus phylogenomics—towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008;58:2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- 4.de Las RB, Marcobal A, Muñoz R. Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiol. 2006;152:85–93. doi: 10.1099/mic.0.28482-0. [DOI] [PubMed] [Google Scholar]
- 5.Delorme C. Safety assessment of dairy microorganisms: Streptococcus thermophilus. Int J Food Microbiol. 2008;126:274–277. doi: 10.1016/j.ijfoodmicro.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Delétoile A, Passet V, Aires J, Chambaud I, Butel MJ, Smokvina T, Brisse S. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res Microbiol. 2010;161:82–90. doi: 10.1016/j.resmic.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felis GE, Dellaglio F. Taxonomy of Lactobacilli and Bifidobacteria. Curr Issues Intest Microbiol. 2007;8:44–61. [PubMed] [Google Scholar]
- 10.Fink WL. Microcomputers and phylogenetic analysis. Science. 1986;234:1135–1139. doi: 10.1126/science.234.4780.1135. [DOI] [PubMed] [Google Scholar]
- 11.Friis C, Wassenaar TM, Javed MA, Snipen L, Lagesen K, Hallin PF, Newell DG, Toszeghy M, Ridley A, Manning G, Ussery DW. Genomic characterization of Campylobacter jejuni strain M1. PLoS One. 2010;5:e12253. doi: 10.1371/journal.pone.0012253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallin PF, Binnewies TT, Ussery DW. The genome BLASTatlas—a GeneWiz extension for visualization of whole-genome homology. Mol Biosyst. 2008;4:363–371. doi: 10.1039/b717118h. [DOI] [PubMed] [Google Scholar]
- 13.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guédon E, Monnet V, Renault P, Kleerebezem M. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev. 2005;29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaenhammer TR, Azcarate-Peril MA, Altermann E, Barrangou R. Influence of the dairy environment on gene expression and substrate utilization in lactic acid bacteria. J Nutr. 2007;137(Suppl 2):748S–750S. doi: 10.1093/jn/137.3.748S. [DOI] [PubMed] [Google Scholar]
- 16.Lagesen K, Ussery DW, Wassenaar TM. Genome Update: the thousandth genome—a cautionary tale. Microbiol. 2010;156:603–608. doi: 10.1099/mic.0.038257-0. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, O’Sullivan DJ. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev. 2010;74:378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes. Stand Genomic Sci. 2009;1:63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picozzi C, Bonacina G, Vigentini I, Foschino R. Genetic diversity in Italian Lactobacillus sanfranciscensis strains assessed by multilocus sequence typing and pulsed-field gel electrophoresis analyses. Microbiol. 2010;156:2035–2045. doi: 10.1099/mic.0.037341-0. [DOI] [PubMed] [Google Scholar]
- 21.Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol. 2003;37:105–118. doi: 10.1097/00004836-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 23.Schleifer KH, Ludwig V. Phylogenetic relationships of lactic acid bacteria. In: Wood BJB, Holzapfel WH, editors. The Genera of Lactic Acid Bacteria. Glasgow: Chapman & Hall; 1995. pp. 7–17. [Google Scholar]
- 24.Snipen L, Ussery DW. Standard operating procedure for comparing pan-genome trees. Stand Genomic Sci. 2010;2:135–141. doi: 10.4056/sigs.38923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/S0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 26.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinforma. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ussery DW, Kiil K, Lagesen K, Sicheritz-Pontén T, Bohlin J, Wassenaar TM. The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 2009;6:140–157. doi: 10.1159/000235768. [DOI] [PubMed] [Google Scholar]
- 29.Vebø HC, Solheim M, Snipen L, Nes IF, Brede DA. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS One. 2010;5:e12489. doi: 10.1371/journal.pone.0012489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O’Toole PW. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 31.Vesth T, Wassenaar TM, Hallin PF, Snipen L, Lagesen K, Ussery DW. On the origins of a Vibrio species. Microb Ecol. 2010;59:1–13. doi: 10.1007/s00248-009-9596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional description of 63 core genes conserved in all analysed genomes. (DOC 35 kb)
(PDF 2570 kb)
(PDF 274 kb)
(PDF 4228 kb)
(PDF 191 kb)