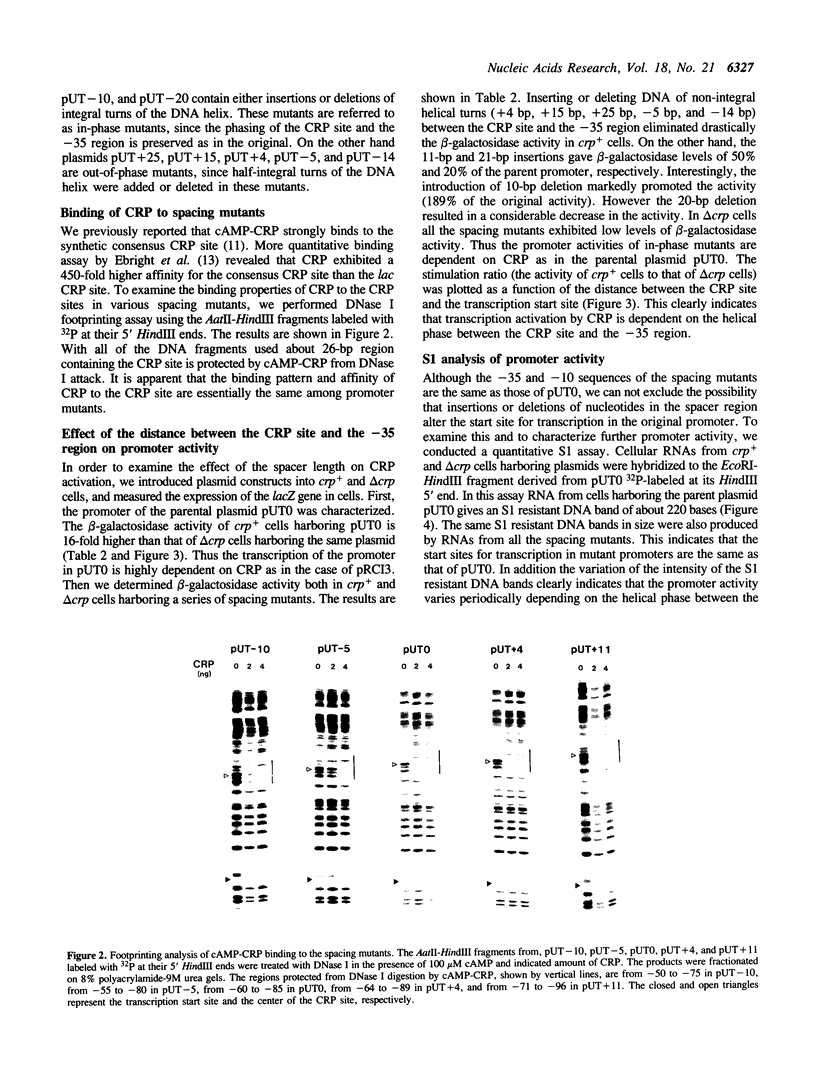
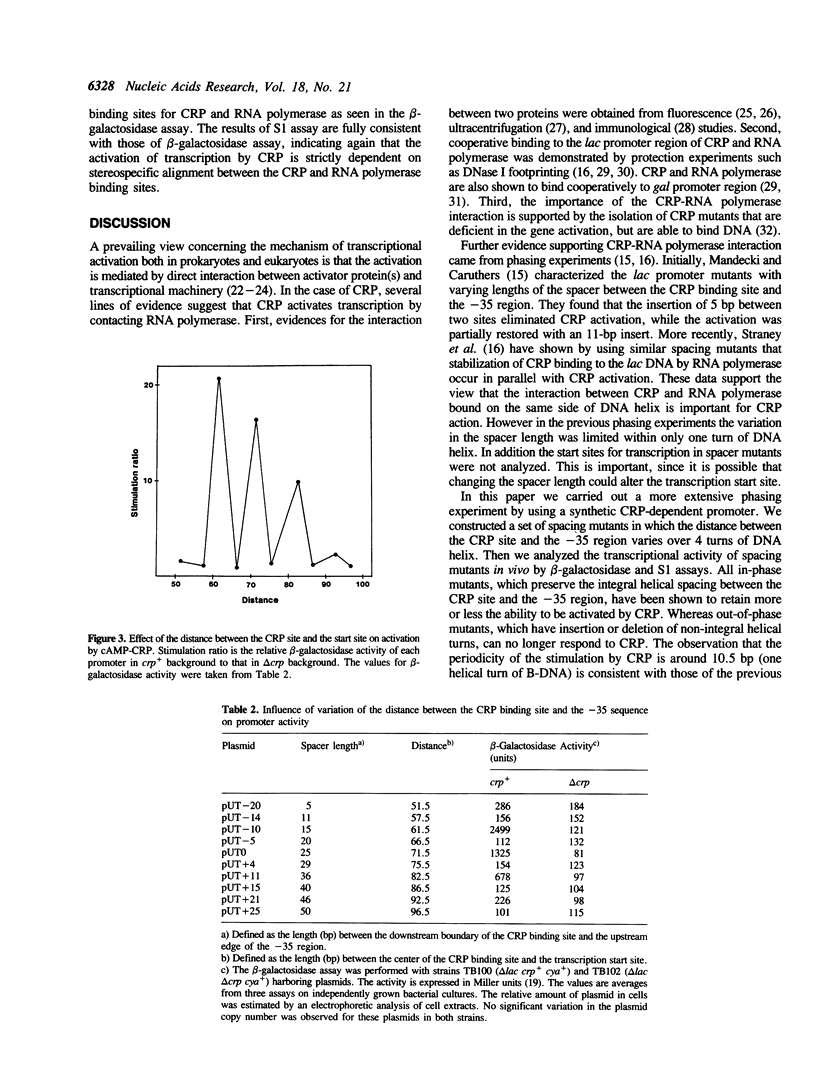
Abstract
A plasmid carrying a CRP-dependent promoter fused to the lac structural genes was manipulated to construct a set of spacing mutants that have varying lengths between the CRP binding site and the -35 region. The lengths of the spacer were changed over 45 bp by inserting or deleting nucleotides. DNase I footprinting analysis revealed that the spacer length did not affect the binding of cAMP-CRP to the CRP site. The effect of the spacer length on transcription activation by cAMP-CRP was tested in vivo by beta-galactosidase and quantitative S1 assays with crp+ and delta crp cells harboring plasmids. Insertions or deletions of non-integral helical turns, which displace the CRP site onto the opposite face of DNA helix compared to the original promoter, eliminated completely the activation of transcription. In contrast, changing the spacer length by integral helical turns allowed the promoter to respond to CRP, although the degree of activation varied with the length of the spacer. We conclude that stereospecific positioning of CRP and RNA polymerase on the DNA helix is strictly required for CRP action. The data support a model that CRP stimulates transcription by directly contacting RNA polymerase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Hanamura A., Tobe T. Semisynthetic promoters activated by cyclic AMP receptor protein of Escherichia coli. Gene. 1989 Dec 21;85(1):91–97. doi: 10.1016/0378-1119(89)90468-x. [DOI] [PubMed] [Google Scholar]
- Aiba H., Krakow J. S. Isolation and characterization of the amino and carboxyl proximal fragments of the adenosine cyclic 3' ,5'-phosphate receptor protein of Escherichia coli. Biochemistry. 1981 Aug 4;20(16):4774–4780. doi: 10.1021/bi00519a038. [DOI] [PubMed] [Google Scholar]
- Bedouelle H. Mutations in the promoter regions of the malEFG and malK-lamB operons of Escherichia coli K12. J Mol Biol. 1983 Nov 15;170(4):861–882. doi: 10.1016/s0022-2836(83)80192-2. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Blazy B., Takahashi M., Baudras A. Binding of CRP to DNA-dependent RNA polymerase from E. coli: modulation by cAMP of the interactions with free and DNA-bound holo and core enzyme. Mol Biol Rep. 1980 Mar 31;6(1):39–43. doi: 10.1007/BF00775753. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Cannon W., Woodcock J. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol Microbiol. 1987 Sep;1(2):243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- Chapon C., Kolb A. Action of CAP on the malT promoter in vitro. J Bacteriol. 1983 Dec;156(3):1135–1143. doi: 10.1128/jb.156.3.1135-1143.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P., Gicquel-Sanzey B. Cloning and sequence of the crp gene of Escherichia coli K 12. Nucleic Acids Res. 1982 Feb 25;10(4):1363–1378. doi: 10.1093/nar/10.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley M. C., Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J Bacteriol. 1982 Aug;151(2):942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989 Dec 25;17(24):10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilen E., Pampeno C., Krakow J. S. Production and properties of the alpha core derived from the cyclic adenosine monophosphate receptor protein of Escherichia coli. Biochemistry. 1978 Jun 27;17(13):2469–2473. doi: 10.1021/bi00606a001. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Gaston K., Kolb A., Busby S. Binding of the Escherichia coli cyclic AMP receptor protein to DNA fragments containing consensus nucleotide sequences. Biochem J. 1989 Jul 15;261(2):649–653. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Bacterial gene regulation from distant DNA sites. Cell. 1989 Apr 21;57(2):193–195. doi: 10.1016/0092-8674(89)90955-0. [DOI] [PubMed] [Google Scholar]
- Göransson M., Forsman P., Nilsson P., Uhlin B. E. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol Microbiol. 1989 Nov;3(11):1557–1565. doi: 10.1111/j.1365-2958.1989.tb00141.x. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Irani M. H., Orosz L., Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983 Mar;32(3):783–788. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- Irwin N., Ptashne M. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8315–8319. doi: 10.1073/pnas.84.23.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakow J. S., Pastan I. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2529–2533. doi: 10.1073/pnas.70.9.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Schleif R. F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989 Jan;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Francklyn C., Hamilton E. P. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Maeda S., Ozawa Y., Mizuno T., Mizushima S. Stereospecific positioning of the cis-acting sequence with respect to the canonical promoter is required for activation of the ompC gene by a positive regulator, OmpR, in Escherichia coli. J Mol Biol. 1988 Aug 5;202(3):433–441. doi: 10.1016/0022-2836(88)90276-8. [DOI] [PubMed] [Google Scholar]
- Mandecki W., Caruthers M. H. Mutants of the lac promoter with large insertions and deletions between the CAP binding site and the -35 region. Gene. 1984 Nov;31(1-3):263–267. doi: 10.1016/0378-1119(84)90219-1. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Minchin S. D., Austin S., Dixon R. A. Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NTRC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J. 1989 Nov;8(11):3491–3499. doi: 10.1002/j.1460-2075.1989.tb08514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Shigesada K., Kimizuka F., Aiba H. Regulatory effect of a synthetic CRP recognition sequence placed downstream of a promoter. Nucleic Acids Res. 1988 Aug 11;16(15):7315–7332. doi: 10.1093/nar/16.15.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkney M., Hoggett J. G. Binding of the cyclic AMP receptor protein of Escherichia coli to RNA polymerase. Biochem J. 1988 Mar 15;250(3):897–902. doi: 10.1042/bj2500897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Spassky A., Busby S. Studies with the Escherichia coli galactose operon regulatory region carrying a point mutation that simultaneously inactivates the two overlapping promoters. Interactions with RNA polymerase and the cyclic AMP receptor protein. FEBS Lett. 1987 Jul 13;219(1):189–196. doi: 10.1016/0014-5793(87)81214-0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Queen C., Rosenberg M. A promoter of pBR322 activated by cAMP receptor protein. Nucleic Acids Res. 1981 Jul 24;9(14):3365–3377. doi: 10.1093/nar/9.14.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Vidal-Ingigliardi D., Richet E. A complex nucleoprotein structure involved in activation of transcription of two divergent Escherichia coli promoters. J Mol Biol. 1989 Feb 5;205(3):471–485. doi: 10.1016/0022-2836(89)90218-0. [DOI] [PubMed] [Google Scholar]
- Reitzer L. J., Movsas B., Magasanik B. Activation of glnA transcription by nitrogen regulator I (NRI)-phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase. J Bacteriol. 1989 Oct;171(10):5512–5522. doi: 10.1128/jb.171.10.5512-5522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y. L., Garges S., Adhya S., Krakow J. S. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4138–4142. doi: 10.1073/pnas.85.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky A., Busby S., Buc H. On the action of the cyclic AMP-cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. EMBO J. 1984 Jan;3(1):43–50. doi: 10.1002/j.1460-2075.1984.tb01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender W. Cyclic adenosine 3':5'-monophosphate receptor protein: interaction with E. coli RNA polymerase. Biochem Biophys Res Commun. 1980 Sep 16;96(1):320–325. doi: 10.1016/0006-291x(80)91217-6. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Straney S. B., Crothers D. M. Synergy between Escherichia coli CAP protein and RNA polymerase in the lac promoter open complex. J Mol Biol. 1989 Mar 5;206(1):41–57. doi: 10.1016/0022-2836(89)90522-6. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. Y., Nath K., Wu C. W. Conformational transitions of cyclic adenosine monophosphate receptor protein of Escherichia coli. A fluorescent probe study. Biochemistry. 1974 Jun 4;13(12):2567–2572. doi: 10.1021/bi00709a015. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]