Abstract
The WRKY transcription factors function in plant growth and development, and response to the biotic and abiotic stresses. Although many studies have focused on the functional identification of the WRKY transcription factors, much less is known about molecular phylogenetic and global expression analysis of the complete WRKY family in maize. In this study, we identified 136 WRKY proteins coded by 119 genes in the B73 inbred line from the complete genome and named them in an orderly manner. Then, a comprehensive phylogenetic analysis of five species was performed to explore the origin and evolutionary patterns of these WRKY genes, and the result showed that gene duplication is the major driving force for the origin of new groups and subgroups and functional divergence during evolution. Chromosomal location analysis of maize WRKY genes indicated that 20 gene clusters are distributed unevenly in the genome. Microarray-based expression analysis has revealed that 131 WRKY transcripts encoded by 116 genes may participate in the regulation of maize growth and development. Among them, 102 transcripts are stably expressed with a coefficient of variation (CV) value of <15%. The remaining 29 transcripts produced by 25 WRKY genes with the CV value of >15% are further analysed to discover new organ- or tissue-specific genes. In addition, microarray analyses of transcriptional responses to drought stress and fungal infection showed that maize WRKY proteins are involved in stress responses. All these results contribute to a deep probing into the roles of WRKY transcription factors in maize growth and development and stress tolerance.
Keywords: WRKY transcription factor, maize, phylogenetic analysis, expression profile, development
1. Introduction
The WRKY family is one of the 10 largest transcription factor families in higher plants and extends all over the green lineage.1 WRKY proteins possess one or two unique DNA-binding domain with nearly 60 amino acids composed of the absolutely conserved signature WRKYGQK followed by a novel zinc-finger-like motif C2H2 (C–X4–5–C–X22–23–H–X–H) or C2HC (C–X7–C–X23–H–X–C). As a transcription factor, WRKY factors act in concert with other components of the transcriptional machinery and direct the temporal- and spatial-specific expression of the designated genes, thereby ensuring proper cellular responses to both internal and external stimuli.1–3 The stimuli may be from abiotic, such as cold, heat, UV-B, drought, wound, etc., or may be from biotic, such as bacteria, fungi and viruses. As well, WRKY proteins have been implicated in the regulation of developmental processes such as trichome,4 seed development,5 embryogenesis,6 leaf senescence,7,8 dormancy,9 plant growth10 and metabolic pathways.4,11 Since the first WRKY cDNA, SPF1, was cloned from sweet potato (Ipomoeabatatas),12 more and more WRKY genes in various plant species have been experimentally identified, including wild oat (Avena fatua; ABF1),13 parsley (Petroselinum crispum; PcWRKY1, 2, 3),14,15 Arabidopsis thaliana (ZAP1),16 orchardgrass (Dactylis glomerata),17 tobacco (Nicotiana tabacum),18–21 chamomile (Matricaria chamomilla),22 rice (Oryza sativa),23 sugarcane (Saccharum hybrid cultivar),24 bittersweet nightshade (Solanum dulcamara),25 potato (Solanum tuberosum),26,27 wheat (Triticum aestivum)28 and grape (Vitis vinifera).29 Zea mays (hereafter called maize) is not only a vital crop, but also serves as an important model system for basic biological research.30,31 Due to its wide ecological potential and excellent characters, maize roots all over the world under a wide range of environmental conditions, occupying 167 million hectares worldwide and producing 872 million tons of grains in July, 2011 according to the USDA (http://www.fas.usda.gov/psdonline/). Recently, the maize B73 genome sequence is now available to public,32 providing a good opportunity to study WRKY transcription factor family extensively.
To date, researchers are still active in this field to reveal the underlying molecular mechanism of WRKY proteins in plant growth and development, as well as to elucidate a huge interaction web involving in response to stresses which is mediated by this transcription factor family.33–38 The WRKY proteins in model plant A. thaliana (hereafter called Arabidopsis) have set a good example for later researchers to study the transcription factor family. In Arabidopsis, 72 characterized genes can be categorized into three groups based on the number of WRKY domains and the pattern of the zinc finger motif according to Somssich and colleagues.39 The first group contains two WRKY domains (N-terminal and C-terminal), whereas the other two groups have only one domain. Group III differs in the pattern of the zinc finger motif because it has the zinc finger motif of C2HC rather than C2H2 in the other groups. Group II proteins can be further subdivided into five subgroups according to the amino acid motifs outside the WRKY domain. All these preliminary knowledge might contribute to our understanding of maize WRKY gene family.
In this study, we identified 136 WRKY proteins encoded by 119 WRKY genes from the maize B73 genome and named them from ZmWRKY1 to ZmWRKY119 based on their loci on corresponding chromosomes. Considering of the different transcripts produced by the same gene, totally 136 putative proteins are all named. These 136 WRKY proteins in maize can be classified into three groups in terms of the phylogenetic tree which is constructed using the WRKY domain peptide sequences. The phylogenetic comparisons of maize, Arabidopsis, O. sativa (hereafter called rice), Hordeum vulgare (hereafter called barley), Physcomitrella patens (hereafter called P. patens) and ancestral eukaryotic organisms reveal the origin and evolutionary expansion of the WRKY family. The maize chromosome sequence information derived from databases is applied to map the 136 WRKY transcripts to their corresponding gene locus in the 10 haploid chromosomes in order to investigate the duplication events occurred on maize chromosomes.
Nowadays, a large number of maize microarray experiments are accessible via different public resources such as GEO or Plexdb, and allow for in silico expression analyses of maize genes. The microarray data of genome-wide gene expression atlas of the maize inbred line B73 offer the chance to understand the distinct expression patterns of the WRKY family under stresses response and growth and development. All these favourable conditions enable us to further our research and get a deep understanding of the roles of WRKY family members in maize.
2. Materials and methods
2.1. Identification and annotation of WRKY family in maize
Since the amended maize B73 genome sequence is now publicly available, we can download the genome sequence (ZmB73_5b_FGS_genes.fasta.gz) and the proteome sequence (ZmB73_5b_FGS_translations.fasta.gz) from the Maize Genome Sequence Project (http://ftp.maizesequence.org/current/filtered-set/). Then, local BLASTP program was run to search the complete WRKY members. The proteome sequences are used as a database for local BLASTP search. The Arabidopsis WRKY proteins are used as query sequences. The e-value for BLASTP was set at 1e−10 to obtain the final data set of WRKY proteins. After removing the overlapping genes manually, a total of 136 protein sequences are eventually identified as WRKY proteins and named based on their exact loci on chromosomes. The putative orthologues were assigned from Arabidopsis, rice and H. vulgare with significance under 1e−20 from National Center of Biotechnology Information (NCBI).
2.2. Phylogenetic analysis of WRKY family
The WRKY family protein sequences of rice (O. sativa japonica) and Arabidopsis were downloaded from TIGR rice database (http://www.tigr.org/) and TAIR (The Arabidopsis Information Resource; http://www.arabidopsis.org/), respectively. Sequences of barley and P. patens were extracted from NCBI. Maize sequences were identified by local BLASTP program as described above. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 540 with pairwise distance and Neighbor-Joining algorithm. The evolutionary distances were computed using the p-distance method and were used to estimate the number of amino acid substitutions per site. The reliability of each tree was established by conducting 1000 bootstrap sampling steps.
2.3. Mapping WRKY genes on maize chromosomes
The maize databases were used in a BLAST-based search of the entire maize genomic sequence to confirm the physical locations of all WRKY genes. The Genome Pixelizer software was used for a graphical display of the WRKY loci in each pair of corresponding maize chromosomes (http://atgc.org/GenomePixelizer/41).
2.4. Microarray-based expression analysis
Transcriptome data of genome-wide gene expression atlas of the maize inbred line B73 made by NimbleGen microarray technology, together with the normalized expression data using RMA (robust multi-chip average) algorithm, were downloaded from the publicly available databases such as MaizePLEX (http://www.plexdb.org/plex.php?database=Corn) and GEO (http://www.ncbi.nlm.nih.gov/geo/). The RMA data which were log2-transformed beforehand were loaded into R and Bioconductor for expression analysis (http://www.bioconductor.org/42). The package Limma43,44 was used to model the systematic parts of data by fitting a linear model in the function lmFit. The model was specified by a design matrix. The empirical Bayes approach, in the function eBays, was used to moderate the standard errors of the estimated log-fold changes and shrink the standard errors to a common value. Then, the gplots45 package was used to make the heat map.
3. Results and discussion
3.1. Identification and nomenclature of 136 maize WRKY proteins
A keyword search against the NCBI and UniProt protein sequence databases returned 34 and 31 previously annotated maize WRKY protein sequences, respectively, by the year 2011; among which, 28 proteins are derived from the maize B73 inbred line (Supplementary Table S1). We greatly appreciate the foregoing efforts made by Alexandrov et al.46 and other scientists, who have named these sequences which were derived from a large-scale cDNA sequencing or from experiments individually. However, the annotations are so disordered and redundant, for they are named neither in chromosome order nor in the group order and the same numbers occur repeatedly, which make research work difficult to go ahead. A uniform nomenclature with consecutive numbering may help avoid confusion and should help communicate in the scientific community. Thus, we renamed the 136 protein sequences which we have identified through an overall search of the complete genome sequence from ZmWRKY1 to ZmWRKY119 based on the exact position of their corresponding genes on chromosomes 1–10 and from top to bottom, while the variant proteins produced from the same locus are recognized by the same name added 1, 2 or 3 behind (Supplementary Table S2). We use the similar numbering system as proposed for bZIP47 and R2R3-MYB,48 provides a unique identifier for each WRKY gene. Genes in different genomes that evolved from a common ancestral gene by speciation can be considered as orthologous genes,49 which retain the similar functions during evolution.50 As a good plant for experiments, Arabidopsis, together with monocotyledonous plants rice and barley, is used to assign orthology for maize, which is valuable for functional analysis of the WRKY family members in maize. The putative orthologues are identified according to their e-value (under 1e−20) and the topology of phylogenetic tree.
3.2. Evolutionary analysis of the WRKY transcription factor family
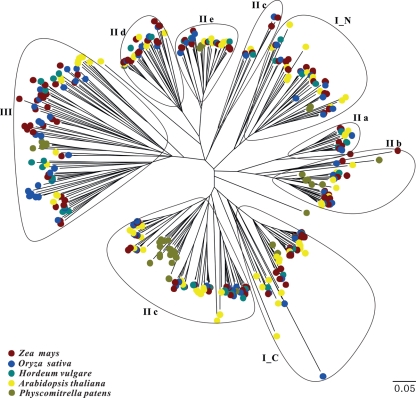
Exposed to a wide variety of biotic and abiotic stresses connected with plant sessile, autotrophic lifestyle, the WRKY family appears to have been under positive selection pressure during evolution. To study the evolutionary origin of maize WRKY family and classify the newly discovered members, multiple sequence alignment is performed using the complete domain sequences derived from Arabidopsis (71),39 maize (136), rice (82),51,52 barley (36)53 and P. patens (36). Because of the difference between the C- and the N-terminal WRKY domain sequences of Group I, which will be described later, we separate them as two independent domains. Then the alignment result is used to generate a phylogenetic tree (Fig. 1). The result shows a well-organized classification according to that in Arabidopsis.39 Here, we call them Group I, IIa–e and III.
Figure 1.
Unrooted Neighbor-Joining phylogenetic tree of WRKY domains in five plants. The tree is reconstructed using WRKY domain sequences from the complete WRKY gene families in A. thaliana (yellow), Z. mays (B73, brown), O. sativa (blue), H. vulgare (green) and P. patens (khaki). The evolutionary distances are computed using the p-distance method and are in the units of the number of amino acid substitutions per site.
The result of phylogenetic analysis implies that domain gain and loss is a divergent force for expansion of the WRKY gene family. It has been reported that WRKY transcription factors have their evolutionary origin in ancient eukaryotes with the most basal WRKY genes identified in two non-photosynthetic organisms, the unicellular protist Giardia lamblia and the slime mold Dictyostelium discoideum.54 Each ancestral eukaryotic organism has single WRKY protein which belongs to Group I because of the two WRKY domains, suggesting that the proteins of this group evolved early and represent the ancestral form. Except AtWRKY10, all WRKY proteins of Group I in Arabidopsis have two WRKY domains. But in monocotyledons, the loss of WRKY domain seems to be more common. For example, Supplementary Fig. S1 shows that Group I contains 16 proteins having only one domain, among which 9 proteins possess the C-terminal domain and the rest with the N-terminal domain only. In addition, some proteins with two domains are clustered into Group III according to the phylogenetic tree, which are OsWRKY41, OsWRKY61, OsWRKY63, OsWRKY81 and HvWRKY25. As the phenomenon occurs in monocotyledons exclusively, the result shows that there exists a divergence between monocot, and demonstrates that Group III proteins emerged later during evolution and maybe evolved from Group I.
The early researches have reported that only the C-terminal domain in Group I has the sequence-specific DNA-binding activity with the W-box, and N-terminal WRKY domain shows weaker binding activity.3,16,55 The first solution structure of the WRKY domain was AtWRKY4_C (PDB: 1WJ2), obtained by NMR spectroscopy, which confirmed the reports described above.56 Further, DNA titration experiment proved that the region corresponding to the conserved sequence WRKYGQK in WRKY4-C is directly involved in DNA binding. In contrast, the N-terminal WRKY domain might participate in the binding process or alternatively provide an interface for protein–protein interactions that coincide with the function of some zinc-finger-like domains.57 In this scenario, we can reasonably assume that the N-terminal WRKY domain seems to be more variable during evolution, so it may be deleted from the sequence or evolve into another pattern to achieve additional functions. Meanwhile, it can be of no question that the single WRKY domains of Groups II and III family members are more similar to the C-terminal WRKY domain of Group I proteins for their common function of DNA binding.
A large number of WRKY proteins exist in the five species, suggesting that they play crucial roles in plant developmental and physiological processes (Table 1).58 The evidence shows that the rapid duplication of WRKY genes occurred before the divergence of monocots and dicots.54 It might be the environmental pressures that impelled WRKY family to enlarge in order to adapt to new or changing environments. In G. lamblia and D. discoideum, only a single WRKY protein has been discovered. And then, in bryophyte, the family member of P. patens has increased to 37 by duplication. There is no related report about the members of Group III in P. patens ever before. However, on the basis of the complete genome sequencing and the topology of the phylogenetic tree, five proteins are found to group into Group III. To angiosperm, monocotyledonous plants develop a larger family of WRKY proteins than dicotyledonous plants. One hundred and thirty-six maize WRKY proteins form the largest WRKY family, while those of rice ranks second, only 71 WRKY members in Arabidopsis. With plants from low to higher and from aquatic environment to terrestrial environment, plants suffering severer stresses shut down basic growth functions, while WRKY proteins may act synergistically with other family members to promote plant development and reinforce resistance to environment stressors by modulating the expression of genes. As an important transcription factors, the rapid duplication of WRKY family members may contribute to the increase in adaptability and the establishment of signal transduction webs to go through the adversity.
Table 1.
Number of WRKY domains and proteins (numbers in parenthesis) of Arabidopsis, rice, maize, barley and P. patens in the WRKY Groups I–III
| WRKY group | AtWRKY | OsWRKY | ZmWRKY | HvWRKY | PpWRKY |
|---|---|---|---|---|---|
| I_N | 13 | 9 | 22 | 4 | 3 |
| I_C | 14 | 11 | 21 | 5 | 3 |
| IIa | 3 | 4 | 7 | 4 | 0 |
| IIb | 8 | 7 | 11 | 1 | 6 |
| IIc | 18 | 19 | 29 | 10 | 18 |
| IId | 7 | 6 | 14 | 5 | 4 |
| IIe | 8 | 8 | 17 | 2 | 0 |
| III | 14 | 26 | 31 | 12 | 5 |
| Total | 85 (71) | 90 (78) | 152 (136) | 43 (39) | 40 (37) |
3.3. Chromosome mapping and genetic analysis
There are 10 chromosomes in maize genome and the length of each chromosome varies from each other. The first chromosome is the longest with 301.0 Mb, and the last is the shortest which is only half of the longest. According to the physical and genetic framework of the maize B73 genome, the 10 chromosomes of the maize genome are structurally diverse and have undergone dynamic changes in chromatin composition.30,32 Mapping genes to individual chromosome can facilitate the elimination of redundant data, and provide a powerful analytical method to conduct genetic research. In order to map 119 WRKY genes to maize chromosomes, the physical location of each gene was required. Many searches are performed in the interest of finding gene orientation and genetic distance of corresponding WRKY genes against maize-related databases. Finally, the software GenomePixelizer was used to visualize the distribution of all WRKY genes on maize 10 chromosomes. The genes are presented by small colour boxes, and the different colours mean different groups and subgroups of the WRKY gene family.
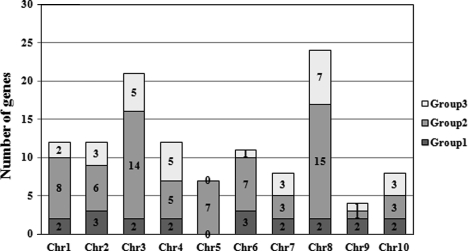
The physical distribution of WRKY genes in the maize B73 genome illustrates the genetic events which result in the diversity and complexity of this gene family. The study of chromosome map (Supplementary Fig. S2) along with the histogram (Fig. 2) comes to the conclusion that the WRKY genes are dispersedly distributed across all the chromosomes in maize. The highest number of WRKY genes is located on Chromosome 8 (24 genes), accounting for 20.17% of the total. The chromosome having least number is Chromosome 9, with only four genes belonging to three different groups, accounting for just 3.36% of the total. The members of three groups are distributed on all chromosomes except Chromosome 5 on which WRKY genes of Group II are located exclusively. Without regard to Chromosome 5, the distribution of Group I genes in each chromosome is more even than that of Groups II and III, with the number of genes ranging from 2 to 3. The formation of this distribution pattern implies that some genetic events of this gene family have taken place during evolution.
Figure 2.
Histogram showing the number and distribution of WRKY genes of three groups on 10 chromosomes.
The analysis of genetics and genomics has offered insights into the generation and the evolution of WRKY genes. Evidences have shown that the dramatic variations in size and distribution of the WRKY gene family are influenced by many processes, including gene duplication owing to large-scale genome events such as polyploidy, segmental duplications and tandem duplication.59,60 An important fact is that maize genome has undergone several rounds of genome duplication, including an ancient paleopolyploid event ∼70 million years ago (mya)61 and an additional whole-genome duplication event ∼11 mya.31,62,63 Furthermore, many studies have revealed that transcriptional factors are more likely to retain after duplication.64–66 All these suggest that genome duplication might be a major mechanism for expansion of WRKY family in maize. Our inference can be confirmed by another gramineous plant, rice, in which the WRKY family comprises ∼77.7% duplicated regions.67 Most WRKY genes show clustering in maize genome. Here, we adopted the standpoint proposed by Meyers et al.68 to define a cluster that the numbers or sizes of clusters changed little when the maximum number of intervening open-reading frames (ORFs) increased to 20 or even 50. Therefore, two or more WRKY genes that occurred within a maximum of 40 ORFs were identified as a cluster in this study. Totally, 47 WRKY genes forming 20 gene clusters distribute dispersedly on the overall chromosomes except Chromosomes 5 and 9 (Supplementary Table S3). The remaining 72 genes are singletons. Strikingly, Chromosome 8 not only owns the most of WRKY genes but also possesses nearly half of gene clusters. In general, we can classify the clusters into three kinds. In the first kind, eight gene clusters form monophyletic tandem duplications with non-WRKY genes intervening, including seven clusters in Group III and one in subgroup IIa. In the second kind, seven clusters are made up of four subgroups except IIa, forming three types of gene pairs. Among them, three are II-b, c pair type; three II-c, e pair and one II-d, e pair. The third kind is mixed by two groups. Beyond that, the rest singletons maybe occurred by random events.
According to Xu et al.,69 if two closed related genes which are located within the same chromosomal region apart from each other fewer than 20 genes can be reckoned as tandem duplication. In this scenario, 16 tandem duplications in our study are inside the gene cluster regions. Interestingly, ZmWRKY78 and ZmWRKY91 are the same gene distributed on Chromosomes 7 and 8, respectively, which may suggest the occurrence of segmental duplication event in maize WRKY family during evolution. The increased number of WRKY genes correlates closely with the frequency of duplication events, and these duplications may be responsible for the maintenance of transcriptional regulation activity. More copies of one gene will contribute to offset the effects of mutations and help the plants to survive adversity.
3.4. Expression analysis of WRKY factors in global transcriptome at different developmental stages and in specific organs
Researches have revealed a multiple roles of WRKY factors in response to abiotic stresses, including drought and salt, which is regarded as an ancestral role of WRKY proteins,4 and biotic stresses such as bacteria and fungi. Elucidating the functions of WRKY family members require great endeavour. Microarray is one of the useful global transcriptome analysis technologies which provides an opportunity to understand the patterns of gene expression. Many expression data generated by microarrays are publicly available in microarray databases, such as GEO and ArrayExpress, which provide valuable resources for gene discovery and functional characterization. We mine the gene expression data under drought stress shown in Supplementary Fig. S3 and during Ustilago maydis infection shown in Supplementary Fig. S4.
The gene expression profiling of 31 ZmWRKYs was compared between the drought-tolerant line Han21 and the drought-sensitive line Ye478 (Supplementary Fig. S3). Interestingly, we can notice that the gene expression levels of the drought-tolerant line Han21 in generally change less than that of the drought-sensitive line Ye478 and recovered more quickly when the seedlings were re-watered. We can see that the expression of ZmWRKY115 is apparently decreased under drought stress, which may be a response to the increased ABA level resulting from drought. Simultaneously, some genes show a cumulative trend under drought, such as ZmWRKY 15.1, −46, −52 and –77. Interestingly, ZmWRKY 90 in the drought-sensitive line Ye478 tend to be expressed at higher mRNA abundances under any conditions. Meanwhile, many WRKY proteins are recognized as pathogen-induced transcription factors and active in immune response such as AtWRKY18, AtWRKY22, AtWRKY29, AtWRKY53, AtWRKY54 and AtWRKY70. Supplementary Figure S4 displays the expression levels of maize WKRY genes during infection with U. maydis. Compared with the mock stimulus, most of the ZmWRKYs exhibit a gradual increase in expression levels with the prolonging of infection time. For example, the ZmWRKY78 and 91 are two orthologues of AtWRKY53 which are activated by the pathogen-triggered SA signalling pathway, showing an obvious increasing trend during the infection with U. maydis. AtWRKY70 functions as an activator of SA-dependent defence genes and a repressor of JA-regulated genes, and the expression levels of its orthologue, ZmWRKY115, are also apparently increased upon the fungal infection, implying a parallel function in defence response.
It is of great significance to study the expression patterns and regulative mechanisms of WRKYs in stress responses. However, in the work described here, we mainly focus on the spatial and temporal specific expression patterns of maize WRKY genes in order to identify their roles in developmental regulation, as the expression of WRKY genes was detected in a wide variety of plant species and is involved in plant growth and development.5,70–71 In this case, we mined microarray data that record the gene expression levels of 60 distinct tissues representing 11 major organ systems and varying developmental stages of the maize (Supplementary Table S4). The organ systems included the germinating seed, primary root, whole seedling, stem and shoot apical meristem, internodes, cob, tassel and anthers, silk, leaf, husk and seed.
A total number of 131 probes detected in R could be assigned to 131 corresponding ZmWRKY transcripts (Supplementary Table S5). The remaining five transcripts with no detectable expression signal are ZmWRKY25.1, ZmWRKY25.2, ZmWRKY25.3, ZmWRKY32 and ZmWRKY102. There are three major reasons for why their signals cannot be detected. First, the different versions of the official maize genome sequence are used in the study. The version of B73 maize genome that are used to map probe set design sequences on the microarray is 5a (RefGen_v2), whereas the version we used to identify the complete WRKY family is 5b (RefGen_v2). The second plausible explanation is that these genes are very specific to organs or developmental stages which are not covered in these experiments so that no signal intensities are detected on the microarray. Last, it is also possible that these genes could be stress-inducible or pseudogenes. In order to investigate the temporal and spatial transcription patterns of the WRKY genes in maize life cycle, a hierarchy cluster was performed to visualize global transcription profile of the WRKY genes across the 60 developmental stages, and the results were illustrated as a heat map (Supplementary Fig. S5).
It can be seen from the heat map that all of the 131 detected transcripts produced by 116 genes are involved in many biological processes, and expressed in all tissues, but their expression levels are distinct. Most of the genes appear to be invariable and lowly expressed among all tissues. Interestingly, the reference gene encoding E2 enzyme is the most stably and highly expressed across maize life, for which is known as a component in the ubiquitin-dependent protein degradation pathway. The stable gene expression across all tissues can be regarded as constitutive expression, and we can infer that many maize WRKY genes were expressed at low level, which may work synergistically with other family proteins during plant growth and development.
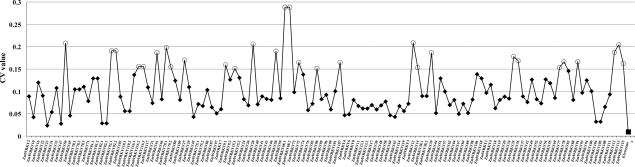
In order to investigate the differential expression patterns, especially fluctuating patterns, in different developmental stages and diverse tissues, we calculate the coefficient of variation (CV value; CV = sd/mean, where sd and mean represent the standard deviation and mean expression level of a gene across all the tissues, respectively) of each gene in 60 tissues accurately (Fig. 3), and the results show that the CV values of these genes range from 2 to 28% (Supplementary Table S6). Those with a CV value of <15% are considered to be the least expression variability.73 Consequently, total 102 transcripts are stably expressed across all tissues. In contrast, the CV values of the rest 29 transcripts are >15%, which indicate the existence of stage-specific gene expression during development. As we know that WRKY factors can act as either negative or positive regulators in gene expressions thus we are more concerned about the dynamic changes in gene expression during the development of major maize organs. Therefore, we highlight the genes with a CV value of >15% and explore their expression levels among the distinct organs in order to find the organ-specific genes in maize (data see Supplementary Table S5).
Figure 3.
Line graph showing the CV of the signal intensities of 131 WRKY transcripts across 60 developmental stages. Normalized signal intensities of 131 ZmWRKY transcripts are displayed (according to MaizePlex), and corresponding ZmWRKY transcripts are presented on the x-axis with the order according to the groups. The transcripts with CV ≥15% are represented by a tiny hollow circle. The reference gene which encodes E2 enzyme is shown as a small square.
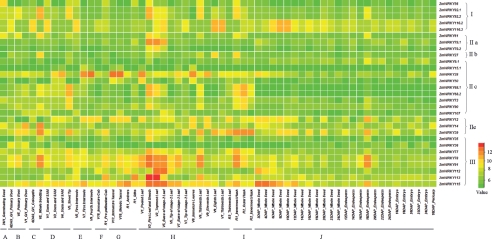
The 11 organ systems can be further subdivided into single tissues which represent the gradual developmental process to some extent. Figure 4 shows the expression levels of the 29 maize WRKY genes in 60 tissues with a CV value of >15%. We ordered the transcripts based on their groups in order to find differential expression between groups. And as expected, genes in the same group appear to share the similar expression patterns as a whole, while about half of 29 WRKY gene expressions show that variation could be generated by different organ types. Expression of ZmWRKY92 appears to be accumulated in the late-development leaves stage. Both ZmWRKY116.2 and ZmWRKY116.3 are highly expressed in the anther and silk, and also highly increased in early whole seed after pollination and before the formation of the endosperm and embryo. Notably, ZmWRKY28 in IIc is remarkably expressed in the stem and SAM, internode, and cob and tassel, and its expression level is gradually changed in consistent with the development of the relevant organs. ZmWRKY68 is highly expressed in late-development leaf and husk. It is notable that there is no gene detected in IId with a CV value of >15%. ZmWRKY39 in subgroup IIe is highly expressed in the primary root, and late leaf and husk. However, the WRKY genes in Group III display a leaf-specific gene expression. Besides, the expression of ZmWRKY78 and ZmWRKY91 may also be specific in the pericarp while ZmWRKY115 perhaps plays a special role in early whole seed. In addition to the highly expressed genes in specific organs, the genes with significant low expression levels may also have effects on the development of given organs. Most of the genes are likely to be stably expressed at low levels in the development of the endosperm and embryo, suggesting that they are not stage-specific and tissue-specific expression.
Figure 4.
Heat map showing the signal intensities of 29 ZmWRKY transcripts with CV ≥15% in 11 specific organs. The alphabet represents specific developmental stages: A, germinating seed; B, primary root; C, whole seedling; D, stem and SAM; E, internode; F, cob; G, tassel and anthers; H, leaves; I, husk; J, seed.
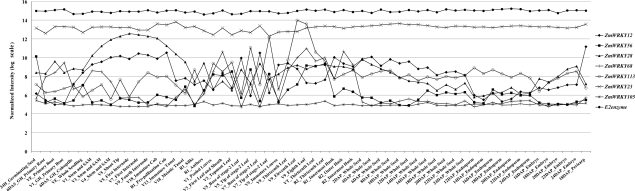
At the same time, we also care about the most variable gene expressions with a CV value of >20%. The line graph in Fig. 5 will do a great help to observe the interest genes and analyse their dynamic changing processes clearly. We use E2 enzyme as an external reference and ZmWRKY23 and ZmWRKY105 as internal references to show the constitutively high expression and constitutively low expression, respectively. It is very interesting that ZmWRKY12 and ZmWRKY28 form an arc line, respectively, revealing their important roles in the development of the stem and shoot apical meristems, and the internode and cob, and ZmWRKY12 is also highly expressed in the pericarp. ZmWRKY56 has the highest expression level in the germinating seed, while ZmWRKY113 shows a fluctuating pattern of expression during the development of the leaf.
Figure 5.
Line graph of the signal intensities of six interest ZmWRKY transcripts with CV ≥20% in 11 specific organs, as well as three reference transcripts.
4. Conclusions
Data acquisition of WRKY transcription factor family is the initial step to go on our analysis of the gene family. Meanwhile, numbering and annotating the identified genes are also of great importance for classification and function characterization. However, a big block stand on our way is the existing identifiers in databases. In database like NCBI, the data of maize WRKY are submitted from large-scale sequencing or experiments individually. The numbering of these data is disorder and redundant ineluctably, and also the data are from several different maize lines, so the numbering system may be diverse and lineage-specific. As the maize B73 genome has been sequenced, we propose to number from chromosomes 1 to 10 and from top to bottom, which can help avoid confusion and promote communication. On this occasion, we give the 136 putative proteins uniform identifier.
Among the 136 WRKY proteins we have identified, 27 proteins belong to Group I, 78 proteins are contained in Group II and 31 proteins are members of Group III. The classification system is based on the features of WRKY domains which can be regarded as evolutionary unit. The sequence divergence usually goes with functional difference in the course of evolution. And it is the divergence of both expression and protein sequence that makes the lineage-specific expansion. Thus, we can conclude that proteins from different groups may accomplish diverse functions.
The comparative analysis of orthologues may help us obtain the information of the evolutionary relationships among the gene family members and help us predict the potential functions of putative proteins. The high-throughput screening method identified that AtWRKY53 is expressed at a very early time point of leaf senescence.8 Not only AtWRKY53, AtWRKY4, -6, -7 and -11 in Arabidopsis play a role in leaf development.39 Among the WRKY genes with the CV value of >15%, ZmWRKY116.2, -116.3 are orthologues of AtWRKY4, and ZmWRKY77, -78, -91 are orthologues of AtWRKY53. However, their functions seem to diverge from each other. As seen in the heat map, ZmWRKY77, -78 -91 are highly expressed in the leaf and present a gradual cumulative trend, implying a regulatory role of these genes in leaf development, especially in the early stage of leaf senescence. While the specific expression of ZmWRKY116.2 and -116.3 emerge mainly in reproductive-related tissues, such as the anther and silk. In addition, ZmWRKY116.2 and -116.3 are also orthologues of AtWRKY44 which are associated with the tannin and mucilage production in the seed coat.4,73 Thus, both genes in maize are expressed at a high level in the early development of the whole seed but not in the development of the endosperm and embryo. Endogenesis abscisic acid (ABA) is crucial in adversity responses as well as growth and development. During the late maturity, the seed enters into the dormant phase due to the increase of ABA. While after imbibition, ABA content usually rapidly decreases and the seed begins to germinate. This ABA-dependent progress of the seed germination and post-germination growth was mediated by AtWRKY2.72 In maize, ZmWRKY56 is the orthologue of AtWRKY2. Coincidently, ZmWRKY56 is highly expressed in the germinating seed after 24 h imbibition, suggesting a similar function of these two orthologues. It has been proved that AtWRKY40 functions as a central negative regulator of ABA signalling in seed germination and post-germination,36 while its orthologues in maize seem insensitive to drought stress in our study.
By means of the microarrary-based data mining and homologous analysis, we can obtain much useful information about the putative functions of the WRKYs in maize. It is of great importance to elucidate the biological functions of these transcription factors and provide us deeper understanding in molecular mechanisms of corn growth development and adversity resistance.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
The project was supported by the Natural Science Foundation of Fujian Province, China (Grant No. B0810040).
Supplementary Material
Acknowledgements
We are grateful to the providers who submitted the microarray data to the public expression databases which can be applied freely.
References
- 1.Ulker B., Somssich I.E. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–8. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulgem T., Rushton P.J., Schmelzer E., Hahlbrock K., Somssich I.E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–99. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson C.S., Kolevski B., Smyth D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–75. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:17531–6. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagacé M., Matton D.P. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2004;219:185–9. doi: 10.1007/s00425-004-1253-2. [DOI] [PubMed] [Google Scholar]
- 7.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–49. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinderhofer K., Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001;213:469–73. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- 9.Pnueli L., Hallak-Herr E., Rozenberg M., et al. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002;31:319–30. doi: 10.1046/j.1365-313x.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen C., Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129:706–16. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willmott R.L., Rushton P.J., Hooley R., Lazarus C.M. DNase1 footprints suggest the involvement of at least three types of transcription factors in the regulation of alpha-Amy2/A by gibberellin. Plant Mol. Biol. 1998;38:817–25. doi: 10.1023/a:1006084104041. [DOI] [PubMed] [Google Scholar]
- 12.Ishiguro S., Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994;244:563–71. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 13.Rushton P.J., Macdonald H., Huttly A.K., Lazarus C.M., Hooley R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 1995;29:691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- 14.Rushton P.J., Torres J.T., Parniske M., Wernert P., Hahlbrock K., Somssich I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–700. [PMC free article] [PubMed] [Google Scholar]
- 15.Cormack R.S., Eulgem T., Rushton P.J., Kochner P., Hahlbrock K., Somssich I.E. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim. Biophys. Acta. 2002;1576:92–100. doi: 10.1016/s0167-4781(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 16.De Pater S., Greco V., Pham K., Memelink J., Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–31. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrova K.S., Conger B.V. Isolation of two somatic embryogenesis-related genes from orchardgrass (Dactylis glomerata) Plant Sci. 2002;162:301–7. [Google Scholar]
- 18.Chen C., Chen Z. Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 2000;42:387–96. doi: 10.1023/a:1006399311615. [DOI] [PubMed] [Google Scholar]
- 19.Hara K., Yagi M., Kusano T., Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. 2000;263:30–7. doi: 10.1007/pl00008673. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z., Yang P., Fan B., Chen Z. An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with the plant defence response. Plant J. 1998;16:515–22. doi: 10.1046/j.1365-313x.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- 21.Yoda H., Ogawa M., Yamaguchi Y., Koizumi N., Kusano T., Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol. Genet. Genomics. 2002;267:154–61. doi: 10.1007/s00438-002-0651-z. [DOI] [PubMed] [Google Scholar]
- 22.Ashida Y., Nishimoto M., Matsushima A., Watanabe J., Hirata T. Molecular cloning and mRNA expression of geraniol-inducible genes in cultured shoot primordia of Matricaria chamomilla. Biosci. Biotechnol. Biochem. 2002;66:2511–4. doi: 10.1271/bbb.66.2511. [DOI] [PubMed] [Google Scholar]
- 23.Kim C.Y., Lee S.H., Park H.C., et al. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant Microbe Interact. 2000;13:470–4. doi: 10.1094/MPMI.2000.13.4.470. [DOI] [PubMed] [Google Scholar]
- 24.MR L. In silico differential display of defense-related expressed sequence tags from sugarcane tissues infected with diazotrophic endophytes. Genet. Mol. Biol. 2001;24:103–11. [Google Scholar]
- 25.Huang T., Duman J.G. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol. Biol. 2002;48:339–50. doi: 10.1023/a:1014062714786. [DOI] [PubMed] [Google Scholar]
- 26.Beyer K., Binder A., Boller T., Collinge M. Identification of potato genes induced during colonization by Phytophthora infestans. Mol. Plant Pathol. 2001;2:125–34. doi: 10.1046/j.1364-3703.2001.00059.x. [DOI] [PubMed] [Google Scholar]
- 27.Dellagi A., Helibronn J., Avrova A.O., et al. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol. Plant Microbe Interact. 2000;13:1092–101. doi: 10.1094/MPMI.2000.13.10.1092. [DOI] [PubMed] [Google Scholar]
- 28.Sun C., Palmqvist S., Olsson H., Boren M., Ahlandsberg S., Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–92. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchive C., Mzid R., Deluc L., et al. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 2007;58:1999–2010. doi: 10.1093/jxb/erm062. [DOI] [PubMed] [Google Scholar]
- 30.Wei F., Zhang J., Zhou S., et al. The physical and genetic framework of the maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaut B.S., Le Thierry d'Ennequin M., Peek A.S., Sawkins M.C. Maize as a model for the evolution of plant nuclear genomes. Proc. Natl Acad. Sci. USA. 2000;97:7008–15. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnable P.S., Ware D., Fulton R.S., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–5. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y.Y., Wang X.F., Wu F.Q., et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–6. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 34.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–71. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Shang Y., Yan L., Liu Z.Q., et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–35. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu H.S., Han M., Lee S.K., et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–47. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- 38.Pandey S.P., Somssich I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–55. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozik A., Kochetkova E., Michelmore R. GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics. 2002;18:335–6. doi: 10.1093/bioinformatics/18.2.335. [DOI] [PubMed] [Google Scholar]
- 42.Team R.D.C. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 43.Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 44.Smyth G.K. Bioinformatics and Computational Biology Solutions using R and Bioconductor. : New York: Springer; 2005. [Google Scholar]
- 45.Warnes G.R. Includes R source code and/or documentation contributed by: Ben Bolker (in alphabetical order), L. B., Robert Gentleman, Wolfgang Huber Andy Liaw, Thomas Lumley, Martin Maechler, Arni Magnusson, Steffen Moeller, Marc Schwartz and Bill Venables, gplots: Various R programming tools for plotting data. 2010. R package version 2.8.0.
- 46.Alexandrov N.N., Brover V.V., Freidin S., et al. Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 2009;69:179–94. doi: 10.1007/s11103-008-9415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nijhawan A., Jain M., Tyagi A.K., Khurana J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–50. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–56. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 49.Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 50.Li W.H., Yang J., Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–7. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Ross C.A., Liu Y., Shen Q.J. The WRKY Gene Family in Rice (Oryza sativa) J. Integr. Plant Biol. 2007;49:827–42. [Google Scholar]
- 52.Xie Z., Zhang Z.L., Zou X., et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–89. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mangelsen E., Kilian J., Berendzen K.W., et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics. 2008;9:194. doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu K.L., Guo Z.J., Wang H.H., Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 55.Maeo K., Hayashi S., Kojima-Suzuki H., Morikami A., Nakamura K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 2001;65:2428–36. doi: 10.1271/bbb.65.2428. [DOI] [PubMed] [Google Scholar]
- 56.Yamasaki K., Kigawa T., Inoue M., et al. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 2005;17:944–56. doi: 10.1105/tpc.104.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackay J.P., Crossley M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 58.Babu M.M., Iyer L.M., Balaji S., Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–20. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 60.Paterson A.H., Bowers J.E., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–8. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanc G., Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–78. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swigonova Z., Lai J., Ma J., et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14:1916–23. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blanc G., Wolfe K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–91. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas B.C., Pedersen B., Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–46. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y., Zhu Z., Ma L., Chen M. The preferential retention of starch synthesis genes reveals the impact of whole-genome duplication on grass evolution. Mol. Biol. Evol. 2008;25:1003–6. doi: 10.1093/molbev/msn052. [DOI] [PubMed] [Google Scholar]
- 66.Ramamoorthy R., Jiang S.Y., Kumar N., Venkatesh P.N., Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008;49:865–79. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- 67.Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–34. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu G., Ma H., Nei M., Kong H. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl Acad. Sci. USA. 2009;106:835–40. doi: 10.1073/pnas.0812043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohashi Y., Oka A., Ruberti I., Morelli G., Aoyama T. Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 2002;29:359–69. doi: 10.1046/j.0960-7412.2001.01214.x. [DOI] [PubMed] [Google Scholar]
- 70.Xing Y., Jia W., Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–51. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- 71.Jiang W., Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9:96. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sekhon R.S., Lin H., Childs K.L., et al. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–63. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 73.Ishida T., Hattori S., Sano R., et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–43. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.