Abstract
Phylogenetic analysis was conducted on 9 kDa non-specific lipid transfer protein (nsLTP) genes from nine plant species. Each of the five classified types in angiosperms exhibited eight conserved cysteine patterns. The most abundant nsLTP genes fell into the type I category, which was particularly enriched in a grass-specific lineage of clade I.1. Six pairs of tandem copies of nsLTP genes on the distal region of rice chromosomes 11 and 12 were well-preserved under concerted evolution, which was not observed in sorghum. The transgenic promoter–reporter assay revealed that both rice and sorghum nsLTP genes of type I displayed a relatively conserved expression feature in the epidermis of growing tissue, supporting its functional roles in cutin synthesis or defence against phytopathogens. For type I, the frequent expression in the stigma and seed are indicative of functional involvement in pistil–pollen interactions and seed development. By way of contrast, several type V genes were observed, mainly in the vascular bundle of the rosette as well as the young shoots, which might be related with vascular tissue differentiation or defence signalling. Compared with sorghum, the highly redundant tissue-specific expression pattern among members of rice nsLTP genes in clade I.1 suggests that concerted evolution via gene conversion favours the preservation of crucial expression motifs via the homogenization of proximal promoter sequences under high selection constraints. However, extensive regulatory subfunctionalization might also have occurred under relative low selection constraints, resulting in functional divergence at the expression level.
Keywords: non-specific lipid transfer protein, cutin synthesis, concerted evolution, gene conversion, subfunctionalization
1. Introduction
Gene family members are paralogous copies created by duplications, which vary in size and genomic organization. Duplication events give rise to multiple gene copies, probably providing raw materials for functional gene innovation. In plants, genome complexity is fundamentally determined by rounds of polyploidy and individual duplications accompanied by gene expansion, thereby implying that the study of evolutionary history is crucial to understanding the functional divergence of gene family members and the manner in which new functions arise.1 There is little doubt that gene duplications have the potential to produce new gene functions in the long-term.2,3 Much less understood are the early stages in the emergence and evolution of gene duplications, and in particular, the fixation of a polymorphic duplicate.1,3 When compared with functional innovation (neofunctionalization), it has been determined that many gene duplicates remain in genomes as the result of passive mechanisms such as subfunctionalization.2 Theoretical studies and data gathered through in silico analyses have advanced our understanding of the functional diversity of gene families and possible retention mechanisms, but large amounts of comparative functional data are required, and data acquisition is usually a very technically challenging proposition.
Non-specific lipid transfer proteins (nsLTPs) are known to be a small multi-gene family in plants and are denominated in accordance with their ability to reversibly bind and transport to hydrophobic molecules in vitro.4 These cationic peptides were originally subdivided on the basis of molecular mass into type 1 (9 kDa) and type 2 (7 kDa), both of which belong to the eight-cysteine motif (8 CM) protein family (Pfam PF00234), with a conserved 8 CM backbone, as follows: C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C.5–7 The cysteine residues are arranged in four disulphide bonds to stabilize a tertiary structure of the hydrophobic cavity, the size plasticity of which allows for the in vitro loading of a broad variety of lipid compounds.4,8 For type 1 LTPs, computational and biochemical analyses have shown that nsLTPs from various plants are capable of accommodating lipids such as phospholipids,4 palmitic acid (C16:0), and the acyl chains of 1,2-dimyristoylphosphatidylglycerol.8 However, less is currently known regarding the lipid-binding properties of type 2 nsLTPs.
The exact functions of nsLTPs remain unclear. Multiple physiological functions that have been suggested include cutin synthesis,9,10 somatic embryogenesis,11 stigma and pollen adhesion,12,13 and plant defence.6,10,14–16 An accumulating body of evidence supports the role of nsLTPs in cuticle synthesis, including their extracellular localization and lipid-binding capability on fatty acids with hydroxyl and cetoxyl groups.9,17 Furthermore, the nsLTP genes from some plants are expressed in the leaf epidermal cell layer, where the waxy cuticle is generally found.9,18 However, more complex expression profiles from individual experiments are frequently observed, such as expression in the xylem, phloem, or stigma, which can lead to disparate and unpredictable gene functions of nsLTPs.11–13
There has been no conserved functional perspective, systematic expression analysis, or clear evolutionary story with regard to members of the nsLTP gene family. Thus, in this study, we conducted a comparative analysis of a cluster of nsLTP genes from rice and sorghum in an effort to address several key questions regarding the evolutionary fate and functional diversity or redundancy of this gene family. A phylogeny analysis was conducted by trying to identify nsLTP genes from nine plant species, covering monocots, dicots, ferns, mosses, and algae. Subsequently, the syntenic gene pairs in rice and sorghum were determined by the genomic organization in conjunction with large-scale duplicated blocks. We combined the analyses with microarray data, transgenic promoter–reporter assay, and semi-quantitative reverse transcription–polymerase chain reaction (RT–PCR) in order to discern the gene functions and evolutionary fate of nsLTP genes. Additionally, the evolutionary mechanisms after duplication for one gene cluster were discussed, considering that rice and sorghum experienced pre-grass whole genome duplication prior to speciation.
2. Materials and methods
2.1. Sequence identification and phylogenetic analysis
Full genome assemblies of nine plant species representing eudicots (Arabidopsis thaliana), magnoliid dicots (Aquilegia coerulea), monocots (Sorghum bicolor, Oryza sativa, and Brachypodium distachyon), ferns (Selaginella moellendorffii), mosses (Physcomitrella patens), and algae (Chlamydomonas reinhardtii and Volvox carteri) were downloaded from the Joint Genome Institute plant genomics database (http://www.Phytozome.net). Protein sequences annotated with the HMMPfam domain PF00234 (plant lipid transfer/seed storage/trypsin–alpha amylase inhibitor) were retrieved as a query and further blast-searched against nine full genome databases to avoid the loss of plant nsLTP genes due to mis- and unannotation, with a cut-off value of e−10. The deduced amino acid sequences of candidate nsLTP genes were manually confirmed to harbour the 8 CM (C … C … CC … CXC … C … C, where X represents any amino acid) and genes lacking the motif and encoding for the putative proline-rich proteins were excluded from further analyses. The protein sequences of the putative 2S-albumins, AtALB119 and alpha amylase inhibitor, RAT120 were then BLAST-searched against the obtained candidate nsLTP proteins to exclude possible cereal storage proteins and inhibitors in the retrieved nsLTP datasets. Additionally, the proteins harbouring the glycosylphosphatidylinositol-anchored motif were not employed. The deduced amino acid sequences of all the nsLTP genes were aligned using the ClustalW program with default parameters. The phylogenetic tree was conducted with MEGA software version 521 using a maximum-like algorithm with 1000 bootstrap replicates.
2.2. Mapping chromosome location and duplicated blocks
Syntenic gene sets against chromosomes of rice and sorghum, and of chromosomes shared between rice and sorghum, were downloaded from the PGDB database (http://chibba.agtec.uga.edu/duplication/). The pan-grass and pre-grass syntenic gene sets were downloaded from the CoGe database (http://synteny.cnr.berkeley.edu/CoGe/). The collinear nsLTP genes that matched in the syntenic gene sets were selected for further study. The genomic localization and collinearity of nsLTP genes in/between the genomes of rice and sorghum were visualized using Circos software.22
2.3. Expression analysis and co-expression analysis based on microarray dataset
The expression of nsLTP genes during developmental stages presented as heat maps was generated by the Gen-evestigator (http://www.genevestigator.ethz.ch) webtool with default parameters. A total of 39 rice and 233 Arabidopsis GEO (Gene Expression Ominbus) datasets of high quality were employed. Genes for which probes were not found included Os11g02389, Os11g02369, Os12g02340, and Os06g06340 of group I and Os04g33930 of group IV; these were excluded for further study, as was Os11g02350, due to low specificity.
To conduct co-expression analysis, the CEL files of Affymetrix GeneChip genome arrays of O. sativa were manually downloaded from the GEO datasets of the NCBI database (http://www.ncbi.nlm.nih.gov/geo/), gathering a total of 914 GSM samples. The microarray CEL files were normalized via the RMA method using R software (http://www.r-project.org/). The similarity between the expression profiles of genes across the microarray dataset was evaluated via Pearson's correlation coefficient. In order to determine the cut-off of the co-expressed gene, we surveyed the distribution of similarity of 124 750 random pairs, resulting in an r = 0.792 (α = 0.01).23 The functional enrichment analysis of co-expressed genes was conducted via agriGO tools (http://bioinfo.cau.edu.cn/agriGO/) with default parameters.
2.4. nsLTP promoter::uidA fusion construction and transgenic plant generation
A total of 23 nsLTP promoters, including 15 of rice and 8 of sorghum, were utilized for a β-glucuronidase (GUS) reporter assay in Arabidopsis. Approximately 1 kb upstream of nsLTP genes was amplified from the genomic DNAs of O. sativa (cv. Niponbire; OsLTP) and S. bicolor (cv. BTX623; ScLTP) and cloned into a binary vector of pBI121 without a native CaMV35S promoter. The construction of promoter::uidA fusions and their transformation into plants were conducted as previously described.24
2.5. Histochemical GUS analyses
T2 plants of 3–10 independent transgenic lines harbouring each construct were selected for GUS expression analysis following the plant whole life cycle. Histochemical analyses were conducted with T2 plants according to the procedures previously described.9 The T3 lines harbouring pBI121 vectors with or without the 35s promoter were used as positive and negative controls, respectively.
2.6. Abiotic stress treatments and semi-quantitative RT–PCR
Seeds of rice and sorghum were allowed to germinate on moisture filter papers and then grown in containers as previously described.24 After 36 h, the water was replaced with 0.44% Murashige and Skoog medium (Duchefa Biochemie B.V., Haarlem, Netherlands) and the plants were grown for 10 days with a photoperiod of 14/10 h at a temperature of 28°C/25°C (day/night) before abiotic stress treatments. Treatments with types of abiotic stresses such as drought, salinity, cold, and wounding were conducted as previously described.24 RNA isolation and semi-quantitative RT–PCR were carried out as described.24 The actin genes for rice and sorghum were employed as an internal control with specific primers (Supplementary Table S1).
3. Results
3.1. NsLTP gene family in plants
The protein sequences of the family annotation (Pfam 00234: Plant lipid transfer/seed storage/trypsin–alpha amylase inhibitor) were first retrieved from the full genome datasets of the nine plant species. Because the plant LTPs were clustered together with several other proteins, the obtained sequences were examined manually for the existence of the 8 CM and seed storage proteins, and trypsin–alpha amylase inhibitors were excluded as described elsewhere.7,25 The protein sequences of alpha amylase/trypsin inhibitors are usually large molecules with 10 cysteine residues.26 Hybrid proline-rich proteins harbour an additional N-terminal domain at the C-terminal 8 CM.25 Primarily, the sequences of 195 plant full-length nsLTP genes were employed in the construction of a phylogenic tree to evaluate their phylogenic relationship. We determined that the putative 7 kDa nsLTPs from rice and Arabidopsis clustered into one group, with a bootstrap value of 95 (Supplementary Fig. S1), distinct from the other group that harbours the putative 9 kDa nsLTPs. A similar result was also noted in a previous study.7 Although both 7 and 9 kDa nsLTPs harbour the conserved 8 CM pattern, structural and lipid-binding studies have demonstrated that they contain different tertiary structures and lipid-binding spectra. Herein, we excluded the group harbouring 7 kDa nsLTP genes in order to clarify the evolutional mechanism 9 kDa nsLTP genes. Finally, a total of 115 full-length 9 kDa nsLTP-related genes retrieved from seven plant species, including 31 of O. sativa, 16 of S. bicolor, 14 of B. distachyon, 24 of A. thaliana, 10 of A. coerulea, 14 of P. patens, and 5 of S. moelendorffii were employed for further analysis.
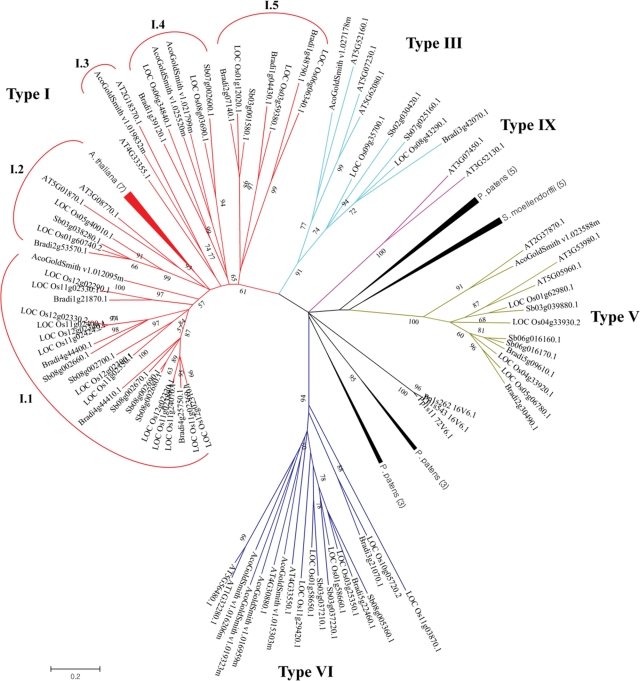
The phylogenetic tree was constructed using the maximum-likelihood method and the solidity of the nodes was assessed by 1000 bootstrap resampling repetitions. Initially, the organization of the tree supports eight independent groups, whereas the nsLTPs from P. patens and S. moelendorffii formed several branches without significant bootstrap values and thus were employed as out-groups. Based on the position in the tree and the criterion of more than 30% sequence identity in one group, we classified the nsLTP genes from angiosperms into five groups with bootstrap values ranging from 61 to 100 (Fig. 1). Boutrot et al.7 previously defined the nsLTPs from Arabidopsis, rice, and wheat into nine types. On the basis of the comparison between the previous dataset and ours, the five groups classified here were in agreement with the nsLTPs of type I, III, V, VI, and IX. Although the type I appeared paraphyletic with a bootstrap value of only 61, the included nsLTPs shared the conserved cysteine motif in the same position, containing nine residues between Cys1 and Cys2, and 19 residues between the conserved Cys4 and Cys5. In fact, the nsLTPs used to classify each group evidenced variations in the number of flanking amino acids between cysteine residues, reflecting possible changes in catalytic activity (Supplementary Fig. S2). The genes of type I were subsequently divided into five clades, in which clade I.1 contained the largest numbers of gene copies among the clades or groups, only in grass lineage. Five nsLTP genes of P. patens and S .moellendorffii were located at the same branch as that of the type V genes, and were classified together as type D by Edstam et al.27; however, no significant bootstrap value was observed in our tree, and they contain a different cysteine pattern from the type V.
Figure 1.
Unrooted phylogenetic tree of plant nsLTP family. The 9 kDa related nsLTP genes identified from seven land plant species were employed for pairwise alignment. The amino acids of the 8 CM region were used for phylogenetic tree construction using the maximum-likelihood algorithm with 1000 bootstrap replicates. Bootstrap values of <50 were hidden. Compressed branches were noted by species name followed by the gene number included in the branch.
3.2. Genomic organization of nsLTP genes in rice and sorghum
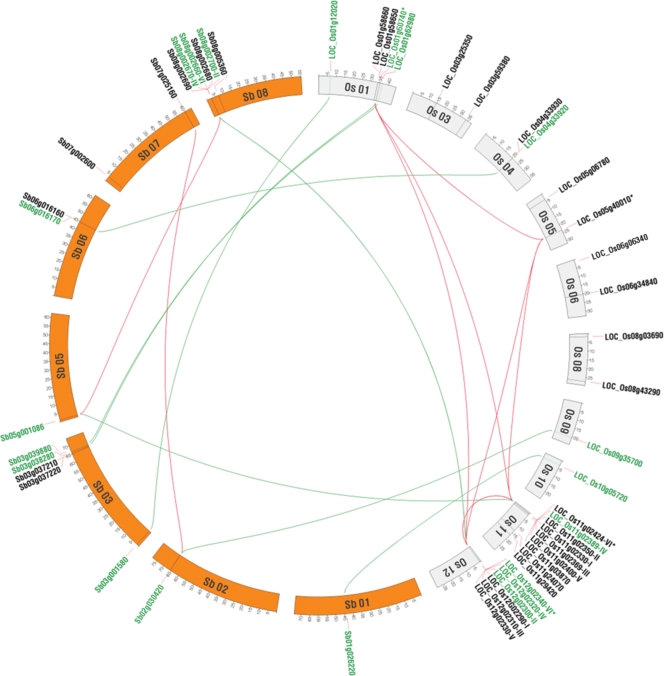
The physical locations of 32 OsLTPs and 17 SbLTPs including a partial gene (Sb05g001086) were assigned to 10 of 12 rice chromosomes and 7 of 10 sorghum chromosomes, respectively (Fig. 2). In clade I.1, six tandem genes on the distal region of rice chromosome 11 were found to be collinear with those in the counterpart of rice chromosome 12 without any loss. By way of contrast, five nsLTP tandem genes were detected on chromosome 8 of sorghum, while only one gene (Sb05g001086) was found in the paralogous chromosome 5 (Fig. 2). The gene (Sb05g001086) contained a partial nsLTP sequence encoding a truncated protein, and thus was not included in our phylogenetic analysis. One of the tandem duplicate pairs (Os11g02424 and Os12g02340) on rice chromosome 11 and 12, respectively, was also collinear with Os01g64070 and Os05g40010 in clade I.2. In group II, Os09g35700 was orthologous with Sb02g030420, which was collinear with Sb07g025160. The orthologous gene pairs between rice and sorghum were also observed in group IV (Sb03g033980 vs Os01g62980, Sb06g016170 vs Os04g33920) and group V (Sb01g026220 vs Os10g05720).
Figure 2.
Genomic organization and colinearity of the nsLTPs genes in rice and sorghum. Chromosomal localization of nsLTP genes was indicated in each species. The orthologous pairs between rice and sorghum were connected in green. The collinear paralogs detected in synteny blocks were connected in red. Detailed information for gene pairs is indicated after the gene locus name.
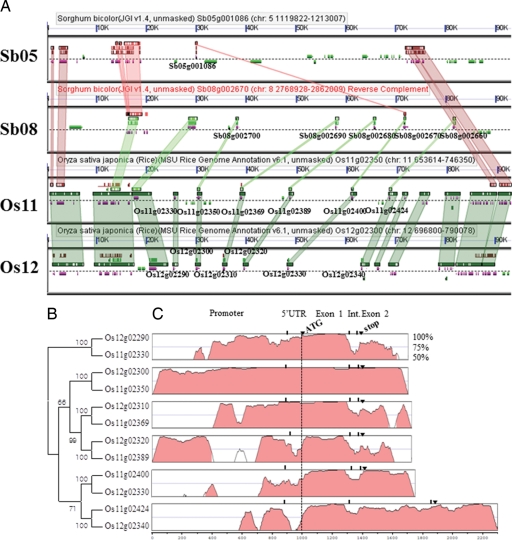
To clarify the relationships among the duplicated gene pairs in clade I.1, detailed genomic visualization of this gene cluster was conducted via a comparison between rice and sorghum. Highly conserved gene order at the locus containing the nsLTP genes was identified between rice chromosomes 11 and 12 and sorghum chromosome 8 (Fig. 3A). The synteny block on sorghum chromosome 8 contained 5 nsLTPs and two nearly collinear genes located towards the centromere (Fig. 3A), while this genome fragment was inverted, perhaps as the result of DNA inversion and rearrangement in the sorghum genome after whole genome duplication. Although only three out of five copies were defined as orthologous with those OsLTP genes in the database (Fig. 2), a more detailed genome comparison by setting the parameter ‘high sequence similarity > 200 bp’ revealed single lines between all five SbLTPs and five pairs of the collinear OsLTP genes, reinforcing the hypothesis of a well-conserved orthologous relationship (Fig. 3A). The collinear gene orders were also found to exist between orthologous chromosome 5 in sorghum and 11 in rice in the region including the nsLTP gene-rich locus, whereas only one partial nsLTP (Sb05g001086) remained in the sorghum genome. The further analysis by the VISTA plot revealed that each of the collinear gene pairs displayed a near-identical homology including both coding regions and at least partial upstream or downstream sequences (Fig. 3C). In particular, Os12g02300 and Os11g02350 evidenced striking sequence identity throughout the whole genomic region covering 1000 bp upstream of the start codon and 300 bp downstream of the stop codon.
Figure 3.
Collinearity and sequence similarity of OsLTPs on the distal region of rice chromosome 11 and 12. (A) Comparison of orthologous and paralogous genomic regions including nsLTP tandem repeats in rice and sorghum. Sorghum bicolor chromosome 8 is displayed in reverse (red). Gene models with collinearity are coloured in purple, while those without collinearity are coloured in grey. The nsLTP gene names are indicated under the gene models. The high gene sequence similarity is depicted in rectangles up or under genes and connecting the collinear genes. This experiment can be reproduced with distance, algorithm changes from database (http://synteny.cnr.berkeley.edu/CoGe/GEvo.pl). (B) Neighbour-joining tree of OsLTP genes at the nucleotide level. The branch containing Os12g02290 and Os11g02330 was selected as a root. One thousand repeats were used for bootstrap evaluation. (C) Similarity of the OsLTP genomic and surrounding sequences between each collinear gene pair. VISTA conservation tracks were generated using the region 1000 bp upstream of the start codon, OsLTP gene exon 1, intron, exon 2 and 300 bp downstream stop codon. The gene pair names for each alignment were shown in phylogeny. The nsLTPs on chromosome 12 were used as a reference for each VISTA alignment. The criterion for sequence similarity is 75%.
3.3. Microarray-based expression profile of OsLTPs
The rice nsLTP genes included in type I were responsible for the most ubiquitous and broadest expression covering the seedling, shoot, inflorescence, and seed, but were rare in the root (Supplementary Fig. S3). The organ-specific expression dynamics revealed the redundant but distinct expression pattern of these genes throughout the entire life cycle in rice. For example, the expressions of Os12g02300, Os11g02330, Os12g02290, Os01g60740, and Os08g03690 were particularly strong in the embryo, whereas Os11g02424 and Os03g59380 expressions were high in the anther. A distinct expression pattern was noted in two type IV genes: Os05g06780 and Os01g62980 were expressed in the crown and shoot, but not in the young seedling, inflorescence, or seed. Only trace expression signals of Os04g33920 were detected in the embryo and crown. No type III genes and only a few genes of type VI were expressed over various tissues.
To obtain additional clues regarding the functions of nsLTPs, co-expression analysis was conducted by employing microarray data. Several OsLTPs in clade I.1 appear to be frequently co-expressed together, including Os11g02400, Os11g02389, Os11g02424, Os11g02369, and Os12g02310; functional enrichment tests identified these OsLTPs as being involved with transport, establishment of localization, and localization (Supplementary Tables S3 and S4). Furthermore, co-expressed gene members related to ATP binding/synthesis, oxidation reduction, and hydrolase were observed abundantly throughout the nsLTP gene family, and were generally associated with primary metabolic and metabolic processes on the functional enrichment test. Os03g59380 in clade I.5 was highly co-expressed with a dozen MADS box transcription factor gene members.
3.4. Tissue-specific expressions of nsLTP promoter::udA fusions in type I
Based on our rice microarray data, two abundantly expressed types (I and V) were chosen for further analysis, using a promoter::uidA fusion strategy in transgenic Arabidopsis. In clade I.1, the transgenic plants for eight OsLTP gene promoters were successively obtained, including three collinear pairs—i.e. Os11g02350 vs Os12g02300, Os11g02369 vs Os12g02310, and Os11g02389 vs Os12g02320—of the duplicate blocks between rice chromosomes 11 and 12. Five sorghum nsLTP gene promoters in the syntenic region of chromosome 8 were employed for analysis. Most nsLTP promoters in clade I.1, with the exception of Sb08g002700, drove GUS activities related principally with the epidermis of various tissues such as the germinated seedling (Fig. 4A1–12), root tip (Fig. 4I1–2), young leaf including guard cell (Fig. 4B1–12 and C1–12), shoot (Fig. 4D1–12 and E1–12), flower bud including ovary, stigma, and anther (Fig. 4F1–12 and G1–12), young silique including abscission zone (Fig. 4H1–12) and seed, including the embryo, endosperm, and integument layer (Fig. 4I3–12). Following the expression dynamics of this gene cluster, we divided this expression module into 12 unique sub-expression motifs, including the cotyledon, hypocotyl, highly active in young leaf, highly active in old leaf, root tip, wound sites, anther, ovary, stigma, silique abscission zone, young silique, and seed (Supplementary Fig. S4). At the tissue-specific level, these genes displayed an overlapping but complementary expression pattern. The most conserved expression was found in the meristematic region of the rosette (highly active in young leaf), wound shoot, and abscission zone (Fig. 4B1–12, C1–12, and H1–12, Supplementary Fig. S4). By way of contrast, only Os11g02400 and Os12g02340 were found to be active in the root tip (Fig. 4I1–2). In rice, a markedly overlapping expression pattern between rice collinear gene pairs (Os11g02350 vs Os12g02300, and Os11g02369 vs Os12g02310) was observed, which also shared relatively longer proximal promoter regions with a high degree of sequence homology (Fig. 3C and Supplementary Fig. S4). However, Os11g02389 evidenced only trace expression in the meristematic region of the rosette, stigma, and abscission zone, when compared with the collinear gene of Os12g02320, which was widely expressed. This expression pattern in relation to the epidermal cells of young growing tissues was also conserved in three sorghum paralogous genes (Sb08g002670, Sb08g002680, and Sb08g002690). Divergent expression was noted for Sb08g002660, which did not evidence the strongest expression in the young leaves, but did evidence strong expression in old leaves (Fig. 4B2), distinct from the majority of the other homologues. Additionally, the GUS activities of the Sb08g002700 promoter were localized in the vascular bundles of rosettes and young shoots (Fig. 4A13, B13, C13, D13, E13, F13, G13, and H13), which differed markedly from those of the collinear rice genes (Os11g02350 vs Os12g02300).
Figure 4.
Tissue-specific expressions of rice and sorghum nsLTP promoter::uidA fuisions in clade I.1. T2 plants of transgenic Arabidopsis were used for activity observation during the plant whole life cycle. The collinear gene pairs are indicated by a vertical line before the gene names. A1–13, germinated seedling; B1–13, 10-day to 2-week-old seedling; C1–13, leaf and leaf sections; D1–13, shoot and sections; E1–13, wound shoot; F1–13, flower bud; G1–13, flower and sections; H1–13, silique; I1–2, root tip; I3–7, seed section. Abbreviations: cot, cotyledon; pI, primary leaf, Hy, hypocotyls; tri, thricom; epi, epidermis; gua, guard cell; sti, stigma; ant, anther; ova, ovary; az, abscission zone; emb, embryo; end, endosperm; int, integument; pol, pollen; va, vascular. Bar = 50 μm.
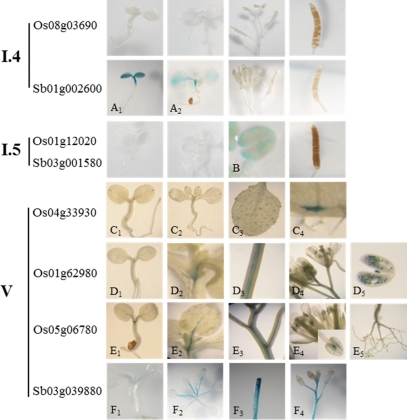
In clade I.4, no GUS activity was observed for Os08g03690, whereas Sb01g002600 displayed apparent activity only in the cotyledon at the germination stage (Fig. 5A). The promoters of both Os01g12020 and Sb03g001580 in clade I.4 drove expression in the anther (Fig. 5B), revealing an expressional conservation. The expression of Os01g12020 in the anther was previously reported.28
Figure 5.
Tissue-specific expressions of rice and sorghum nsLTP promoter::uidA fusions in clade I.3, I.4, and type V. T2 transgenic Arabidopsis was used for activity observation during the whole plant life cycle.
3.5. Tissue-specific expressions of nsLTP promoter::udA fusions in type V
Distinct expression patterns of members of type V, which included Os01g62980, Os05g06780 and the orthologous gene of Sb03g039880, were expressed principally in the vascular tissues of the rosette (including hypocotyl and root) and young shoot (Fig. 5C1–F4). Strong expression was noted in the meristematic region and was reduced as the tissues aged. GUS activities were also evident in the anther, including pollen, driven by the promoters of Os01g62980 and Os05g06780, which were not demonstrated via microarray analysis.
3.6. Transcriptional responses of rice and sorghum nsLTP genes against abiotic stresses
A total of 22 OsLTP and 10 SbLTP genes of type I and type V were employed for semi-quantitative RT–PCR analysis and their amplicons were confirmed via sequencing. However, seven genes (Os11g02330, Os11g02389, Os03g59380, Os08g03690, Os01g12020, Os04g033930, and Sb03g001580) were not amplified, even after 45 PCR cycles. The lack of amplification of Os11g02330 and Os04g033930 was inconsistent with the microarray and GUS assay data (Figs 3 and 4). This may be attributable to the cross-hybridization of microarray probes, low expression level in native plants, or unsuitable primer design.
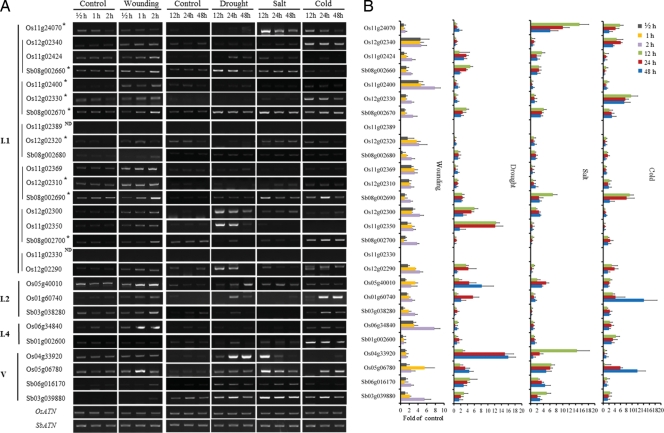
Figure 6 shows that most of the genes in clade I.1 evidenced induced expression against wounding stress, with the exception of Os11g024070. When compared with other rice genes in clade I.1, Os01g024070 evidenced a minimal relationship and a lack of wounding-mediated induction, but was induced by salt. The expression of the tandem-repeat genes in rice chromosomes 11 and 12 (clade I.1, except for Os11g024070) appeared to be rarely stimulated in response to drought, salt, and cold. By way of contrast, the five orthologous sorghum nsLTP genes were more broadly and frequently induced, in which Sb08g002670 and Sb08g002690 could be induced by all four of the abiotic stresses. Sb08g002680, a tandem copy, could not be induced by drought, salt, or cold. Among the detected genes in clade I.1, four OsLTPs (36.3%) and four SbLTPs (80.0%) were abundantly expressed, whereas four OsLTPs (36%) and three SbLTPs (60.0%) were stimulated by drought. Additionally, three OsLTPs (27.2%) and three SbLTPs (60%) were stimulated by salt, and four OsLTPs (60%) and three SbLTPs (60%) were stimulated by cold. The collinear OsLTP gene pairs in clade I.1 did not evidence a significantly parallel expression pattern, as most of these genes were not stimulated by the tested stresses. By way of contrast, both Os12g02300 and Os11g02350 were induced by wounding and drought.
Figure 6.
RT–PCR analysis of the expressions of nsLTP genes against abiotic stresses. The stress treatments were applied by subjecting 10-day-old seedlings to wounding, drought (polyethylene glycol, 25%), salt (NaCl, 250 mM), or cold (4°C) stresses for variable time periods from 0 to 48 h. (A) Representative semi-quantitative RT–PCR analysis. The collinear nsLTP gene pairs were indicated by a vertical line before the gene names. OsATN, Oryza sativa actin gene; SbATN, Sorghum bicolor actin gene. *Highly expressed genes amplified with <28 PCR cycles. ND, not detected. (B) Statistical analysis of the semi-quantitative RT–PCR shown as mean ± SEM for three to four independent experiments.
In clade I.2 and I.4, most genes were also induced by wounding, except for Sb01g002600, which was expressed only in cotyledons, according to the results of a promoter–reporter assay. Os05g40010 was induced by all four abiotic stresses, which differed from the rice gene pairs in type I.1. Os01g60740 appeared to lose the salt-induced expression. Sb03g038280 was stimulated only by cold. The orthologous genes of Os05g06780 and Sb03g039880 in type V could both be induced by all the four stresses. Both of Os04g33920 and Sb06g016170 lost the expression induced by cold, and Os04g33920 was not stimulated by wounding.
4. Discussion
4.1. Phylogeny and nsLTP gene family radiation
Among the nine species examined, the lack of nsLTP genes in the two algal genomes of C. reinhardtii and V. carteri probably suggests an ancient origin of the 9 kDa nsLTP gene in embryophytes after divergence between green algae and land plants, which might be consistent with the emergence of the cuticle during land plant evolution. Edstam et al.27 also found no nsLTP genes in an examination of four green algae of Chorophyta and one red algae of Charophyta, but identified two types of nsLTP genes in Marchantiophyta (liverwort). Some bryophytes, including moss, liverwort, and hornwort, harbour a thin layer of cuticle on their surface as well as on their reproductive organs.29,30 However, the presence of a cuticle in P. patens has never been confirmed.
Boutrot et al.7 previously conducted a phylogenetic analysis of nsLTPs from wheat (122), rice (52), and Arabidopsis (49), classifying them into nine types with distinct cysteine patterns. In this study, we have classified the 9 kDa nsLTP genes of angiosperm into five groups, which should be the previously named type I, III, V, VI, and IX, according to the cysteine pattern and the gene members. For instance, Os11g02379.1 and Os11g02379.2, which were previously identified as type I, were replaced by Os11g02369 and Os11g02389 following database update. We also identified additional nsLTPs of Os03g25350 and Os11g03870 in type VI. However, the previously identified type VIII (Os06g49770) and type Y (Os03g44000, Os07g27940, and Os11g34660) were not employed in our analysis, as the peptide length in these genes exceeded 120 amino acids. The five classified types in our study harboured identical Arabidopsis nsLTPs members in the previous analysis conducted by Boutrot et al.,7 whereas additional AtLTP genes such as At1g77470, At1g52415, At2g16592, At2G13295, At3F29152, and At4g12825 were found among these five types. Using the 8 CM method, Liu et al.31 identified 135 Solanaceae nsLTPs, 52.59% of which were classified into type X, a possible subtype of type I with a similar cysteine pattern. In our analysis, the type I nsLTPs were composed of those of monocots, magnoliid dicots, and eudicots. Five sub-clades were grouped into type I, while the positions in the phylogenetic tree and the low bootstrap values indicate that several ancestors might exist in this group prior to diversification into angiosperms. Additionally, the author argued that some monocot- or dicot-specific types of nsLTPs, e.g. type IX, might have evolved before the monocot-dicot split. The results of our comparative phylogenetic analysis also demonstrated the Arabidopsis-specific type IX and several independent branches of nsLTPs from fern (S. moellendorffii) and moss (P. patens). These results imply that the nsLTP genes with different cysteine patterns emerged, subsequently expanded, and became evolutionarily fixed in angiosperms or single species. In the present study, our interest laid principally with the nsLTP genes in Poaceae, such as rice and sorghum. On the basis of the phylogenetic tree (Fig. 1), clusters of the nsLTP genes in grass lineage have continuously expanded in type I, V, and VI, indicating rounds of duplications with different gene loss in each species. The preservation of a large number of young gene copies in clade I.1 is interesting with regard to functional innovation and evolutionary mechanisms occurring after duplication.
4.2. Concerted evolution of rice nsLTPs in clade I.1
Poaceae dates back to 50–70million years, after the shared pre-grass whole genome duplication event on the last common ancestor.32 The distal regions of rice chromosomes 11 (5.44 Mb) and 12 (4.27 Mb) were thought to have resulted from a recent (5–7million years ago) duplication,33 but a comparison with the sorghum genome indicates their existence before sorghum and rice diverged about 50million years ago,32 thus suggesting its ancestral nature. Illegitimate recombinations such as gene conversion may have occurred frequently at this segmental region between the homeologous chromosomes, allowing for rounds of genomic sequence homogenization, and leading to a high level of concerted evolution of this genomic segment on rice chromosomes 11 and 12.32,33
As described elsewhere,35 six tandem nsLTP gene pairs on the distal region of rice chromosomes 11 and 12 were presently found to be in collinear order. However, only five gene copies were preserved in the corresponding region of sorghum chromosome 8, and only one truncated gene in chromosome 5. Even though this segmental duplication was supposed to have appeared at least 50million years ago, the highly similar sequences between each collinear gene pair enable them to fall within the same phylogenic branch (Fig. 3B), as the alignment of coding sequences displayed an identity of >98%. The high sequence similarity between the collinear gene pairs on the genomic locus included both the coding and flanking regulatory sequences (Fig. 3C). Because non-coding regions generally evolve rapidly after duplication, the high sequence homology at this locus between rice chromosomes 11 and 12 strongly supports the existence of gene conversion events, as previously described,34 which means that this cluster of nsLTP genes on rice chromosomes 11 and 12 have undergone concerted evolution for more than 50million years. However, the sorghum genome fragment the nsLTP genes appears to be inverted on chromosome 8, and thus may have escaped the chance of recombination with the homoeologous region on chromosome 5.
4.3. Expression and possible functions of nsLTPs
According to our analysis of the public rice microarray data, the most abundant nsLTP expression can be attributed to type I nsLTPs, especially in clade I.1, which might have undergone concerted evolution. The systematic examination of transgenic promoter activities of this type nsLTPs also evidenced a broad expression profile during the entirety of the plant life cycle. In clade I.1, an expression feature related with the epidermis of aerial growing tissues, which was highly conserved in the young leaf/leaf primordia, wound site, and abscission zone, was observed for most rice and sorghum genes, even though the expression dynamics in each organ/tissue were also apparent. This expression feature is supported by previous studies, including nsLTPs, throughout dicot and monocot plants.5,13,18,23,36 By accessing the public microarray data in Arabidopsis, it was determined that similar expression patterns could be represented by the gene cluster in clade I.2, in which at least four genes, including AT2G15050, AT3G51600 (AtLTP5), AT2g38530 (AtLTP2), and At2g38540 (AtLTP1), should have preserved expression motifs in the shoot apical meristem or leaf primordial (Supplementary Fig. S4). This result is consistent with the recent transgenic promoter analysis including AtLTP1-6.13
The epidermal expression of young aerial growing tissue has been noted as one of the evidences correlating nsLTP to cutin synthesis, where the cuticular wax is usually localized.18 As anticipated, the parallel distribution of the cuticle and nsLTPs were detected at the abscission zone, guard cell, ovary epidermis, anther, stigma, embryo, and testa.30 Plants evolved the cuticle and stomata at the same time, and the presence of these structures as early as that of the gametophytes of moss and hornworts has been confirmed. Herein, both our phylogenic analysis and the observed expression conservation in type I support the association between nsLTPs and cutin synthesis. Additionally, the epidermis-specific expression could also indicate the involvement of nsLTPs in plant defence to protect young growing tissues against phytopathogen attack, as these peptides evidence intrinsic antimicrobial activities and their involvement in plant defence mechanisms has been demonstrated.6,14 Such a role was particularly consistent with the fact that most of the type I nsLTP genes were wounding-inducible, as revealed by both our transgenic promoter and RT–PCR analysis (Figs 4E1–13 and 6A).
The expression patterns in stigma and pollen were also observed in clade I.1, especially for Os11g02389 and Sb08g002680, which conferred rare expression in floral organs but strong in the stigma, which indicated the possible function of pollen–pistil interaction (Fig. 4G5 and 7). An LTP like protein SCA from the lily was previously suggested to play a role in pollen tube adhesion and guidance, which functions to form an adhesive matrix with pectin that guides pollen tubes to the ovules.12,13 On the basis of the lipid-binding cavity, another model was suggested wherein LTP could exert a cell wall-loosening effect on the cells of the secretory zone of the stigma. It could also be proposed that the relative conserved expression in young growing tissues may also imply the function involved in cell expansion, as plant growth is largely attributable to the cell expansion that occurs after cell division from the meristem.37
The expression in grain was observed primarily in the embryo, endosperm, and integumentary layer (Fig. 4I3–7), as was also detected in another study of wheat nsLTP promoters.38 As the cuticle does not exist in the endosperm, a simple functional involvement of these LTPs in cutin synthesis seems modest. In endospermic seeds, such as cereals, the endosperm generally accumulates significant quantities of starch and lipid reserves and is hydrolyzed to fuel post-germination growth. Therefore, the properties of spectrum transport in lipids imply that nsLTPs may play endosperm lipid-recycling role during seed maturation and germination. In Euphorbia lagascae, several nsLTPs were abundantly observed and correlated with programmed cell death response during endosperm degradation.39 This hypothesis is consistent with our co-expression data showing that abundant genes related to oxidation reduction and hydrolase were observed. Another possibility is that these peptides may function similarly to seed storage proteins, such as 2S-albumins and protease inhibitors, as these evolutionarily related proteins could also harbour 8 CM motif sharing structures similar to those of LTPs.40 In the present study, clade I.1 is in a grass lineage and harbours the largest nsLTP duplicates. Our examination of public microarrays indicated that these young duplicates in clade I.1 contributed to the broadest expression in rice but to clade I.2 in Arabidopsis, in which the functional significance was unknown. One possible functional shift might involve seed development. According to our microarray analysis, only two genes in Arabidopsis were apparently expressed in the embryo and no gene was observed to be expressed in endosperm; this differs profoundly from the strong expression observed in rice and sorghum (Fig. 4, Supplementary Figs S3 and S5).
By way of contrast, the expression of the nsLTP genes of type V was observed primarily in relation with the vascular bundles, which was functionally unclear. Similar expressions of nsLTP genes were previously suggested to be involved in vascular differentiation.41 Furthermore, the functions related to vascular specificity were demonstrated in the study of DIR1, a 7 kDa nsLTP, which releases a systemic signal responsible for systemic acquired resistance (SAR) in the vascular system.15 A mutation of the AZI1 gene, which is induced by azelaic acid, also resulted in the specific loss of systemic immunity in priming defences. In these studies, it was supposed that nsLTPs could bind to signal molecules and translocate SAR signals at long distance.15,16 Interestingly, our promoter–reporter approach demonstrated that Sb08g002700 in clade I.1 also evidenced expression in vascular tissue (Fig. 4E13), which indicates the possible functional redundancy between type I and type V nsLTPs.
4.4. Possible retention mechanisms of the OsLTP genes under concerted evolution
Compared with sorghum, the concerted evolution of the genomic segment between rice chromosomes 11 and 12 resulted in the retention of six pairs of tandem nsLTP copies in clade I.1. As previously hypothesized, rounds of inter-locus gene conversion between homeologous chromosome segments might be responsible for this concerted evolution status.32 How to interpret the functional redundancy/divergence and evolutionary mechanisms of these co-evolved genes after duplication remains unanswered.
Gene conversion mediates the unequal transfer of genetic information between highly similar paralogous counterparts, and the key role of gene conversion in maintaining the high level of sequence homogeneity between the tandemly repeated ribosomal RNA (rRNA) genes in eukaryotes has been widely reported.42,43 In the present study, the highly similar sequences between OsLTPs paralogs were observed in both coding and non-coding regulatory regions, which differ in extant according to gene pairs. With a cluster of highly conserved nsLTP genes preserved, we propose that the functional significance of the concerted evolution should be apparent at the expression level. All of the tested OsLTP gene promoters in clade I.1 showed that cuticle-related expression was highly conserved in the young leaves/shoots, wound site, and abscission zone, even though two genes (Os12g02340 and Os11g02389) lost the most expression motifs; however, two sorghum genes (Sb08g002660 and Sb08g002700) lost this expression feature, indicating a more divergent regulatory system. One piece of evidence supporting this notion is that Os11g02350 and Os12g02300 maintained almost identical tissue-specific expression in epidermal cells of the youngest aerial tissue, wound site, ovary, stigma, sillique, and seed, whereas the ortholog of Sb08g002700 was expressed principally in the vascular bundle of the rosette and shoot. Furthermore, the Sb08g002660 did not evidence most profound expression in young growing leaves; instead, extensive activities were in old leaves (Fig. 4B2). These lines of evidence indicate that the gene conversion between nsLTP homeologs in rice favoured the preservation of crucial expression motifs. Several nsLTP promoter deletion analyses have revealed that the proximal promoter region close to the start codon should be essential to drive the epidermis-specific expression.18,44,45 On a genetic basis, gene conversion should help to preserve the sequence of the proximal promoter region under high selective constraints, which only occur between highly similar sequences, and always at >92%.42 This is probably reasonable in light of our result that most orthologous pairs could retain high sequence similarity at the proximal promoter region at different extents, resulting in slow evolution with a relatively non-divergent tissue-specific expression of OsLTPs in clade I.1. Thus, the sequence homogenization process driving concerted evolution may preserve sequence similarity by removing deleterious mutations that slow the evolution of these genes and the crucial expression pattern.
However, the expression preservation effect of concerted evolution could not explain all of our data. Indeed, the sequence homogenization induced by illegitimate recombination again results in redundancy in gene pairs, which may lead to relatively rapid mutation accumulation. The rapid evolution of this gene cluster could be explained by classical evolutionary mechanisms and was supported by the results of genetic-level studies.1,3 Although several expression motifs were well conserved in our promoter–reporter assay, large dynamics were also observed, including the fact that the expression in the anther, ovary, silique, and seed were frequently lost by individual genes in clade I.1 (Fig. 4), which implies a regulatory subfunctionalization process. Unlike the proximal promoter region essential to drive the conserved expression, the further upstream sequences were under relatively relaxing selection constraints, wherein other cis-motifs might be localized to affect the expression at different organs. This hypothesis is further supported by our RT–PCR analysis against abiotic stresses. When compared the sorghum orthologs, the rice homeologous gene pairs in clade I.1 extensively lost inducibility under drought, salt, and cold stresses. Although a total of 13 OsLTP genes and 5 SbLTP genes were preserved in clade I.1, the numbers of highly expressed genes and genes in response to drought, salt, and cold were comparable between rice and sorghum. Therefore, we suggest that the preservation of greater numbers of nsLTP genes in rice by concerted evolution also evidenced functional redundancy, and might have resulted in extensive regulatory subfunctionalization under relatively relaxed selection pressures.
5. Conclusions
The diversity of 9 kDa nsLTPs appear to have undergone long-term evolution in land plants and expanded in angiosperms after diving into different groups with fixed cysteine positions. In rice and sorghum, most nsLTP genes of type I evidenced a conserved expression feature related to the epidermal cells of young aerial growing tissues, supporting the notion of functional involvement in cuticle synthesis or plant defence against phytopathogens. Broad expression of this type was also observed frequently in the stigma, implying functional involvement in pollen–pistil interaction. In clade I.1, a grass-specific lineage, a possible functional shift is proposed in seed development, as strong expression in embryo and endosperm was detected in both rice and sorghum. By way of contrast, a vascular tissue-related expression pattern was primarily observed in type V, with a possible functional involvement in signal transduction. In rice, the major expression of OsLTPs in type I is attributed to the concerted evolution of six duplicated gene pairs on the distal region of rice chromosomes 11 and 12, whereas only five full gene copies were preserved on the orthologs chromosomes in sorghum. The gene conversion event possibly responsible for this concerted evolution status in rice resulted in the preservation of crucial expression under high selection constraints; however, extensive regulatory subfunctionalization followed by functional divergence was suggested to have occurred under relative low selection constraints in members of this gene cluster.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2009-0064150) to CSJ.
Supplementary Material
References
- 1.Kondrashov F.A. Gene dosage and duplication. In: Dittmar K., Liberles D., editors. Evolution after Gene Duplication. Wiley-Blackwell, New Jersey; 2010. pp. 57–64. [Google Scholar]
- 2.Conant G.C., Wolfe K.H. Turning a hobby into a job: how duplicated genes find new functions. Nature Rev. Genet. 2008;9:938–50. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 3.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 4.Vergnolle C., Arondel V., Jolliot A., Kader J.C. Phospholipid transfer proteins from higher plants. Methods Enzymol. 1992;209:522–30. doi: 10.1016/0076-6879(92)09063-9. [DOI] [PubMed] [Google Scholar]
- 5.Kader J.C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:627–54. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho A.D.O., Gomes V.M. Role of plant lipid transfer proteins in plant cell physiology: a concise review. Peptides. 2007;28:1144–53. doi: 10.1016/j.peptides.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Boutrot F., Chantret N., Gautier M.F. Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics. 2008;9:86. doi: 10.1186/1471-2164-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin D.H., Lee J.Y., Hwang K.Y., Kim K.K., Suh S.W. High resolution crystal structure of the non-specific lipid transfer protein from maize seedlings. Structure. 1995;3:189–99. doi: 10.1016/s0969-2126(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 9.Pyee J., Yu H., Kolattukudy P.E. Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 1994;311:460–8. doi: 10.1006/abbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- 10.Lee S.B., Go Y.S., Bae H.J., et al. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen, Alternaria brassicicola. Plant Physiol. 2009;150:42–54. doi: 10.1104/pp.109.137745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sterk P., Booij H., Schellekens G.A., Van Kammen A., De Vries S.C. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 1991;3:907–21. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S.Y., Jauh G.Y., Mollet J.C., et al. A lipid transfer–like protein is necessary for lily pollen tube adhesion to an in vitro Stylar Matrix. Plant Cell. 2000;12:151–64. doi: 10.1105/tpc.12.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae K., Gonong B.J., Kim S.C., et al. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010;61:4277–90. doi: 10.1093/jxb/erq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blein J.P., Coutos-Thevenot P., Marion D., Ponchet M. From elicitins to lipid-transfer proteins: a new insight in cell signaling involved in plant defence mechanisms. Trends Plant Sci. 2002;7:293–6. doi: 10.1016/s1360-1385(02)02284-7. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado A.M., Doerner P., Dixon R.A., Lamb C.J., Cameron R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- 16.Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- 17.Thoma S., Hecht U., Kippers A., Botella J., De Vries S., Somerville C. Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 1994;105:35–45. doi: 10.1104/pp.105.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiderdoni E., Cordero M.J., Vignols F., et al. Inducibility by pathogen attack and developmental regulation of the rice Ltp1 gene. Plant Mol. Bio. 2002;49:683–99. doi: 10.1023/a:1015595100145. [DOI] [PubMed] [Google Scholar]
- 19.Guerche P., Tire C., de Sa F.G., de Clercq A., Van Montagu M., Krebbers E. Differential expression of the Arabidopsis 2S-albumin genes and the effect of increasing gene family size. Plant Cell. 1990;2:469–78. doi: 10.1105/tpc.2.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strobl S., Maskos K., Wiegand G., Huber R., Gomis-Rüth F.X., Glockshuber R. A novel strategy for inhibition of α-amylases: yellow meal worma-amylase in complex with the Ragi bifunctional inhibitor at 2.5 Å resolution. Structure. 1998;6:911–21. doi: 10.1016/s0969-2126(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S., Dudley J., Nei M., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzywinski M., Schein J., Birol I., et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yim W.C., Lee B-M., Jang C.S. Expression diversity and evolutionary dynamics of rice duplicate genes. Mol. Genet. Genomics. 2009;281:483–93. doi: 10.1007/s00438-009-0425-y. [DOI] [PubMed] [Google Scholar]
- 24.Wang H.W., Kwon H.J., Yim W.C., et al. Expressional diversity of wheat nsLTP genes: evidence of subfunctionalization via cis-regulatory divergence. Genetica. 2010;138:843–52. doi: 10.1007/s10709-010-9467-7. [DOI] [PubMed] [Google Scholar]
- 25.Dvořáková L., Cvrckova F., Fischer L. Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genomics. 2007;8:412. doi: 10.1186/1471-2164-8-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautier M.F., Alary R., Joudrier P. Cloning and characterization of a cDNA encoding the wheat (Triticum durum Desf.) CM16 protein. Plant Mol. Biol. 1990;14:313–22. doi: 10.1007/BF00028768. [DOI] [PubMed] [Google Scholar]
- 27.Edstam M.M., Viitanen L., Salminen T.A., Edqvist J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant. 2011;4:947–64. doi: 10.1093/mp/ssr019. [DOI] [PubMed] [Google Scholar]
- 28.Huang M.D., Wei F.J., Wu C.C., Hsing Y.I., Huang A.H. Analyses of advanced rice anther transcriptomes reveal global tapetum secretory functions and potential proteins for lipid exine formation. Plant Physiol. 2008;149:694–707. doi: 10.1104/pp.108.131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominick J., Paolillo D.J., Jr. The swimming sperms of land plants. BioScience. 1981;31:367–73. [Google Scholar]
- 30.Jeffree C.E. The fine structure of the plant cuticle. In: Riederer M., Müller C., editors. Annual Plant Reviews, Vol 23: Biology of the Plant Cuticle. Oxford: Blackwell; 2006. pp. 11–110. [Google Scholar]
- 31.Liu W.F., Huang D.W., Liu K., Hu S.N., Yu J., Gao G. Discovery, identification and comparative analysis of non-specific lipid transfer protein (nsLtp) family in Solanaceae. Geno. Proteo. Bioinfo. 2010;8:229–37. doi: 10.1016/S1672-0229(10)60024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson A.H., Bowers J.E., Bruggmann R., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–6. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 33.The Rice Chromosomes 11 and 12 Sequencing Consortia. The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3:20. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Tang H., Bowers J.E., Paterson A.H. Comparative inference of illegitimate recombination between rice and sorghum duplicated genes produced by polyploidization. Genome Res. 2009;19:1026–32. doi: 10.1101/gr.087288.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang C.S., Jung J.H., Yim W.C., Lee B.M., Seo Y.W., Kim W. Divergence of genes encoding non-specific lipid transfer proteins in the Poaceae family. Mol. Cells. 2007;24:215–23. [PubMed] [Google Scholar]
- 36.Tanaka H., Watanabe M., Watanabe D., Tanaka T., Machida C., Machida Y. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development. 2001;128:4681–9. doi: 10.1242/dev.128.23.4681. [DOI] [PubMed] [Google Scholar]
- 37.Jeroen N., Feron R., Huisman B.A.H., Fasolino A., Hilbers C.W., Derksen J. Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell. 2005;17:2009–19. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutrot F., Meynard D., Guiderdoni E., Joudrier P., Gautier M.F. The Triticum aestivum non-specific lipid transfer protein (TaLtp) gene family: comparative promoter activity of six TaLtp genes in transgenic rice. Planta. 2007;225:843–62. doi: 10.1007/s00425-006-0397-7. [DOI] [PubMed] [Google Scholar]
- 39.Eklund D.M., Edqvist J. Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiol. 2003;132:1249–59. doi: 10.1104/pp.103.020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jose-Estanyol M., Gomis-Ruth F.X., Puigdomenech P. The eight cysteine motif, a versatile structure in plant proteins. Plant. Physiol. Biochem. 2004;42:355–65. doi: 10.1016/j.plaphy.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Motose H., Sugiyama M., Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–8. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- 42.Chen J.M., Cooper D.N., Chuzhanova N., Ferec C., Patrinos G.P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–75. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 43.Eickbush T.H., Eickbush D.G. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–85. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H.C., Creech R.G., Jenkins J.N., Ma D.P. Cloning and promoter analysis of the cotton lipid transfer protein gene Lpt3. Biochim. Biophys. Acta. 2000;1487:106–11. [PubMed] [Google Scholar]
- 45.Canevascini S., Caderas D., Mandel T., Fleming A.J., Dupuis I., Kuhlemeier C. Tissue-specific expression and promoter analysis of the tobacco ltp1 gene. Plant Physiol. 1996;112:513–24. doi: 10.1104/pp.112.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.