Abstract
Semi-mangroves form a group of transitional species between glycophytes and halophytes, and hold unique potential for learning molecular mechanisms underlying plant salt tolerance. Millettia pinnata is a semi-mangrove plant that can survive a wide range of saline conditions in the absence of specialized morphological and physiological traits. By employing the Illumina sequencing platform, we generated ∼192 million short reads from four cDNA libraries of M. pinnata and processed them into 108 598 unisequences with a high depth of coverage. The mean length and total length of these unisequences were 606 bp and 65.8 Mb, respectively. A total of 54 596 (50.3%) unisequences were assigned Nr annotations. Functional classification revealed the involvement of unisequences in various biological processes related to metabolism and environmental adaptation. We identified 23 815 candidate salt-responsive genes with significantly differential expression under seawater and freshwater treatments. Based on the reverse transcription–polymerase chain reaction (RT–PCR) and real-time PCR analyses, we verified the changes in expression levels for a number of candidate genes. The functional enrichment analyses for the candidate genes showed tissue-specific patterns of transcriptome remodelling upon salt stress in the roots and the leaves. The transcriptome of M. pinnata will provide valuable gene resources for future application in crop improvement. In addition, this study sets a good example for large-scale identification of salt-responsive genes in non-model organisms using the sequencing-based approach.
Keywords: transcriptome characterization, Illumina sequencing, salt-responsive genes
1. Introduction
High salinity is one of the most severe environmental stresses causing crop yield reduction. The ability of plants to develop salt tolerance is a trait governed by multigenes and is a determining factor in their growth and productivity.1 Among the intertwining physiological and molecular mechanisms, the regulation of gene expression plays a pivotal role in plant salt tolerance,2,3 and it has therefore stimulated increasing efforts on global expression profiling for various plant species over the past decade.
The early insights into transcriptome responses were primarily obtained from the glycophytic model plants and major crops such as Arabidopsis,4,5 rice,6,7 and maize,8,9 in which hundreds of potential salt-responsive genes were identified and analysed to determine their expression patterns under salt treatments. The studies on glycophytes also revealed the involvement of several signal transduction pathways in salt stress response, including the salt overly sensitive (SOS) pathway, the ABA signalling pathway, the ethylene signalling pathway, and the Ca2+-dependent signalling pathway.10–13 These pathways constitute a dynamic network and work collectively to combat ionic disequilibrium, osmotic imbalance, and other deleterious effects of salt stress. However, most glycophytes can only endure relatively low concentrations of salt. For example, Arabidopsis cannot survive NaCl concentrations higher than 200 mM.14 Therefore, the transcriptome analyses of these glycophytes would miss the information about some key genes or pathways that are related to salt tolerance.
To broaden the knowledge on the regulatory network for plant salt tolerance, the focus of recent transcriptomic research has been gradually shifting from the salt-sensitive glycophytes to the salt-tolerant halophytes. Salt cress (Thellungiella halophila) is the most commonly surveyed halophyte because its high nucleotide sequence identity with Arabidopsis cDNA facilitates the use of molecular and genetic tools that are available for Arabidopsis. Comparisons of the expression profiles between these two species demonstrated that they possessed both shared and divergent responses to salt stress, and the constitutive overexpression of many genes that were stress-inducible in Arabidopsis might contribute to the stress tolerance of salt cress.15,16 These findings are partly in accordance with the hypothesis that salt tolerance mechanisms are largely conserved in halophytes and glycophytes, and that the large variations in their tolerance arise from subtle differences in the regulation of the same basic set of genes.17 In other words, the formation of salt tolerance is a cumulative process with stepwise changes in gene regulation. Therefore, the salt-induced response of the transitional species between glycophytes and halophytes may provide new insights into the acquisition of salt tolerance. Nevertheless, the transcriptomic profiles of such transitional species are still not documented.
Millettia pinnata is a semi-mangrove (also called ‘mangrove associate’) that can grow in either freshwater or seawater habitats. A recent study regarding the differentiation between true mangroves and semi-mangroves based on their leaf traits and salt contents proposed M. pinnata to be a glycophyte with moderate salt tolerance.18 Unlike other mangrove species, M. pinnata does not exclude salt via ultra-filtration system at its root surface, excrete salt through glands on its leaf surface, or develop succulence leaf. However, this plant can still endure salinity up to 3%.19,20 In the absence of specialized morphological and physiological traits, the adaptation of M. pinnata may be more exclusively attributed to the alternation in gene regulation, which makes M. pinnata extremely attractive for investigating on its transcriptomic adjustments under salt stress.
As an outbreeding diploid (2n = 2x = 22) legume, M. pinnata has high levels of seed oil and has received much attention on its applications in biodiesel.21–23 In addition, its biomedical properties have also been investigated because it was traditionally known as a medicinal plant.24,25 However, the genetic background of this non-model species remains largely unknown. To date, only a few genes, such as the phytochrome A (PHYA) gene, the maturase (matR) gene, and the ribosomal RNA gene, have been sequenced from M. pinnata, and none of these genes have been shown to be directly responsible for its salt tolerance.26–28 This situation might now be changed with the introduction of next-generation massively parallel sequencing technologies developed by Roche/454 Life Sciences and Illumina.29,30 These sequencing-based approaches do not rely on the prior knowledge of genomic sequences, hence they are particularly useful in quantifying transcriptomes for non-model organisms.31
In this study, we initiated a comprehensive transcriptome analysis of M. pinnata based on the Illumina sequencing platform. Our goals were to (i) set up a data set with abundant transcript sequences from the root and leaf tissues of M. pinnata, (ii) screen out genes with differential expression under seawater and freshwater treatments as candidate salt-responsive genes, and (iii) generalize the common and tissue-specific patterns of transcriptomic responses to salt stress in M. pinnata. The characterization of the transcriptome changes in this transitional species may provide useful complements to the understandings of molecular mechanisms underlying plant salt tolerance. The candidate genes identified in this study can be applied to molecular breeding for plants with enhanced salt tolerance.
2. Materials and methods
2.1. Plant growth and salt treatments
The seeds of M. pinnata were obtained from the Dongzhaigang National Nature Reserve in Hainan, China. The field-collected seeds were surface-sterilized with a 75% ethanol solution, rinsed with sterilized distilled water, and sown in sterilized cover soil for germination. The seedlings were grown in plastic pots under a 12/12 h photoperiod at 25°C (day) and 18°C (night). The volume of each plant pot is ∼800 ml. For the salt treatment, a group of 3-week-old plants were watered by seawater with a salinity of 3% (w/v, ∼500 mM NaCl). The treatments were initiated at 8:00 a.m., at which time each individual plant was watered with 300 ml of seawater to field capacity. Meanwhile, a group of control plants were watered with an equal amount of freshwater (distilled water).
2.2. Sample collection and RNA preparation
The roots and leaves were sampled at 2, 4, and 8 h after the application of seawater or freshwater. At each time point, the primary root with some lateral roots and three leaves at the shoot apex were simultaneously collected from each individual plant, and were separately frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. The total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer's instructions. The quality of RNAs was determined with a 2100 Bioanalyzer RNA Nanochip (Agilent Technologies). For each sample, at least 20 µg of total RNAs was sent to the Beijing Genomics Institute for Illumina sequencing (commercial service). For the reverse transcription–polymerase chain reaction (RT–PCR) and real-time PCR experiments, the total RNAs were prepared following the aforementioned procedures.
2.3. Sequencing and assembly
For a given tissue, equal amounts of total RNAs from the plants treated for 2, 4, and 8 h were pooled in a combined sample. Totally, four combined samples were collected and denoted as MpRs, MpRf, MpLs, and MpLf according to the tissues (Root or Leaf) and the treatments (seawater or freshwater) of their sampling sources. For each combined sample, mRNAs were purified using oligo(dT)-attached beads and fragmented into small pieces (100–400 bp). The cleaved RNA fragments were then primed with random hexamers and submitted to the synthesis of the first-strand and second-strand cDNAs. The synthesized cDNAs were ligated with paired-end adaptors. Then, the cDNA fragments with 200 bp (±20 bp) size were selected by agarose gel electrophoresis and enriched by PCR amplification. Finally, four cDNA libraries were constructed for sequencing on an Illumina GA IIx. All sequence data have been deposited in the Short Read Archive (SRA) at the NCBI database under the project accession number SRA046342.1.
After sequencing, the raw sequence data were first purified by trimming adapter sequences and removing low-quality sequences. The resulting clean reads were assembled using the SOAPdenovo program (version 1.04; http://soap.genomics.org.cn/soapdenovo.html) with K-mer size set to 29 bp.32 The clean reads were split into smaller pieces, the K-mers, and were conjoined into contigs using the de Bruijn graph. Next, the contigs from the same transcript were identified by paired-end reads and were connected into scaffolds. Then the gap fillings were carried out using the pair-end information to retrieve read pairs with one read well aligned on the contigs and another read located in the gap region. The resulting scaffolds with least Ns were defined as unigenes. The unigenes assembled by short reads from four samples were further clustered into all-unigenes in a comprehensive transcriptome using TGI Clustering Tool (TGICL) with the parameters of 50 bp overlap and a minimum of 90% identity.33
2.4. Data analysis
The sequence orientations of the all-unigenes were determined by BLASTx against the NCBI non-redundant (Nr) protein database, the Swiss-Prot protein database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, and the Cluster of Orthologous Groups (COG) database. The incongruent results from different databases were settled under a priority order of Nr, Swiss-Prot, KEGG, and COG. For the rest all-unigenes that were unaligned to the above databases, ESTScan was used to predict their sequence orientations.34
For assignments of gene descriptions, the all-unigenes were searched against the Nr database using BLASTx with an E-value cut-off of 1.0e–5. The BLAST results were parsed by a Perl script (available on request). The results of only the best hit were extracted. In practice, the E-values for more than 85% of the annotated sequences were ≤1.0e–10. Based on their Nr annotations, the all-unigenes were assigned GO annotations using Blast2GO (http://www.blast2go.rog/),35 followed by functional classification using the WEGO software.36 Besides, the putative metabolic pathways for the all-unigenes were assigned by performing BLASTx against the KEGG pathway database with the E-value cut-off of 1.0e–5.
The RPKM (reads per kilobase per million reads) values were applied to measure the gene expression levels.37 For a given all-unigene, four RPKM values were generated by mapping the reads from the four subtranscriptomes to it using SOAP2 with a maximum of three mismatches.38 Only the reads mapped to a single all-unigene were used to estimate the expression level. The differentially expressed genes (DEGs) between the seawater- and freshwater-treated samples were identified using an R package named ‘DEGseq’.39 The GO enrichment analysis and KEGG pathway enrichment analysis for the DEGs were both performed by conducting hypergeometric tests with the whole M. pinnata transcriptome set as the background.
2.5. RT–PCR and real-time PCR analyses
We conducted RT–PCR for 100 DEGs using the M. pinnata actin and 18S rRNA genes as the controls. The first-strand cDNAs were synthesized from 1 µg of total RNAs using the PrimeScript® 1st Strand cDNA Synthesis Kit (TAKARA). The gene-specific primers were designed based on the sequencing results using the Primer Premier software (version 6.0). Then, the 20 µl of PCR samples containing 1 µl of first-strand cDNAs and 1 µl of primers were subjected to 35 cycles of 30 s denaturing at 94°C, 30 s annealing at the temperature that was altered for different DEGs, and 30 s extending at 72°C. The PCR products were electrophoresed on a 1% agarose gel.
The real-time PCR was performed on an ABI PRISM 7300 Sequence Detection System (Applied Biosystems) using 1 µl of first-strand cDNAs and SYBR® Premix Ex Taq™ (TAKARA). The thermal cycling conditions were 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 31 s at 60°C. All of the reactions were performed in triplicate. The relative expression levels for each gene were calculated using the 2−ΔΔCT method with normalization to the internal control gene. The LSD-t test of one-way analysis of variance was performed to determine the significant differences in expression levels between the treated samples at 1, 2, 4, and 8 h and those at 0 h using the SPSS software package (version 13.0).
3. Results
3.1. Illumina sequencing and de novo assembly
Our preliminary experiments showed that the young leaves of the M. pinnata plants treated with seawater began to wilt at 2 h, wilted to the greatest extent at 4 h, and almost completely recovered by 8 h after the treatments. Meanwhile, the control plants showed no symptoms during the course of the freshwater treatments. This result indicates that M. pinnata may suffer severe salt stress at 4 h, but it may overcome this stress by 8 h. We performed high-throughput sequencing of four samples, i.e. MpRs and MpRf from the roots, MpLs and MpLf from the leaves. For a broad survey of the salt-responsive genes, a combined cDNA library was constructed for each sample, which was represented by a transcript mixture with equal amounts of total RNA from 2, 4, and 8 h-treated plants. The Illumina sequencing was then performed separately for four cDNA libraries and generated four sub-transcriptomes with 75-bp raw reads. After filtration of low-quality and adapter sequences, nearly 192 million clean reads were remained for the four sub-transcriptomes. The percentage of Q20 bases for the clean reads in the four sub-transcriptomes were all ∼93% (Supplementary Table S1). In total, these clean reads constitute ∼12 GB of sequence data.
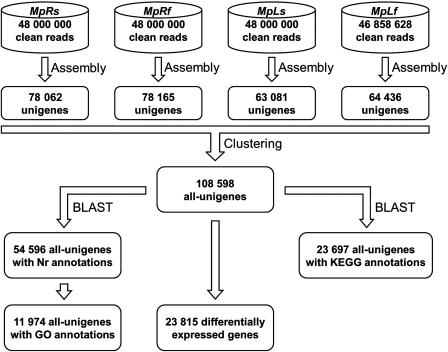
The clean reads from the MpRs, MpRf, MpLs, and MpLf sub-transcriptomes were assembled into 248 011, 238 722, 194 806, and 194 020 contigs, respectively (Supplementary Table S2). The average contig sizes of the four sub-transcriptomes were all above 200 bp. The following step used the paired-end reads to identify the contigs from the same transcript, and these contigs were merged into 130 027, 127 341, 101 737, and 104 087 scaffolds with an average size over 350 bp (Supplementary Table S2). The scaffolds were further processed by a gap-filling step, which resulted in 78 062, 78 165, 63 081, and 64 436 unigenes for MpRs, MpRf, MpLs, and MpLf, respectively (Fig. 1). The numbers of unigenes obtained from the root samples were larger than those from the leaf samples, whereas the mean lengths of the unigenes were longer in the latter samples than those in the former ones (Table 1).
Figure 1.
Flowcharts of the transcriptome analyses for M. pinnata. The steps include sequence assembly and clustering, Nr annotation, GO annotation, KEGG analysis, and identification of DEGs as candidate salt-responsive genes.
Table 1.
Statistics for the unigenes of M. pinnata
| Number of unigenes |
|||||
|---|---|---|---|---|---|
| Length of unigenes (bp) | MpRs | MpRf | MpLs | MpLf | All |
| 200–500 | 54 617 | 53 061 | 41 787 | 41 700 | 70 455 |
| 500–1000 | 16 231 | 16 819 | 13 930 | 14 539 | 21 028 |
| 1000–1500 | 4403 | 4930 | 4348 | 4604 | 8184 |
| 1500–2000 | 1677 | 1914 | 1800 | 2036 | 4356 |
| ≥2000 | 1134 | 1441 | 1216 | 1557 | 4575 |
| Total | 78 062 | 78 165 | 63 081 | 64 436 | 108 598 |
| Mean length (bp) | 497 | 521 | 537 | 558 | 606 |
| N50 (bp) | 589 | 634 | 674 | 708 | 887 |
| Total length (bp) | 38 794 023 | 40 744 474 | 33 850 579 | 35 949 682 | 65 814 388 |
MpRs, MpRf, MpLs, and MpLf represent four sub-transcriptomes that were derived from two tissues (Root or Leaf) under two treatments (seawater or freshwater). The last column presents the statistics for the all-unigenes clustered by the unigenes from four sub-transcriptomes.
Under the clustering criteria with a minimum of 50-bp overlap and 90% identity, the unigenes from all four sub-transcriptomes were further clustered into 108 598 all-unigenes to form a comprehensive transcriptome of M. pinnata (Fig. 1). The gene number showed a substantial increase from the sub-transcriptome to the comprehensive transcriptome, and the increase was more prominent for the genes with larger sizes (Table 1). The length of the all-unigenes ranged from 200 to 10 843 bp, with the mean length and the N50 value rises to 606 and 887 bp, respectively. As a result of the increase in both gene number and gene length, the total length of the all-unigenes extended to 65.8 Mb after clustering (Table 1). A total of 49 836 (45.9%) all-unigenes were clustered by at least two unigenes, with a maximum of 31 unigenes per all-unigenes, whereas the rest 58 762 (54.1%) were corresponding to single unigene from any of the four sub-transcriptomes. The average sequencing depth calculated by realigning all usable sequencing reads to the all-unigenes were ∼30-folds for each sample. The quality of these all-unigenes was evaluated by two analyses: (i) the random distribution of reads in the all-unigenes showed that the all-unigenes were evenly covered by the reads from each sub-transcriptome with relatively fewer reads in the 3′ ends of them (Supplementary Fig. S1), and (ii) the gap distribution of the all-unigenes indicated that the majority of them displayed a ratio of gap length to gene length <5% (Supplementary Fig. S2).
3.2. Gene annotation and functional classification
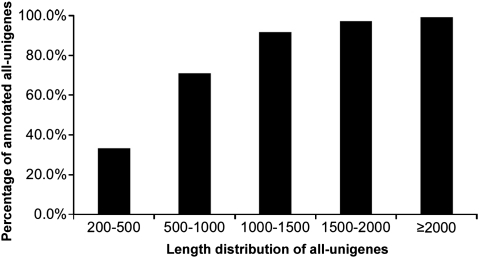
First, the all-unigenes of M. pinnata were assigned putative gene descriptions based on the BLAST search against the NCBI non-redundant (Nr) protein database. Out of the 108 598 all-unigenes, 54 596 (50.3%) showing significant similarity with proteins in the Nr database were assigned Nr annotations (Fig. 1). Overall, these all-unigenes matched 24 281 unique protein accessions, which indicated that our sequencing project had generated a substantial number of genes for M. pinnata. Only 33.2% of the all-unigenes with a length <500 bp had BLAST hits in the Nr database. The proportion of all-unigenes with BLAST hits increased markedly for those with larger sizes (Fig. 2). It seemed that longer all-unigenes were more likely to have Nr annotations. The E-value distribution of the top hits in the Nr database also revealed that a larger proportion of all-unigenes longer than 500 bp had strong homology (E-value ≤1.0e–50) (Supplementary Fig. S3).
Figure 2.
Length distribution of the all-unigenes with Nr annotations. In this study, only 33% of the all-unigenes shorter than 500 bp had BLAST hits in the Nr database, whereas more than 80% of the all-unigenes over 500 bp did.
The majority of these annotated all-unigenes (54 266 out of 54 596) displayed the highest homology to genes from plants (Table 2). A quarter of them retrieved an annotation from Arabidopsis, partly due to the fact that the Arabidopsis genome annotation is the most advanced and complete for any higher plant to date in the Nr database. Besides, there were also a quarter of annotated all-unigenes with matched accessions from its legume relatives (Table 2). The remaining <1% of annotated all-unigenes were annotated with sequences from the non-plant sources. Interestingly, we found 12 all-unigenes with an Nr annotation from Phytophthora sojae, which is a plant pathogen that causes soybean stem and root rot.40 Accordingly, we verified that all of these 12 all-unigenes were clustered by unigenes from the root-derived sub-transcriptome. This observation demonstrated a good representation of the comprehensive transcriptome in which even trivial amounts of pathogen transcripts were detected.
Table 2.
Annotation sources for the all-unigenes of M. pinnata
| Annotation sources | Number of matched all-unigenes |
|---|---|
| Ten species with the most abundant matches | |
| Arabidopsis thaliana | 15 308 |
| Oryza sativa | 5739 |
| Glycine max | 5198 |
| Arabidopsis lyrata | 4551 |
| Medicago truncatula | 4414 |
| Populus trichocarpa | 3463 |
| Vitis vinifera | 2491 |
| Zea mays | 1067 |
| Lotus japonicus | 705 |
| Ricinus communis | 685 |
| Ten legume genus with the most abundant matches | |
| Glycine | 5220 |
| Medicago | 4722 |
| Lotus | 722 |
| Pisum | 683 |
| Phaseolus | 550 |
| Vigna | 312 |
| Cicer | 197 |
| Trifolium | 157 |
| Arachis | 152 |
| Vicia | 108 |
| nr_plants | 54 266 |
| nr_nonplants | 330 |
‘nr_plants’ and ‘nr_nonplants’ indicate the annotation sources from the NCBI non-redundant sequences for plants and non-plant species, respectively.
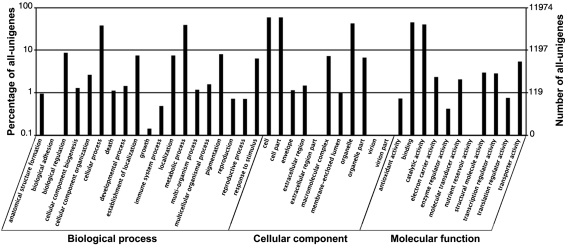
Following their Nr annotations, we mapped the all-unigenes into the records of the GO database and retrieved GO annotations for 11 974 of them (Fig. 1). These all-unigenes were assigned GO terms from the three main categories, including 6407 all-unigenes with terms from ‘Biological process’, 7095 all-unigenes with terms from ‘Cellular component’, and 8299 all-unigenes with terms from ‘Molecular function’ (Fig. 3). Among them, 3228 all-unigenes had an assignment in all three categories. The remaining all-unigenes failed to obtain a GO term, largely due to their uninformative (e.g. ‘unknown’, ‘putative’, or ‘hypothetical’ protein) descriptions. Within the ‘Biological process’ category, the two most abundantly represented lineages were ‘metabolic process’ and ‘cellular process’. There were also a large number of genes involved in ‘biological regulation’, ‘pigmentation’, ‘localization’, and ‘response to stimulus’. In the ‘Molecular function’ category, ‘binding’ and ‘catalytic activity’ were predominantly represented. The former was mainly represented by genes for ‘nucleotide binding’ and ‘protein binding’, while the latter was mainly represented by genes with ‘transferase activity’, ‘hydrolase activity’, and ‘kinase activity’. In the ‘Cellular component’ category, most of all-unigenes were located in ‘cell part’. In contrast, rare all-unigenes were sorted into ‘extracellular region part’ or ‘virion part’.
Figure 3.
GO classification of M. pinnata transcriptome. The results are summarized in three main categories as follows: biological process, cellular component, and molecular function. In total, 11 974 all-unigenes have been assigned GO terms. In some cases, one all-unigene has multiple terms.
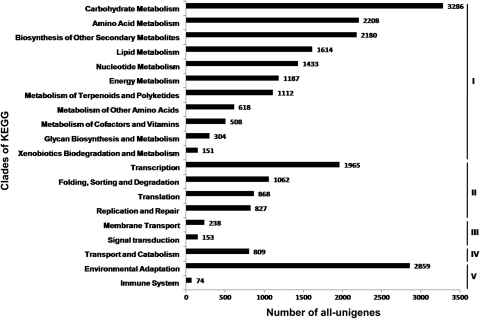
Finally, we performed the KEGG pathway analysis to assign biological pathways to the all-unigenes. In total, 23 697 out of 108 598 (21.8%) all-unigenes were assigned 124 KEGG pathways (Fig. 1). These pathways belonged to 20 clades under five major KEGG categories, including ‘Metabolism’, ‘Genetic information processing’, ‘Environmental information processing’, ‘Cellular processes’, and ‘Organismal systems’ (Fig. 4). Among them, ‘plant–pathogen interaction’, ‘spliceosome’, ‘biosynthesis of phenylpropanoids’, ‘biosynthesis of plant hormones’, and ‘ribosome’ were the top five pathways most represented by the all-unigenes. These results suggested that the living of M. pinnata was featured by dynamic metabolism as well as active environmental adaptation.
Figure 4.
Number of all-unigenes in each clade of the KEGG pathway maps. The all-unigenes were assigned 124 KEGG pathways within 20 clades under five major categories: Metabolism (I), Genetic information processing (II), Environmental information processing (III), Cellular processes (IV), and Organismal systems (V). In each category, the clades are listed according to their abundancies of all-unigenes.
3.3. Identification and functional classification of DEGs
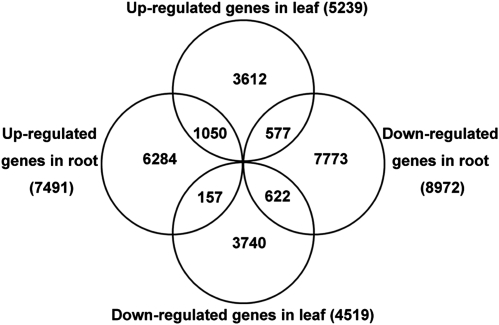
We singled out the DEGs with two criteria for their expression profiles: (i) the average fold change in gene expression level was more than or equal to 2-fold between the seawater- and freshwater-treated samples and (ii) the false discovery rate of the single-sample t-test was <0.1%. Under these criteria, 23 815 out of 108 598 (21.9%) all-unigenes were identified to be differentially expressed in at least one tissue following the salt treatment (Supplementary Table S3; Fig. 1). In this study, DEGs with higher expression levels in seawater-treated samples when compared with freshwater-treated samples were denoted as ‘up-regulated’, while those with lower expression levels in seawater-treated samples were denoted as ‘down-regulated’. Overall, there were much more DEGs in the roots than those in the leaves (Fig. 5). In the root, the number of up-regulated DEGs was less than that of down-regulated DEGs. The situation was reversed in the leaf, in which more up-regulated DEGs than down-regulated DEGs were identified. Only a small portion of DEGs (1050 up-regulated and 622 down-regulated) shared common tendency of expression changes between the roots and the leaves. A still less of DEGs showed a completely opposite tendency of expression changes between the two tissues (577 DEGs up-regulated in the leaf but down-regulated in the root, and 157 DEGs down-regulated in the leaf but up-regulated in the root). The remaining majority of DEGs were exclusively up-regulated or down-regulated in either tissue.
Figure 5.
Number of DEGs in the root and the leaf of M. pinnata. The numbers of DEGs that were exclusively up- or down-regulated in one tissue are shown in each circle. The numbers of DEGs with a common or opposite tendency of expression changes between the two tissues are shown in the overlapping regions. The total numbers of up- or down-regulated genes in each tissue are shown outside the circles.
To further characterize the expression changes in these two tissues, we conducted GO enrichment analysis for the DEGs with the whole transcriptome set as the background. In the root, the significantly overrepresented GO terms of ‘Biological process’ were ‘gene expression’, ‘sulphur metabolic process’, ‘response to oxidative stress’, ‘cellular amino acid derivative metabolic process’, ‘oxidation reduction’, and ‘secondary metabolic process’ (Supplementary Table S4). In the leaf, the significantly overrepresented GO terms of ‘Biological process’ included ‘oxidation reduction’, ‘cellular amino acid derivative metabolic processes’, and ‘cellular aromatic compound metabolic process’. From these results, we noticed that both tissues of M. pinnata tend to mobilize genes related to oxidation reduction and cellular amino acid metabolism. Furthermore, as the salt-induced deleterious effects varied among different parts of a plant, the roots and the leaves also activated some distinct groups of genes (i.e. genes in ‘translation’, ‘sulphur metabolic process’, and ‘response to oxidative stress’ for the root, genes in ‘phenylpropanoid metabolic process’ for the leaf) to overcome them.
The KEGG pathway enrichment analysis for the DEGs also revealed both common and tissue-specific patterns of overrepresentations. The DEGs were enriched in 37 pathways in the root and 31 pathways in the leaf (Supplementary Table S5). Among them, 19 pathways appeared in both tissues and were mostly related to amino acid metabolism, lipid metabolism, energy metabolism, biosynthesis of secondary metabolites, and environmental adaptation. Generally speaking, this finding demonstrated that both tissues of M. pinnata demand more energy and more biological substances to cope with salt stress. In addition to these common pathways, there were some tissue-specific pathways with overrepresented DEGs. For example, in the clade of ‘Energy metabolism’, the leaves of M. pinnata were enriched with DEGs in ‘oxidative phosphorylation’ and ‘photosynthesis’ pathways, and these processes were also employed by the leaves of other plant species under salt stress.41–43 On the other hand, the roots of M. pinnata were enriched with DEGs in the ‘sulphur metabolism’ pathway, which was likely related to their sulphur-rich living conditions.44
3.4. Experimental verification of DEGs
To assess the reliability of our sequencing-based approach in identifying salt-responsive genes, we monitored the expression of candidate DEGs. In the first place, we performed RT–PCR for 100 randomly chosen candidates with specific primers (Supplementary Table S6). We compared gene expression levels at the three time points separately between stressed and control samples. The expression profiles of 70 candidates were basically in agreement with the predictions from the Illumina sequencing results. Six candidates displayed incongruent expression profiles with the predictions. The remaining 24 candidates showed non-specifically amplified products or no products. In other words, 70–92% of the candidates were confirmed by RT–PCR analyses. The confirmed candidates were differentially expressed in at least one of the three time points (Supplementary Fig. S4). In the root, the maximal number of confirmed DEGs occurred at the 4 h time point (Supplementary Table S7). Conversely, the minimal number of confirmed DEGs in the leaf occurred at the 4 h time point, whereas the maximum occurred at the 8 h time point. From these results, we can infer a delay of transcriptional response to salt stress in the leaf, which may be explained by a later salt accumulation in this tissue.9
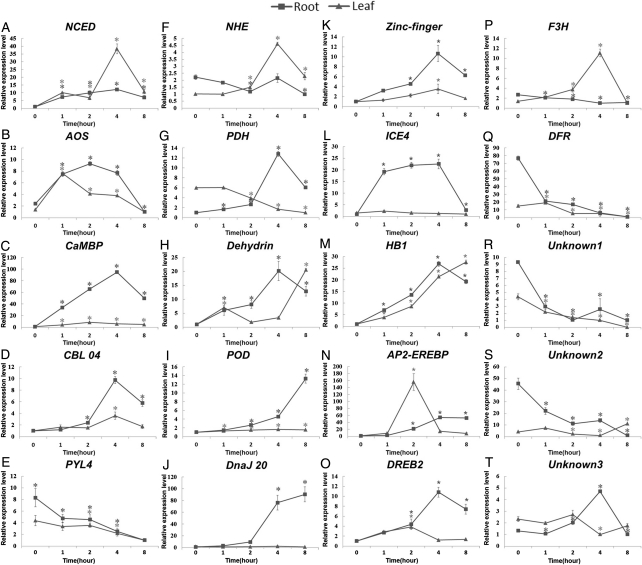
In the second place, we employed real-time PCR to delineate the detailed expression profiles of 20 candidate DEGs. These candidates mainly included genes that were reported to be related to salt stress response in other plant species, such as genes involved in several signal transduction pathways, genes associated with osmotic adjustments or detoxification of reactive oxygen species (ROS), and genes encoding transcription factors (Supplementary Table S8). Besides, there were three candidates with unknown functions, which represented a new resource for gene transfer from M. pinnata to its legume relatives or other crops.
All of these 20 candidates showed significantly differential expression with the original expression level in at least one time point following the seawater treatments (Fig. 6). In other words, all candidates displayed an expression pattern roughly consistent with the prediction based on the Illumina sequencing results. In most cases, the expression changes were more dramatic in the roots than in the leaves. Besides, most of the candidates were up-regulated from the 0 to 4 h time point, and then they gradually restored to the original expression levels. The pace of the rise and fall in gene expression levels was roughly coincident with the wilting and recovering of the young leaves. This phenomenon was also observed in other mangroves,42 and could be explained by referring to the different phases of salt stress response.43 The up-regulation and restoration of the DEGs in both tissues may parallel the phases of ‘salt accumulation’ and ‘osmotic recovery’, and the delay in gene activation in the leaves may result from an extra phase of ‘dehydration’ preceding the above two phases. The leaves wilted during the ‘dehydration’ phase, and then revived gradually during the following two phases. In this sense, these results again demonstrated that a later salt accumulation was responsible for the delay in the transcriptional response in the leaves.
Figure 6.
Relative expression levels of 20 DEGs at a series of time points following the seawater treatments. The relative gene expression levels as expressed by 2−ΔΔCT were determined separately for each tissue and were presented as the mean ± SD. Significant differences (P < 0.01, LSD-t test) between the expression levels at 1, 2, 4, and 8 h time points and those at 0 h are indicated by asterisks.
4. Discussion
Mangroves are a group of plant species that live along the sea coast with the ability to tolerate high salinity.44 The extremely high salinity within their living conditions confers mangroves unique resources for exploiting the dynamic nature of their gene regulation during salt exposure. While the collections of ESTs based on cDNA libraries from several mangrove species have been described,45–47 the transcript data for such a plant group containing diverse species are still scarce. In a recent study using the 454 pyrosequencing platform, 67 375 and 96 989 unisequences with a mean length of 360 and 433 bp were produced for two taxonomically diverse mangroves.48 Compared with these results, our transcriptome for M. pinnata exceed in both the total number and the mean length of the unisequences. The mean length of the all-unigenes in our transcriptome was also longer than those reported for other plant transcriptomics using either the Illumina sequencing technology49,50 or the 454 pyrosequencing technology.51,52
The enhancement in the gene length of our transcriptome may be attributed to various factors, such as the optimized assembly parameters (e.g. the K-mer size). One of the most important reasons for this enhancement might be that the final comprehensive transcriptome was indeed integrated by four sub-transcriptomes, each of which was comparable in gene number and gene length with those in the previous studies. The final unisequences possessed a relatively high depth of average coverage (∼30-fold) by reads from each sub-transcriptome. We also noticed an increase in the number of unisequences from the sub-transcriptome to the comprehensive transcriptome. The ‘extra’ unisequences represented either splice variants or genes with tissue-specific or stress-specific expression. So far, our transcriptome for M. pinnata has presented many more sequences than the sequencing efforts for any other mangrove species have produced.53
As for the GO and KEGG annotations, the aforementioned two mangrove species displayed remarkable similarities to each other.48 They both showed a lack of representation in the GO category ‘response to stimulus’, whereas the transcriptome of M. pinnata was well represented in this category. We suggest that this discrepancy may be partly attributed to the different status of adaptation for each mangrove species. It has been hypothesized that semi-mangroves represent an intermediate during the long-term evolution of mangroves in adaptation to the high-salinity environments.54 Accordingly, while those two mangroves that were ‘completely’ adapted to the extreme conditions did not require so many transcripts for responding to stimulus, M. pinnata with a ‘half’ tolerance may still be in great need of these transcripts. With the exception of this discrepancy, the functional categories were similar between M. pinnata and those two mangroves. For example, all these three species possessed large quantities of transcripts related to energy metabolism.
Based on the comparison of gene expression levels between stressed and control samples using RPKM values, we screened out 23 815 all-unigenes as candidate salt-responsive genes, which took up ∼22% of the total all-unigenes. This percentage was higher than the percentages of salt-induced DEGs derived from other related species, such as Bruguiera gymnorrhiza (another mangrove) and Medicago truncatula (another legume), using the array-based platform.41,55 First, the differences in DEG percentages might reflect the inherent differences between species in their transcriptome profiles. Secondly, it might reflect a greater sensitivity and higher resolution of the sequencing-based platform in identifying DEGs. The sequencing-based approaches offer several advantages over the existing hybridization-based approaches, which often suffer from background and cross-hybridization problems.56 Thirdly, since each sample for Illumina sequencing was pooled by equal amounts of RNAs from 2, 4, and 8 h-treated plants, the RPKM values in each sub-transcriptome actually reflected the average gene expression levels for the three time points. Theoretically speaking, our methods would miss some genes with 2-fold difference in expression levels between stressed and control samples at less than two time points due to the diluting effect of the pooled samples. Nevertheless, it could help us to detect genes with more dramatic changes (4-fold or greater) in expression levels at even only one time point, which might be more closely related to salt stress response. Lastly, we still should not neglect the possibilities of falsely discovered DEGs through the sequence-based platform. In our study, both RT–PCR and real-time PCR analyses demonstrated a high reliability of the candidate salt-responsive genes identified by sequencing-based approach.
The DEGs identified in M. pinnata were enriched in the pathways related to protein synthesis and biosynthesis of secondary metabolites. A comparative study between glycophyte and halophyte showed that Arabidopsis adopted a global defence strategy that requires bulk protein synthesis, whereas salt cress mainly activated genes that functioned in protein modification and redistribution.16 It seemed that protein modification might be a faster mechanism for stress response than protein synthesis. In this aspect, M. pinnata was more similar to glycophytes. On the other hand, salt cress showed a salt-induced increase in secondary metabolites,16 which were also found to accumulate in M. pinnata but not in Arabidopsis. Taken together, these two results suggest that M. pinnata can utilize the adjustment mechanisms preferentially adopted by either glycophytes or halophytes.
In conclusion, we characterized a comprehensive transcriptome of a semi-mangrove plant M. pinnata using the Illumina sequencing technology. This transcriptome will not only provide plenty of informative data, but also expand the vision of the genetic basis underlying adaptation for the mangrove community. The candidate salt-responsive genes identified in M. pinnata contain both the previously reported salt-responsive genes and some species-specific genes which will be a new resource for molecular breeding in legumes or other crops. The functional enrichment analyses for the candidate genes showed both common and tissue-specific patterns of transcriptome remodelling under salt treatments. Moreover, the experimental analyses for some candidate genes provided some evidences for a delay in transcriptional responses in the leaves. Our work has demonstrated a high reliability of the sequencing-based approach in identifying stress-responsive genes in non-model organisms and established a connection between glycophytes and halophytes in the views of transcriptomic adjustments to salt stress.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the National Basic Research Program of China (2012CB114200), the Startup Project of Marine Biotechnology from Shenzhen University, and the National Transgenic Research Project (2009ZX08004-006B).
Supplementary Material
Acknowledgements
We appreciate Mr Shichuan Li from the Dongzhaigang National Nature Reserve for assistance in collecting the seeds of M. pinnata. We are also grateful to three anonymous editors from the Nature Publishing Group Language Editing for improving the text.
References
- 1.Flowers T.J. Improving crop salt tolerance. J. Exp. Bot. 2004;55:307–19. doi: 10.1093/jxb/erh003. doi:10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- 2.Hasegawa P.M., Bressan R.A., Zhu J.K., Bohnert H. J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–99. doi: 10.1146/annurev.arplant.51.1.463. doi:10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. doi:10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki M., Narusaka M., Ishida J., et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–92. doi: 10.1046/j.1365-313x.2002.01359.x. doi:10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y.Q., Deyholos M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006;6:25. doi: 10.1186/1471-2229-6-25. doi:10.1186/1471-2229-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabbani M.A., Maruyama K., Abe H., et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA get-blot analyses. Plant Physiol. 2003;133:1755–67. doi: 10.1104/pp.103.025742. doi:10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walia H., Wilson C., Zeng L.H., Ismail A.M., Condamine P., Close T.J. Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol. Biol. 2007;63:609–23. doi: 10.1007/s11103-006-9112-0. doi:10.1007/s11103-006-9112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Miyazaki S., Kawai K., Deyholos M., Galbraith D.W., Bohnert H.J. Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol. Biol. 2003;52:873–91. doi: 10.1023/a:1025029026375. doi:10.1023/A:1025029026375. [DOI] [PubMed] [Google Scholar]
- 9.Qing D.J., Lu H.F., Li N., Dong H.T., Dong D.F., Li Y.Z. Comparative profiles of gene expression in leaves and roots of maize seedlings under conditions of salt stress and the removal of salt stress. Plant Cell Physiol. 2009;50:889–903. doi: 10.1093/pcp/pcp038. doi:10.1093/pcp/pcp038. [DOI] [PubMed] [Google Scholar]
- 10.Gong Z.Z., Koiwa H., Cushman M.A., et al. Genes that are uniquely stress regulated in salt overly sensitive (SOS) mutants. Plant Physiol. 2001;126:363–75. doi: 10.1104/pp.126.1.363. doi:10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L.J., Chen X.L., Ren H.Y., et al. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta. 2007;226:815–25. doi: 10.1007/s00425-007-0528-9. doi:10.1007/s00425-007-0528-9. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y.R., Chen S.Y., Zhang J.S. Ethylene signaling regulates salt stress response: an overview. Plant Signal. Behav. 2008;3:761–3. doi: 10.4161/psb.3.10.5934. doi:10.4161/psb.3.10.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romeis T., Ludwig A.A., Martin R., Jones J.D.G. Calcium-dependent protein kinases play an essential role in a plant defence response. Embo J. 2001;20:5556–67. doi: 10.1093/emboj/20.20.5556. doi:10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inan G., Zhang Q., Li P., et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004;135:1718–37. doi: 10.1104/pp.104.041723. doi:10.1104/pp.104.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taji T., Seki M., Satou M., et al. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–709. doi: 10.1104/pp.104.039909. doi:10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Q.Q., Li P.H., Ma S.S., Rupassara S.I., Bohnert H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–39. doi: 10.1111/j.1365-313X.2005.02587.x. doi:10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. doi:10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang L.M., Mu M.R., Li X.F., Lin P., Wang W.Q. Differentiation between true mangroves and mangrove associates based on leaf traits and salt contents. J. Plant Ecol. 2010 doi:10.1093/jpe/rtq1008. [Google Scholar]
- 19.Singh K., Jha M.N. Trees, shrubs and grasses on saline soils of Indo-Gangetic plains. Indian Forester. 1993;119:630–47. [Google Scholar]
- 20.Singh K., Yadav J. Effect of soil salinity and sodicity on seedling growth and mineral compsition of Pongamia pinnata. Indian Forester. 1999;125:618–22. [Google Scholar]
- 21.Karmee S.K., Chadha A. Preparation of biodiesel from crude oil of Pongamia pinnata. Bioresour. Technol. 2005;96:1425–9. doi: 10.1016/j.biortech.2004.12.011. doi:10.1016/j.biortech.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Sharma Y.C., Singh B., Korstad J. High yield and conversion of biodiesel from a nonedible feedstock (Pongamia pinnata) J. Agr. Food Chem. 2010;58:242–7. doi: 10.1021/jf903227e. doi:10.1021/jf903227e. [DOI] [PubMed] [Google Scholar]
- 23.Scott P.T., Pregelj L., Chen N., Hadler J.S., Djordjevic M.A., Gresshoff P.M. Pongamia pinnata: an untapped resource for the biofuels industry of the future. Bioenergy Res. 2008;1:2–11. doi:10.1007/s12155-008-9003-0. [Google Scholar]
- 24.Essa M.M., Subramanian P., Suthakar G., Manivasagam T., Dakshayani K.B. Protective influence of Pongamia pinnata (Karanja) on blood ammonia and urea levels in ammonium chloride-induced hyperammonemia: antihyperammonemic effect of the leaf extract. J. Appl. Biomed. 2005;3:133–8. [Google Scholar]
- 25.Badole S.L., Bodhankar S.L. Investigation of antihyperglycaemic activity of aqueous and petroleum ether extract of stem bark of Pongamia pinnata on serum glucose level in diabetic mice. J. Ethnopharmacol. 2009;123:115–20. doi: 10.1016/j.jep.2009.02.018. doi:10.1016/j.jep.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Lavin M., Eshbaugh E., Hu J.M., Mathews S., Sharrock R.A. Monophyletic subgroups of the tribe Millettieae (Leguminosae) as revealed by phytochrome nucleotide sequence data. Am. J. Bot. 1998;85:412–33. doi:10.2307/2446334. [Google Scholar]
- 27.Hu J.M., Lavin M., Wojciechowski M.F., Sanderson M.J. Phylogenetic analysis of nuclear ribosomal ITS/5.8S sequences in the Tribe Millettieae (Fabaceae): Poecilanthe-Cyclolobium, the core Millettieae, and the Callerya group. Syst. Bot. 2002;27:722–33. [Google Scholar]
- 28.Shi S.H., Huang Y.L., Zeng K., et al. Molecular phylogenetic analysis of mangroves: independent evolutionary origins of vivipary and salt secretion. Mol. Phylogenet. Evol. 2005;34:159–66. doi: 10.1016/j.ympev.2004.09.002. doi:10.1016/j.ympev.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Bennett S.T., Barnes C., Cox A., Davies L., Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–82. doi: 10.1517/14622416.6.4.373. doi:10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 30.Margulies M., Egholm M., Altman W.E., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins L.J., Biggs P.J., Voelckel C., Joly S. An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform. 2008;21:3–14. doi:10.1142/9781848163324_0001. [PubMed] [Google Scholar]
- 32.Li R.Q., Zhu H.M., Ruan J., et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009 doi: 10.1101/gr.097261.109. doi/10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pertea G., Huang X.Q., Liang F., et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–2. doi: 10.1093/bioinformatics/btg034. doi:10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 34.Iseli C., Jongeneel C.V., Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:138–48. [PubMed] [Google Scholar]
- 35.Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. doi:10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 36.Ye J., Fang L., Zheng H., et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–7. doi: 10.1093/nar/gkl031. doi:10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. doi:10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 38.Li R., Yu C., Li Y., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. doi:10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 39.Wang L., Feng Z., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8. doi: 10.1093/bioinformatics/btp612. doi:10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 40.Tyler B.M. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007;8:1–8. doi: 10.1111/j.1364-3703.2006.00373.x. doi:10.1111/j.1364-3703.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 41.Miyama M., Hanagata N. Microarray analysis of 7029 gene expression patterns in burma mangrove under high-salinity stress. Plant Sci. 2007;172:948–57. doi:10.1016/j.plantsci.2007.01.004. [Google Scholar]
- 42.Miyama M., Tada Y. Transcriptional and physiological study of the response of Burma mangrove (Bruguiera gymnorhiza) to salt and osmotic stress. Plant Mol. Biol. 2008;68:119–29. doi: 10.1007/s11103-008-9356-y. doi:10.1007/s11103-008-9356-y. [DOI] [PubMed] [Google Scholar]
- 43.Brinker M., Brosche M., Vinocur B., et al. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 2010;154:1697–709. doi: 10.1104/pp.110.164152. doi:10.1104/pp.110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomlinson P.B. The Botany of Mangroves. Cambridge: Cambridge University Press; 1986. [Google Scholar]
- 45.Mehta P.A., Sivaprakash K., Parani M., Venkataraman G., Parida A.K. Generation and analysis of expressed sequence tags from the salt-tolerant mangrove species Avicennia marina (Forsk) Vierh. Theor. Appl. Genet. 2005;110:416–24. doi: 10.1007/s00122-004-1801-y. doi:10.1007/s00122-004-1801-y. [DOI] [PubMed] [Google Scholar]
- 46.Miyama M., Shimizu H., Sugiyama M., Hanagata N. Sequencing and analysis of 14,842 expressed sequence tags of burma mangrove, Bruguiera gymnorrhiza , Plant Sci. 2006;171:234–41. doi:10.1016/j.plantsci.2006.03.015. [Google Scholar]
- 47.Nguyen P.D., Ho C.L., Harikrishna J.A., Wong M.C.V.L., Rahim R.A. Generation and analysis of expressed sequence tags from the mangrove plant, Acanthus ebracteatus Vahl. Tree Genet. Genomes. 2006;2:196–201. doi:10.1007/s11295-006-0044-2. [Google Scholar]
- 48.Dassanayake M., Haas J.S., Bohnert H.J., Cheeseman J.M. Shedding light on an extremophile lifestyle through transcriptomics. New Phytol. 2009;183:764–75. doi: 10.1111/j.1469-8137.2009.02913.x. doi:10.1111/j.1469-8137.2009.02913.x. [DOI] [PubMed] [Google Scholar]
- 49.Garg R., Patel R.K., Tyagi A.K., Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. doi:10.1093/dnares/dsq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi C.Y., Yang H., Wei C.L., et al. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12:131. doi: 10.1186/1471-2164-12-131. doi:10.1186/1471-2164-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edwards C.E., Parchman T.L., Weekley C.W. Assembly, gene annotation and marker development using 454 floral transcriptome sequences in Ziziphus Celata (Rhamnaceae), a highly endangered, Florida endemic plant. DNA Res. 2011 doi: 10.1093/dnares/dsr037. doi:10.1093/dnares/dsr1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parchman T.L., Geist K.S., Grahnen J.A., Benkman C.W., Buerkle C.A. Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics. 2010;11:180. doi: 10.1186/1471-2164-11-180. doi:10.1186/1471-2164-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dassanayake M., Haas J.S., Bohnert H.J., Cheeseman J.M. Comparative transcriptomics for mangrove species: an expanding resource. Funct. Integr. Genomics. 2010;10:523–32. doi: 10.1007/s10142-009-0156-5. doi:10.1007/s10142-009-0156-5. [DOI] [PubMed] [Google Scholar]
- 54.Liang S., Zhou R.C., Dong S.S., Shi S.H. Adaptation to salinity in mangroves: Implication on the evolution of salt-tolerance. Chinese Sci. Bull. 2008;53:1708–15. doi:10.1007/s11434-008-0221-9. [Google Scholar]
- 55.Li D., Su Z., Dong J., Wang T. An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics. 2009;10:517. doi: 10.1186/1471-2164-10-517. doi:10.1186/1471-2164-10-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. doi:10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.