Abstract
Actin polymerization is typically initiated at specific sites in a cell by membrane-bound protein complexes, and the resulting structures are involved in specialized cellular functions, such as migration, particle uptake, or mitotic division. Here we analyze the potential of the actin system to self-organize into waves that propagate on the planar, substrate-attached membrane of a cell. We show that self-assembly involves the ordered recruitment of proteins from the cytoplasmic pool and relate the organization of actin waves to their capacity for applying force. Three proteins are shown to form distinct three-dimensional patterns in the actin waves. Myosin-IB is enriched at the wave front and close to the plasma membrane, the Arp2/3 complex is distributed throughout the waves, and coronin forms a sloping layer on top of them. CARMIL, a protein that links myosin-IB to the Arp2/3 complex, is also recruited to the waves. Wave formation does not depend on signals transmitted by heterotrimeric G-proteins, nor does their propagation require SCAR, a regulator upstream of the Arp2/3 complex. Propagation of the waves is based on an actin treadmilling mechanism, indicating a program that couples actin assembly to disassembly in a three-dimensional pattern. When waves impinge on the cell perimeter, they push the edge forward; when they reverse direction, the cell border is paralyzed. These data show that force-generating, highly organized supramolecular networks are autonomously formed in live cells from molecular motors and proteins controlling actin polymerization and depolymerization.
Introduction
In connection with specialized areas of the plasma membrane, actin can assemble into structures as different as lamellipodia, filopodia, phagocytic cups, or cleavage furrows. However, the actin system has also a high capacity of self-organization. As a prominent example, we have investigated actin waves that travel freely on the planar, substrate-attached membrane of a Dictyostelium cell (1). The propagating actin structures thus formed are capable of applying force when they reach the cell perimeter; in pushing the border forward, they act as a leading edge.
Different types of wave phenomena in the actin system of motile cells have been reported. Reaction-diffusion waves of actin have been proposed to determine locomotion and shape of fibroblasts, melanoma and Dictyostelium cells (2). The waves formed in Dictyostelium cells under the conditions we have used are nonperiodic, varying in shape, and capable of changing directions (1). Actin-based waves of Hem-1, a constituent of the SCAR/WAVE complex regulating Arp2/3 activity, have been discovered in neutrophils and shown to be directed by chemoattractant toward the front of the cells (3). It is unclear whether these wave phenomena have a common basis. To characterize the three-dimensional organization and dynamics of one type of these waves, we investigated the recruitment of proteins from the cytosol and the pattern they form within a propagating wave. The actin waves addressed in this study are spontaneously generated in migrating cells in association with the Arp2/3 complex (4), but they are more frequently formed under conditions of reduced F-actin content.
Profuse wave formation has been observed in Dictyostelium cells recovering from the inhibition of actin polymerization (1) and in mutant cells defective in a Dictyostelium ortholog of the human lissencephaly gene (5). Mutations in the Dictyostelium gene reduce the F-to-G actin ratio, in addition to impairing the cortical attachment of microtubules. Large wave-like structures are also found in RasG-deficient cells of Dictyostelium (6). A most efficient strategy of increasing the abundance of actin waves in wild-type cells is to depolymerize actin with latrunculin A, a blocker of actin polymerization (1), and to seize a transient state of profuse wave formation during the recovery of actin organization. We have made use of the time window during recovery to determine the dynamics of constituent proteins and their discrete positions within the three-dimensional wave structure. Using dual-emission total internal reflection fluorescence (TIRF) microscopy and confocal spinning-disk microscopy combined with deconvolution, we have localized three actin-binding proteins and an adaptor protein to the waves. These proteins, myosin IB (MyoB), Arp3 as a constituent of the Arp2/3 complex, coronin, and CARMIL, were tagged with green fluorescent protein (GFP) and coexpressed in Dictyostelium cells with red-fluorescent LimEΔ, a construct that served as a reference for filamentous actin structures (4,7).
MyoB is a single-headed motor protein harboring along its tail a site interacting with acidic membrane lipids, a second actin-binding site in addition to its motor domain, and an SH3-domain, which is required for function in vivo (8). The Arp2/3 complex branches actin filaments and is thus responsible for the formation of dendritic actin-filament assemblies (9–12). The adaptor protein CARMIL acts as a bridge between the SH3 domain of MyoB and the Arp2/3 complex (13). Coronins are WD-repeat proteins (14) capable of forming a seven-bladed propeller (15). Both yeast and mammalian type I coronins inhibit the actin-nucleating activity of the Arp2/3 complex, reportedly by binding to its p35 Arc subunit (16,17). Similarly, Dictyostelium coronin favors actin depolymerization in vivo (18), a function involved in cell motility, phagocytosis, and cytokinesis (19,20). This prototypic coronin resembles mammalian members of the coronin-1 subfamily (21).
We show that the formation of actin waves does not require any external signal transmitted through heterotrimeric G-proteins, and their propagation does not depend on SCAR, a regulator upstream of the Arp2/3 complex. Expansion and retraction of the waves is independent of the conventional, double-headed myosin-II. To propagate, actin waves must establish a polarity that determines their direction of propagation. This polarity is reflected in the three-dimensional arrangement of proteins that constitute a wave, and in the reversal of this pattern when a wave changes its direction of propagation. We show by photobleaching that MyoB is recruited to the front, and actin polymerizes de novo at the front of a propagating wave. Based on the discrete positions of proteins within the three-dimensional wave structure, a mechanism of wave propagation by the programmed activation and inhibition of actin polymerization is proposed.
Materials and Methods
Cells of D. discoideum clone AX2–214 were permanently transformed with integrating vectors encoding fluorescent fusion proteins. Clones of the transformants were cultivated and the cells were washed before imaging was done in 17 mM K-Na-phosphate buffer, pH 6.0, as described (1).
For latrunculin A treatment, we used the lowest concentration and incubation time required for actin depolymerization. Latrunculin A (Molecular Probes, Carlsbad, CA) was diluted from a 1 mM stock solution in DMSO with the phosphate buffer and added to the cells at 5 μM final concentration. Depolymerization of actin was checked by microscopic inspection using the mRFP-LimEΔ label to recognize remaining actin clusters and was found in most cells to be complete after 15 min. Subsequently, the drug solution was replaced by phosphate buffer.
Dual-color TIRF microscopy was performed as described (1). For photobleaching, a 405-nm laser was used coupled to an Olympus (Center Valley, PA) FV1000 laser scanning confocal microscope. Z-stacks for three-dimensional reconstructions were obtained by recording optical planes at 200-nm distances using PerkinElmer (Waltham, MA) Ultra View spinning disk equipment.
Details of vector construction, cell culture, fluorescent labeling, live-cell imaging, and photobleaching are given in the Supplemental Experimental Procedures in the Supporting Material.
Results
Patterns of Arp2/3 and actin in propagating waves
The waves we investigated are dense actin assemblies in the cell cortex that are integrated into a loose, highly dynamic network of actin filaments (4). The technique of choice to visualize actin structures in the cortex is TIRF microscopy (22). This technique allows the selective illumination within an evanescent field of the cortical region of Dictyostelium cells up to a distance of 100–200 nm perpendicular to the glass surface on which the cells are spread (23). To relate the patterns of proteins associated with actin waves to actin dynamics, we applied dual-emission TIRF microscopy to cells expressing particular GFP-fusion proteins together with mRFP-LimEΔ, as a probe for filamentous actin. Optimizing the monomeric red fluorescent protein mRFP for brilliant fluorescence yield in Dictyostelium (7) enabled us to use the 488-nm laser line for the excitation of GFP as well as mRFP and thus to excite GFP- and mRFP-labeled proteins in the same evanescent field.
To stimulate the formation of actin waves, we treated the cells with 5 μM latrunculin A for the depolymerization of actin and then allowed repolymerization by washing off the drug. On recovery from the treatment, actin polymerization starts with the formation of mobile clusters and proceeds with the excessive generation of dynamic waves (1) (Movie S1). Because previous work on spontaneously formed waves has shown that the Arp2/3 complex is also organized into waves (4), we first combined the mRFP-LimEΔ probe with GFP-Arp3.
At all stages of wave generation and propagation, actin was associated with the Arp2/3 complex, but there was no strict correlation of fluorescence intensities (Fig. 1 A and Movie S2). By superposition of the Arp3 and actin labels, substructures that were rich in both actin and the Arp2/3 complex could be distinguished from others in which either actin or the Arp2/3 complex was more strongly enriched (Fig. 1, B and C). There was no consistent enrichment of the Arp3 label at the front relative to the bulk of a wave. Often trails of the Arp3 label were retained behind the wave where actin was no longer detectable (Fig. 1 D and Movie S2).
Figure 1.
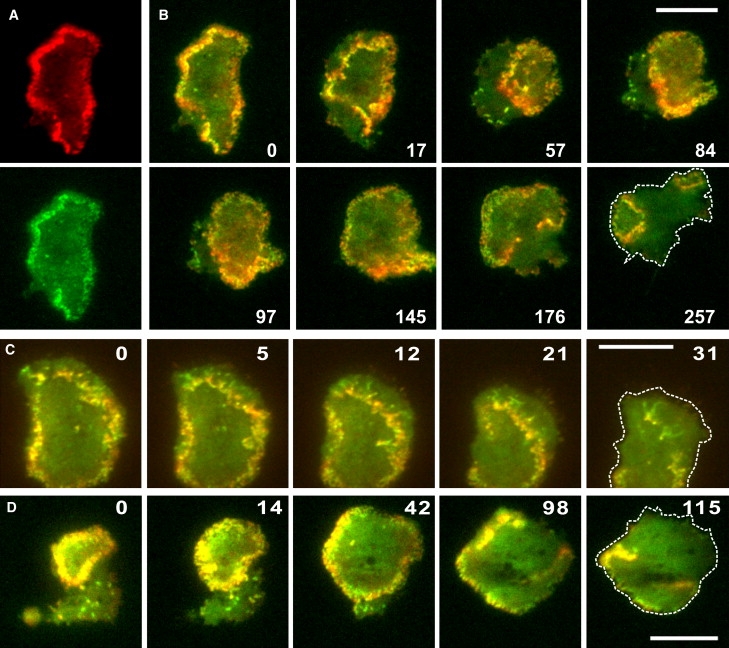
Dynamics of actin waves and recruitment of the Arp2/3 complex. Cells expressing mRFP-LimEΔ to label filamentous actin structures and GFP-Arp3 for incorporation into the Arp2/3 complex were recorded by dual-emission TIRF microscopy during recovery from latrunculin A treatment. (A) Images of the mRFP channel (top) and GFP channel (bottom) of the first frame of the time series shown in B. (B) A wave undergoing contraction, expansion, and splitting. The time series starts with the same frame (0) as shown in A. The two labels are superimposed on each other, yellow indicating merger of the labels. The same cell is shown in Movie S3. (C) Merged images of a collapsing wave, showing substructures more enriched in either actin or Arp2/3 and trailing of the Arp2/3 label behind the wave. (D) Merged images of a circular wave expanding the cell border. This protrusion stops when the wave starts to collapse. In the last panels of each sequence, cell borders are accentuated by dashed lines. Time is indicated in seconds. Bars, 10 μm.
One characteristic of the Arp2/3-associated actin waves is their ability to change direction. The wave shown in Fig. 1 B and Movie S3 moves first inward (frames 0 to 57) and subsequently outward, back to the cell border (frames 84 to 145). At the final stage, this wave breaks off at the rear, and the two free ends translocate inward (Fig. 1 B, frame 176) to form two separate circular waves (frame 257).
Actin waves typically spread on the inner face of the substrate-attached membrane area. However, when a wave front reaches the border of the cell, it can propel the membrane forward in the form of a broad lamellipod (Fig. 1 D, 0- to 42-s frames). In contrast, when a wave separates from the border, this region is paralyzed (Fig. 1 D, 98- and 115-s frames) and may eventually retract (Fig. 1 B, 176- to 257-s frames).
Association of MyoB with the front of actin waves
The question of whether a motor protein is associated with the actin waves was addressed by the use of GFP-tagged myosin-II and myosin-IB. The Dictyostelium genome contains a single gene encoding conventional, filament-forming myosin (24). This double-headed myosin-II proved not to be integrated into the actin waves, and its elimination by gene disruption did not impair wave formation. In particular, myosin-II was not required for the retraction of a wave (Movie S4).
Among the 12 unconventional myosins of Dictyostelium, we used the most abundant one, the single-headed myosin-IB (MyoB) (25,26), as an example. Fig. 2 shows that GFP-tagged MyoB is consistently enriched at the wave front. This is true not only for waves spreading across the substrate-attached area of the cell membrane but also during propulsion of cell edges and, in reverse direction, when a wave detaches from an edge (Fig. 2, A and B, and Movie S5). The distribution of MyoB along the wave profile in the TIRF plane, i.e., close to the bottom surface of the cell, is shown in Fig. 2, C and D. The fluorescence intensity of GFP-MyoB sharply rises at the front of the actin waves and reaches a peak either ahead of or coincident with the actin peak (top and bottom panels, respectively, in Fig. 2, C and D).
Figure 2.
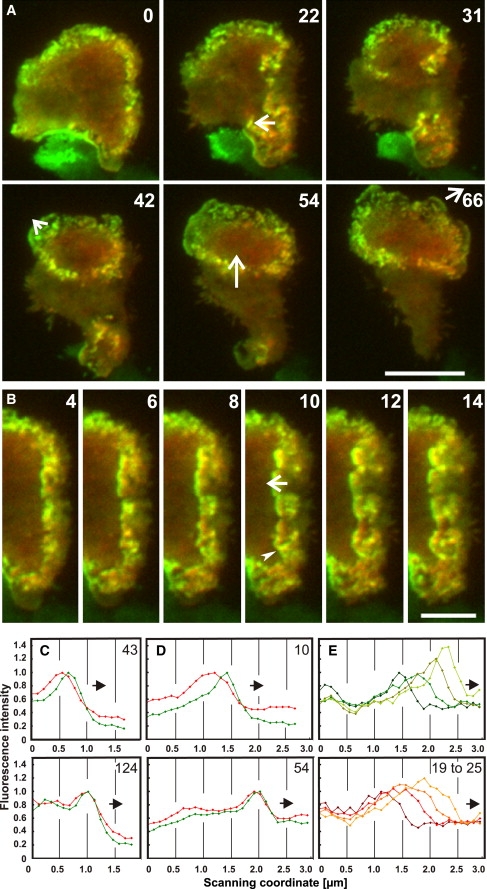
MyoB at the front of waves and their substructures. The GFP-MyoB label (green) is superimposed on the mRFP-LimEΔ label for filamentous actin (red). Cells recovering from latrunculin A treatment were recorded by TIRF microscopy (A,B) and fluorescence profiles quantified by line scans (C–E). (A) A wave showing irregular orientation of substructures during its reversal from collapsing to expanding. (B) A wave front in the same cell propagating from the right to the left and showing partitioning into ensembles led by MyoB. (Arrowhead in the 10-s frame; see also Movie S5 for substructure dynamics). Time is indicated in seconds after the first frame in A. Bars, 10 μm in A and 5 μm in B. (C) Two scans through waves that expand the cell border. (D) Two scans through retracting portions of waves that propagate on the substrate-attached cell surface. (E) Separate propagation of a substructure as indicated in the 22-s frame of A, plotted at 2-s intervals. (Top panel: dark to light green for MyB. Bottom panel: dark to light red for the actin label). Arrows in A and B indicate local direction of wave propagation and position of the scans in C–E. Numbers within panels C–E are seconds after the first frame in A; corresponding scan positions are indicated in A and B. (Position of the 124-s scan is indicated in the 66-s panel of A.) In C–E, the directions of wave propagation are plotted toward the right as indicated by arrows.
MyoB tends to form arrays at the front of a wave (Fig. 2 B, 4- to 10-s frames). The propagation of such an array led by MyoB is plotted in Fig. 2 E. However, MyoB can also move in different directions from the overall direction of wave propagation, for instance, perpendicular or opposite to an expanding wave front (Fig. 2 B, 10- to 14-s frames, and Movie S6). This implies that the overall propagation of a wave results from the predominant direction of its substructures.
CARMIL is an adaptor protein that binds to the SH3-domain of MyoB and links this motor protein to the Arp2/3 complex (13). To figure out whether CARMIL is enriched in the waves together with its binding partners, we imaged GFP-tagged CARMIL by TIRF microscopy. GFP-CARMIL proved to be abundant in the form of diffuse patches outside the waves. It was further enriched within the actin waves, although this enrichment was more delicate than that of actin. The examples shown in Fig. 3 and Movie S7 illustrate the turning of waves from retraction to expansion. During this change of direction a new front is established while the previous one fades out (Fig. 3, 18.5- and 26-s frames). CARMIL remains associated with the waves in all phases of the turning (Movie S8), and z-scans indicate that CARMIL is associated with the waves through their entire depth (data not shown).
Figure 3.
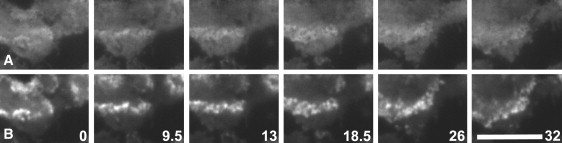
CARMIL in a wave that changes direction. The time series obtained by TIRF microscopy shows part of a cell expressing GFP-CARMIL and mRFP-LimEΔ; numbers indicate seconds. (Top panels) Channel showing the emission of GFP-CARMIL. (Bottom panels) Channel showing the emission of mRFP-LimEΔ. Up to the 13-s frame, the wave is moving toward the top, and subsequently toward the bottom of the panels. Bar, 10 μm.
Coronin marks the back of a wave in a plane close to the membrane
When we localized coronin by the use of TIRF microscopy, we found this protein in a zone at the back of a wave, clearly distinct from the other proteins studied. In the cell shown in Fig. 4 A, a zone enriched in coronin continuously follows the expanding front of filamentous actin. Scanning of fluorescence intensities along a line across the wave profile reveals a peak of coronin ∼1 μm behind the actin front within the plane close to the membrane that is illuminated by the TIRF mode (Fig. 4 B; see also Movies S9 and S10).
Figure 4.
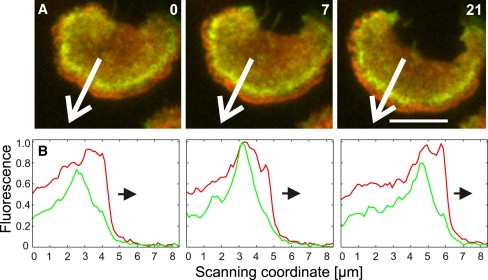
TIRF images localizing coronin to the back of actin waves. The cells coexpress GFP-coronin (green) and mRFP-LimEΔ (red). (A) Three stages of a wave propagating from top to bottom and showing a regular zonal pattern. (B) Scans of fluorescence intensities along the lines indicated in A. These scans quantify fluorescence intensities in a plane close the substrate, which is limited by the z-axis penetration of the evanescent field generated at 488 nm for the excitation of both the GFP and mRFP labels. Within this plane, the actin label (red) sharply rises at the wave front, whereas the increase of coronin fluorescence (green) is less steep. Consequently, coronin peaks at the posterior side of the zone populated by filamentous actin. Time is indicated in seconds. Bar, 5 μm.
Waves propagate in the absence of particular signal transducers
To underscore the independence of actin waves from external signals, we investigated mutant cells deficient in the Gβ-subunit of heterotrimeric G-proteins (27). D. discoideum has a single Gβ subunit, which is essential for the transmission of chemoattractant signals as well as for the induction of phagocytic cups in response to attaching particles (28). As illustrated in Fig. 5 and Movies S11 and S12, Gβ-null cells can generate propagating waves similar to their parents, showing that no signal transmitted through heterotrimeric G-proteins is required.
Figure 5.
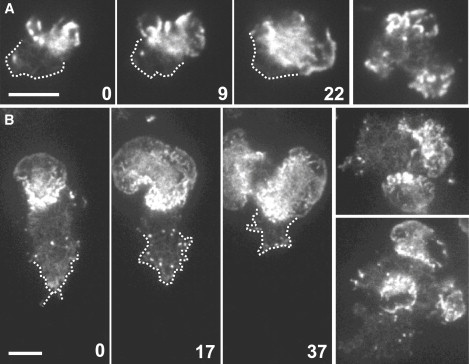
Independence of wave formation from signal transduction through heterotrimeric G-proteins. (A) Parent strain DH1. (B) Gβ-null mutant LW6 (27), both expressing LimEΔ-GFP. Time series on the left show the evolution of single waves; the one in B is split into two. Numbers indicate seconds. Panels on the right exemplify cells with two or three waves. Outside the waves the actin network in the cell cortex is distinguishable. The actin patches in this area are mostly involved in clathrin-dependent endocytosis (51). The two time series are typical in showing retraction of this area when the wave expands, as indicated by dotted lines. Bars, 10 μm.
A common activator of the Arp2/3 complex is the SCAR/WAVE protein, a member of a pentameric complex that links extracellular signals to actin-based cell motility (29). SCAR has been eliminated in Dictyostelium by gene disruption (30,31). This allowed us to examine whether actin waves can propagate in the absence of an intact SCAR complex and, if so, whether Arp2/3 is still recruited to these waves.
TIRF images of SCAR-null cells expressing mCherry-LimEΔ and GFP-Arp3 are shown in Fig. 6, A and B (see also Movies S13 and S14). In these images, the capability of SCAR-null cells to form large, rapidly expanding waves is evident. Velocities of 7 to 9 μm/min indicate that propagation rates are not reduced in the absence of SCAR (Fig. 6 C compared with Fig. 4 B). This is true even for waves propagating on the free substrate surface, which requires protrusion of the plasma membrane. The origin of large waves formed in SCAR-null cells can be traced back to an expanding area densely packed with short-lived actin and Arp2/3 clusters (Fig. 6, B and D). In SCAR-null cells, the Arp2/3 complex is recruited to the propagating waves (Fig. 6 D), and the actin-enriched layer is followed by a zone of coronin (not shown), indicating that the on- and off-regulation of actin polymerization in free-running actin waves does not require the SCAR protein.
Figure 6.
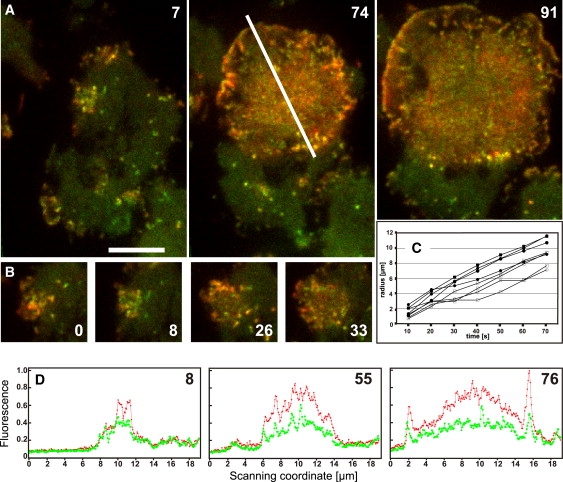
Wave propagation without SCAR. The expression of mCherry-LimEΔ and GFP-Arp3 in SCAR-null cells shows waves propagating with normal speed and their association with the Arp2/3 complex. (A) Merged images of mRFP-LimEΔ (red) and GFP-Arp3 (green) in three stages of an expanding wave. (B) Sequence illustrating origin of this wave from an area of dense and dynamic actin and Arp2/3 assembly. Time is indicated in seconds. Bar for A and B, 5 μm. (C) Velocity of wave expansion in SCAR-null cells. Distance of the actin front from the origin of two waves is determined (open symbols for the one shown in A and B and Movie S13; solid symbols for the one shown in Movie S14). In each wave, expansion has been measured in four directions, either over the cell body (circles), i.e., toward the bottom in A or over the free substrate surface (squares), i.e., toward the top in A. (D) Three scans of fluorescence intensities (in arbitrary units) along the line indicated in A. Red, mCherry-LimEΔ; green, GFP-Arp3. Numbers indicate seconds in accord with A and B. On the left of each panel the wave propagates toward the top in A, on the right toward the bottom.
Photobleaching distinguishes between two mechanisms of wave propagation
Wave propagation may be based on the net transport of actin filaments or on treadmilling, which means de novo polymerization at the front and net depolymerization at the back of the wave. To distinguish between these possibilities, fluorescence recovery after photobleaching (FRAP) was performed on propagating waves.
To define the fluorescence recovery of GFP-actin, cells were colabeled with mRFP-LimEΔ, the reference of wave position used in the previous experiments (Fig. 7 A). In the case of directed transport, the bleached spot is predicted to move with the same speed as the wave. In the case of treadmilling, the spot should stay in place while the wave passes over the bleached area. In the latter case, there are two possibilities, depending on the three-dimensional organization of actin filaments within the waves: 1), at the wave front actin filaments may elongate parallel to the membrane, or 2), new filaments may be nucleated there to grow from the membrane into the cytoplasmic space (Fig. 7 B). In the bleached area, the fluorescence of actin recovered predominantly at the front of the wave (Fig. 7 C). During recovery, the bleached spot of GFP-actin remained stationary, and the recovery was too fast for the actin to be carried with a wave that propagates ∼1 μm in 10 s (Fig. 7 D). Together, these data support a treadmilling mechanism of wave propagation.
Figure 7.
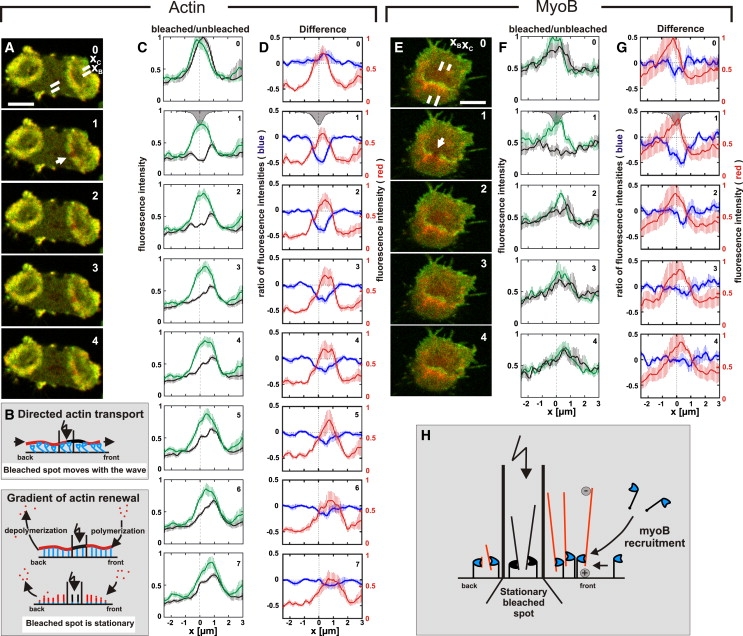
Actin and MyoB dynamics in a propagating wave as revealed by FRAP. (A) FRAP in a cell double-labeled with GFP-actin and mRFP-LimEΔ. Numbers in the panels indicate seconds after beginning of the recording. The 0-s frame was recorded before and the 1-s frame immediately after bleaching. Scans of fluorescence intensities through the bleached area (xB) and the unbleached control area (xC) are indicated in the 0-s frame. The bleached spot is indicated by an arrow in the 1-s frame. The wave propagates from left to right. (B) Diagram illustrating putative modes of wave propagation and their distinction by FRAP. In the top panel actin is assumed to be translocated, for instance, by the motor activity of myosin along the membrane of the cell (black line). In the middle and bottom panels actin is assumed to be membrane-anchored and the wave to propagate in a treadmilling mode. In this case, the wave might propagate by elongation of actin filaments along the membrane (middle) or by the nucleation of new filaments (bottom). (C) Linear scans of fluorescence intensities of GFP-actin along xB (black) and xC (green) in the direction of wave propagation. The gray profile in the 1-s frames circumscribes the actual distribution of fluorescence intensities in the bleached area. Dashed vertical lines indicate the center position of the bleached area in the xB scan or the corresponding laterally shifted position in the xC scan. (D) Blue curves: difference of the fluorescence intensities along the scan through the bleached area (xB) minus the scan through the unbleached area (xC). These curves show the dip caused by bleaching and recovery of the GFP-actin fluorescence within a 7-s period. Red curves: mRFP-LimEΔ label showing wave positions in the unbleached area (xC scan). (E–G) FRAP of a cell double-labeled with GFP-MyoB and mRFP-LimEΔ. The panels E, F, G correspond to panels A, C, D, respectively. In E, the wave is propagating from bottom to top. (H) Diagram illustrating MyoB recruitment. The FRAP data argue against the movement of MyoB along actin filaments in the direction of wave propagation. They indicate that MyoB is recruited from outside to the wave front, probably by diffusion within the cytoplasm and on the membrane (arrows on the right). MyoB heads are colored blue, and actin filaments red, outside the bleached area; their orientation is hypothetical. Fluorescence intensities were recorded by concomitant excitation of GFP and mRFP at 488 nm and separation of the emissions. The plots in C and D represent averages from eight experiments; the plots in F and G from six experiments. Vertical bars indicate standard errors of the mean. Scale bars in A and E, 5 μm.
In parallel experiments, cells expressing GFP-MyoB together with mRFP-LimEΔ were subjected to photobleaching. Fig. 7 E shows a zone enriched in GFP-MyoB at the wave front. After the bleaching, the fluorescence of GFP-MyoB recovered primarily in front of the bleached spot (Fig. 7 F), and this recovery was fast relative to the velocity of wave propagation (Fig. 7 G). As diagrammed in Fig. 7 H, these data indicate that MyoB is exchanged and de novo incorporated into the wave front.
Assuming actin to be polymerized only at the front and depolymerized at the back of a wave, the lifetime of an actin filament would be given by the distance between front and back divided by the velocity of a wave. A front-to-back distance of 2 μm and a velocity of 1 μm per 10 s (Fig. 4) would then correspond to a filament lifetime of ∼20 s. From the data of Fig. 7 D, half-maximal fluorescence recovery within 3 to 4 s is deduced. This rapid recovery implies that the turnover of actin filaments is not restricted to the front and back but continues within the bulk of a wave. The time for half-maximal recovery of GFP-MyoB is even shorter, 1 to 2 s (Fig. 7 G), suggesting rapid exchange of the motor protein not only at the front but also within a wave.
Three-dimensional organization of actin waves
In the two-dimensional protein patterns seen in TIRF images, which compose a layer close to the membrane ∼100 nm in depth, coronin was localized to a zone at the back of the waves (Fig. 4). This zonal pattern could be interpreted to mean that actin polymerizes at the front and depolymerizes at the back of a wave, where the Arp2/3 complex is turned off. However, the FRAP data suggested that actin filaments are recycled throughout the entire wave. This apparent discrepancy has been solved by imaging the three-dimensional protein pattern, again using the mRFP-LimEΔ label as a reference for filamentous actin. The reconstructed profiles from the plasma membrane toward the interior of a cell show coronin to decorate the roof of the waves, i.e., their boundary to the cytoplasmic space, throughout their entire length. However, in the front region the coronin is separated too far from the plasma membrane to be imaged by TIRF.
In Fig. 8 we show the three-dimensional arrangement of coronin relative to actin, reconstructed from deconvolved image stacks (the unprocessed images are shown in Fig. S1). In an x-y plane close to the bottom surface of a cell, a zonal actin-coronin pattern similar to the TIRF images is obtained (Fig. 8 A, top panel). In cross-sections through a cell, the waves are seen to extend by 1–2 μm from the bottom surface into the cytoplasm (Fig. 8 A, middle panel).
Figure 8.
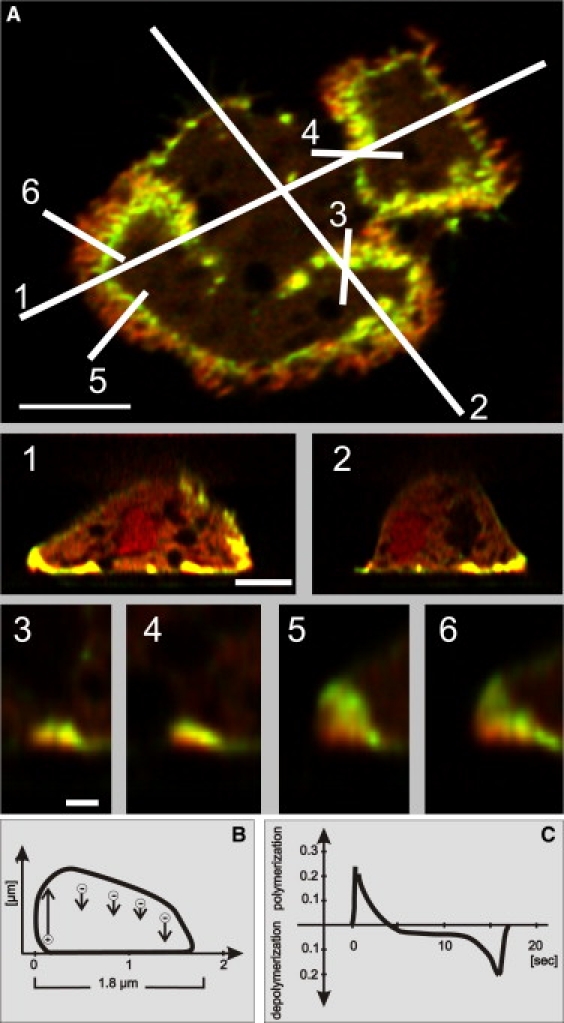
Three-dimensional patterns of coronin distribution in actin waves. (A) The z-stacks were acquired by spinning-disk confocal microscopy from a cell double-labeled with GFP-coronin (green) and mRFP-LimEΔ (red). Three-dimensional protein patterns are constructed from deconvolved images. (Top panels) Confocal plane close to the substrate, showing a zonal actin-coronin pattern similar to the TIRF images of Fig. 4. In the x,y plane of A, lines 1–6 indicate the directions of z-scans shown in the middle and bottom panels. These scans are oriented such that the number of each line in the top panel corresponds to the left-hand side of the frames below. (Middle panels) The z-scans 1 and 2 through the entire cell illustrate localization of the waves marked by actin and coronin on the bottom surface and at the border of the cell. (Bottom panels) Actin-coronin patterns in waves at higher magnification. Scans 3 and 4 show waves propagating on the bottom surface of the cell, and scans 5 and 6 waves in contact with the cell border. Bars, 5 μm for the top and middle panels and 1 μm for the bottom panels. (B) Scheme of a wave modeled according to the regular shape of scan 4 (A). Assuming that the height of the actin layer (including the coronin-enriched zone) reflects the net rate of actin polymerization, there is rapid polymerization going on at the front, followed by continuous net depolymerization up to the back of the wave. (C) The spatial profile of B can be translated into a temporal sequence. Here we assume that the wave propagates with constant velocity of 6 μm/min as in the waves in Figs. 2 and 4. The profile illustrates the sequence of changes in net polymerization or depolymerization of actin at a point on the membrane when a wave passes. These data suggest that a short phase of high-rate actin polymerization turns abruptly into a longer phase of net depolymerization.
In free-running waves, the zone of coronin essentially slopes from the front of a wave, where it is separated from the plasma membrane by an actin layer, to the back where it adjoins the plasma membrane (Fig. 8 A, bottom panels 3 and 4). In waves that meet the lateral border of the cell, the bottom-to-top polarity rolls up by ∼90°, such that actin polymerization is now directed toward the lateral border of the cell where the membrane is pushed forward (Fig. 8 A, bottom panels 5 and 6).
The profile of polymerized actin with the coronin layer on top of free-running waves is consistent with a high rate of actin polymerization at the front and close to the membrane, the sites most distant from the coronin layer. The declining thickness of the actin layer toward the back of the waves suggests that actin is continuously depolymerized along the waves, most likely within the coronin-enriched zone on top of the waves (see Fig. 8 B). Translated into a temporal sequence, these data imply a high rate of actin polymerization at the front of a wave, which converts into net depolymerization ∼5 s later (Fig. 8 C).
In contrast to coronin, MyoB is enriched at the bottom of the waves close to the cell membrane, although it extends further into the cytoplasm at the wave front (Fig. 9 A, panels 1 and 2). This is also seen in the tangential sections through a wave as shown in panels 3 to 5 of Fig. 9 A. MyoB covers the entire height of the wave at the front; just behind the front MyoB becomes concentrated in a layer close to the plasma membrane, and in the middle of the wave most of the space is filled with actin. Substructures of a wave are revealed in scan 6 of Fig. 9 B, which is positioned similar to scan 4 in Fig. 9 A. Along the wave, actin clusters can be seen to be located on top of MyoB clusters, suggesting that myoB is localized to membrane-associated building blocks of polymerized actin structures. At the left and right end of this scan, i.e., at sites where the wave meets the cell border, the MyoB layer is bent around the border, and polymerized actin extends in horizontal direction to the interior of the cell.
Figure 9.
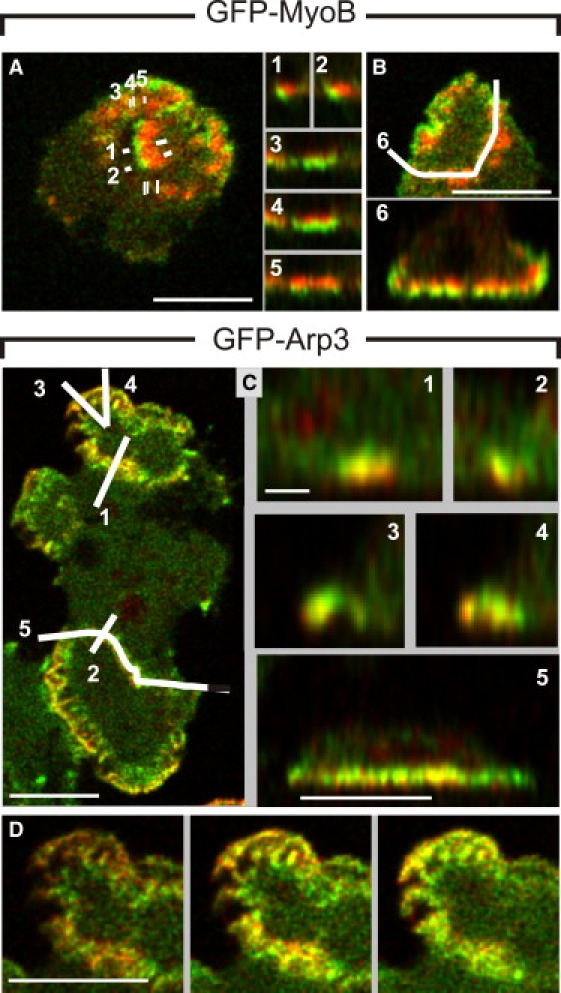
Three-dimensional distribution of MyoB and Arp2/3 in actin waves. Cells double-labeled either with GFP-MyoB or GFP-Arp3 and mRFP-LimEΔ were subjected to spinning disk confocal microscopy, and deconvolved distributions in the z-direction obtained from image stacks. (A) (Left panel) A wave with a clear zonal pattern of MyoB (green) and actin (red) in a plane of focus close to the substrate surface. (Right panels) Z-scans 1 to 5 as indicated in the left panel. Scans 1 and 2 show cross sections through the wave; scans 3 to 5 sections parallel to the wave from the front (3) to the back (5). (B) Wave extending from cell edge to edge (top), and the distribution in the z-direction of MyoB and actin along scan 6 (bottom). (C) A cell double-labeled with GFP-Arp3 and mRFP-LimEΔ. (Left panel) Optical plane showing two waves in which the Arp2/3 complex (green) and actin (red) overlap but do not completely coincide. (Right panels) Z-scans 1 to 5 along the lines indicated on the left. (D) Z-series of three x,y scans through the wave shown on top of the cell in C. These scans are acquired at distances of 200 nm beginning with the left panel close to the substrate-attached cell surface. Bars, 5 μm except 1 μm in scans 1 to 4 of C.
The distribution of the Arp2/3 complex in the z-direction of the wave structure proved to be distinct from the distributions of either MyoB or coronin (Fig. 9 C). In general, the Arp2/3 label becomes stronger, relative to the label of filamentous actin, with increasing distance from the plasma membrane. This is true not only for free-running waves (Fig. 9 C, scans 1, 2, and 5) but also for waves that already arrived at the cell border (Fig. 9 C, scans 3 and 4). Patches with varying actin-to-Arp2/3 ratios are observed throughout the waves, suggesting the participation of additional proteins in constituting the wave structure (Fig. 9 D).
Discussion
Self-organization of actin into dynamic wave structures
In this study we have analyzed the dynamic pattern of proteins constituting waves of polymerized actin in view of the mechanisms of wave propagation. The waves investigated in Dictyostelium cells travel on the cytoplasmic face of the planar substrate-attached membrane (1). When these waves are viewed by TIRF microscopy, they exhibit a zonal structure with MyoB at the front, coronin at the back, and the Arp2/3 complex throughout the wave.
In three dimensions, the waves show patterns characteristic of each protein. MyoB is concentrated close to the membrane and is enriched at the wave front (Fig. 9, A and B). In contrast, the Arp2/3 label actually increases relative to actin toward the interior of the cell (see Fig. 9 C, cross sections 1 to 5, and Fig. 9 D). Coronin forms a layer on top of the waves facing the cytoplasmic space. The distance between the membrane and the zone of coronin shrinks behind the wave front until, at the back of the wave, coronin approaches the membrane (Fig. 8). This slope is the reason that TIRF microscopy visualizes coronin only at the back of a wave.
The discrete positions of actin-associated proteins within a wave are the result of self-organization in the membrane-associated actin system. The FRAP data presented show that bleached spots of actin remain stationary relative to the substrate, which excludes translocation of preestablished actin filaments as a basis of wave propagation. Similarly, the bleached spots of MyoB turned out to be stationary, and unbleached, i.e., newly recruited MyoB was incorporated at the propagating wave front. As shown previously (4), the Arp2/3 complex also resides in a stationary position relative to the substrate.
In summary, it is the de novo assembly of its constituents that forces the wave front to move. This process is comparable to the treadmilling shown in early studies for actin filaments at the leading edge of fibroblasts or in the filopodia of neuroblastoma cells (32,33), and recently also for the Arp2/3 complex in the lamellipodia of melanoma cells (34). However, as long as the waves propagate on the substrate-attached membrane area, there is no membrane fold located in front of them that would serve as a scaffold for the ordered assembly of proteins (1). Furthermore, the waves are generated independently of chemotactic or other signals transmitted through heterotrimeric G-proteins (Fig. 5), and they do not require SCAR upstream of the Arp2/3 complex (Fig. 6).
Force production by actin waves pushing against the cell border
Actin waves are capable of pushing the membrane forward when they hit the cell border (1), which means they produce force as actin does at a leading edge (35). This activity of actin waves can be understood on the basis of the three-dimensional protein pattern presented here. At the cell border, the vertical protein pattern turns by 90°, such that the proposed gradient of actin polymerization is brought into a horizontal direction (Figs. 8 and 9). As a consequence, the force of actin polymerization will be directed toward the lateral border of the cell. Actin waves propagate with velocities ∼6 μm/min, which is in the same order as the migration of an entire cell. Motor proteins of the myosin-I type are not only present at the front of the actin waves, they are intrinsic constituents of leading edges (36,37), where they may play a similar role as in actin waves. The double-headed myosin-II, typically enriched at the retracting tail of a cell, is not a wave constituent and is not required for wave formation (Movie S4).
It appears, therefore, that the structure and dynamics of the free-running waves reflect the state of the actin system at a leading edge. Like actin itself, all three wave constituents are exchanged with the pool of proteins in the cytoplasm or, in the case of MyoB, also with adjacent areas of the plasma membrane. This pattern is the basis of a programmed sequence of controls that organize the activation and inactivation of actin polymerization within periods of a few seconds. We conclude that the machinery for lamellipod protrusion can be gathered together by the self-assembly of proteins with no need of being fixed to a particular membrane site. In a sense, the waves may be viewed as free-running actin assemblies in search of a leading edge or a particle to be phagocytosed.
Toward the modeling of wave structure and dynamics
The proteins shown to be associated with actin waves are functionally linked to each other and connected by physical interactions. The adaptor protein, CARMIL, joins MyoB to the Arp2/3 complex (13). If MyoB molecules are not bound to a membrane, they can bind a second actin filament to a nonmotor (ATP-insensitive) actin-binding site (38,39). This binding might be involved in thick accumulations of MyoB at the wave front (Fig. 9, A and B). Coronins of yeast and mammals not only bind to actin filaments but interact directly with the Arp2/3 complex, thus inhibiting its nucleating activity (17,40).
Based on the fact that within a wave, MyoB and coronin sandwich the actin between them, it is reasonable to assume that MyoB is involved in the initiation of actin polymerization close to the membrane and at the front of a wave, and coronin in the termination of polymerization at the top and back of the wave. A role of MyoB in organizing actin waves can be deduced from its known activities and its exchange between cytosolic and membrane-bound states. The elimination of MyoB slows down cell motility (25,26), indicating that this motor protein facilitates actin-driven membrane protrusion. MyoB interacts with lipids as well as indirectly with membrane-bound proteins and is targeted by this interaction specifically to the plasma membrane (41). In a projection map obtained by cryoelectron tomography from Acanthamoeba MyoB bound to a layer of phosphatidylserine, the actin-binding site of the motor domain is placed 12 nm away from the polybasic lipid-binding site on the tail (42). Because single MyoB molecules move in a nonprocessive way along actin filaments (43), Jontes et al. (42) suggested that plaques of membrane-bound MyoB molecules are held together by tail-tail interactions and cooperate in the movement of actin filaments.
In the scheme of Fig. 10 we take this arrangement into account and assume that actin filaments in the waves are kept in a more or less upright position by the cooperation of membrane-bound motor proteins. This hypothesis is consistent with the absence of a detectable net movement of actin in a direction parallel to the membrane (Fig. 7) and also with the columns of polymerized actin that are organized on the top of MyoB patches accumulated at the membrane (Fig. 9 B). If the actin filaments point with their plus ends to the membrane, MyoB as a plus-end-directed motor would move the filaments off the membrane, allowing continued polymerization by the addition of new actin subunits close to the membrane. In this view, MyoB acts similar to a pulling force proposed to enhance formin-mediated actin polymerization (44).
Figure 10.
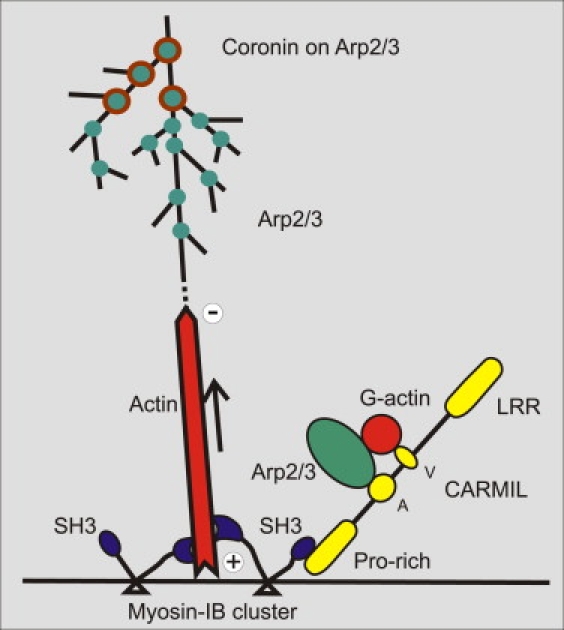
Hypothetical role of MyoB in organizing actin filaments into waves and in recruiting the Arp2/3 complex. Based on the z-scans shown in Fig. 9A, MyoB (blue) is proposed to assist by multiple interactions in organizing the membrane-anchored actin network. MyoB binds with its tail (triangles) to the plasma membrane (42), and its N-terminal motor domain moves toward the plus end of actin filaments. In that way, clusters of MyoB may keep the growing filaments separate from the membrane (arrow), thus allowing subunits to enter. The C-terminal SH3 domain of MyoB interacts with the proline-rich region of the adaptor protein CARMIL (yellow), which links MyoB to the Arp2/3 complex through its acidic domain (A). These interactions of CARMIL as well as the presence of the protein-protein interacting, verprolin-like (V), and leucine-rich-repeat (LRR) sequences are adopted from Jung et al. (13). The actin filaments are branched at sites of Arp2/3 binding (green) until activity of the Arp2/3 complex is inhibited by coronin (brown circles).
A minimal model for propagating actin rings in human lymphoblasts requires only two autocatalytic steps, the nucleation of actin polymerization and the dissociation of actin filaments from the membrane (45). Similarly, wave propagation can be modeled on the basis of cooperative membrane binding and activation of a nucleator, followed by its detachment (46). An intriguing possibility has been the autocatalytic activation of actin nucleation by upstream effectors of the Arp2/3 complex, in particular the pentameric SCAR/WAVE complex (29). In neutrophils, Hem-1, a member of this complex, is seen to move in the form of waves along the substrate-attached membrane (3), with a similar velocity as the actin waves in Dictyostelium (∼4 μm/min vs. 6 μm/min). Hem-1 waves propagate by recruiting Hem-1 to the wave front, similar to the actin waves, which propagate by the polymerization of actin at their front (Fig. 7). Hem-1 waves annihilate on collision (3), as actin waves tend to vanish at the site of collision and thus to fuse into one wave (Movie S9), provided they do not generate new expanding wave fronts at their site of fusion (4,47). Despite these obvious similarities between propagation of a member of the SCAR complex and actin waves, the SCAR protein itself proved to be dispensable for the propagation of actin waves in Dictyostelium. However, these data do not rule out a role of SCAR in wave initiation; in fact, Movies S13 and S14 indicate that in SCAR-null cells only a few of the primordial actin and Arp2/3 clusters are converted into propagating waves.
The autocatalytic progression of actin polymerization may be maintained by the molecular players already known to be recruited to the waves: MyoB can bind to the newly formed actin filaments that are nucleated as a result of Arp2/3 action, and the MyoB molecules thus recruited can, through interaction with CARMIL, engender binding of the Arp2/3 complex, which in turn will initiate another run of actin polymerization. Because of its branching activity, the Arp2/3 complex increases the number of polymerizing actin filaments in exponential progression (12). However, as a caveat against oversimplification, we wish to underscore the multiple control pathways for actin polymerization in Dictyostelium and the plethora of regulatory proteins with overlapping functions. For example, in the myosin-I family of Dictyostelium there are two more members that contain SH3 domains (for review see Soldati (48)), and MyoB even overlaps in function with MyoA, a myosin-I that lacks an SH3-domain on its tail (25,26). Therefore, it seems unlikely that the generation and propagation of actin waves are based on the activity of only the few proteins studied.
Conclusions
Actin waves exemplify the potential of the actin system to self-organize into supramolecular complexes by recruiting proteins from the cellular pool in a defined temporal and spatial pattern. Propagation of these waves appears to be based on a programmed sequence of controls that activate and inactivate actin polymerization within a few seconds. The complexes of proteins constituting a wave are functional as judged from the force they produce for membrane protrusion. The ordered assembly of proteins that constitute an actin wave also holds for other dynamic actin structures formed under various conditions within the environment of a living cell. For instance, spatiotemporal patterns of actin and associated proteins are found at the leading edge of chemotaxing cells, in phagocytic cups (49), and at rocketing phagosomes (50). The data presented here underscore the importance of analyzing the three-dimensional organization of advanced actin structures as a prerequisite for modeling the molecular interactions that underlie their generation and dynamics.
Acknowledgments
We thank Margaret Titus, University of Minnesota, for the GFP-MyoB vector, Robert Insall, CR-UK Beatson Institute for Cancer Research, Glasgow, for the SCAR-null mutant, Markus Fischer, Universität Hamburg, for synthesizing DdmCherry, Ina Weisswange, Ireen König, and Silke Gerwig for expert assistance in imaging, and Emmanuel Burghardt, Gudrun Trommler, and Dirk Wischnewski for providing transformants.
This work was supported by grants to G.G. from the Deutsche Forschungsgemeinschaft (SPP 1128 and SFB 413) and from the Max Planck Society.
Footnotes
Till Bretschneider and Kurt Anderson contributed equally to this work.
Till Bretschneider's present address is Warwick Systems Biology, Coventry House, University of Warwick, Coventry CV4 7AL, UK.
Kurt Anderson's present address is Leader Light Microscopy, Beatson Institute for Cancer Research, Garscube Estate, Glasgow G61 1BD, Scotland, UK.
Annette Müller-Taubenberger's present address is Adolf Butenandt Institute for Cell Biology and Center for Integrated Protein Science Munich (CIPSM), Ludwig Maximilians Universität, D-80336 Munich, Germany.
Supporting Material
References
- 1.Gerisch G., Bretschneider T., Müller-Taubenberger A., Simmeth E., Ecke M. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys. J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vicker M.G. Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp. Cell Res. 2002;275:54–66. doi: 10.1006/excr.2001.5466. [DOI] [PubMed] [Google Scholar]
- 3.Weiner O.D., Marganski W.A., Wu L.F., Altschuler S.J., Kirschner M.W. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:2053–2063. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretschneider T., Diez S., Anderson K., Heuser J., Clarke M. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Rehberg M., Kleylein-Sohn J., Faix J., Ho T.-H., Schulz I. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell. 2005;16:2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuxworth R.I., Cheetham J.L., Machesky L.M., Spiegelmann G.B., Weeks G. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer M., Haase I., Simmeth E., Gerisch G., Müller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 2004;577:227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
- 8.Novak K.D., Titus M.A. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol. Biol. Cell. 1998;9:75–88. doi: 10.1091/mbc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullins R.D., Heuser J.A., Pollard T.D. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egile C., Rouiller I., Xu X.-P., Volksmann N., Li R. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:1902–1909. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goley E.D., Welch M.D. The Arp2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 12.Carlier M.-F., Pantaloni D. Control of actin assembly dynamics in cell motility. J. Biol. Chem. 2007;282:23005–23009. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- 13.Jung G., Remmert K., Wu X.F., Volosky J.M., Hammer J.A., III The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Hostos E.L., Bradtke B., Lottspeich F., Guggenheim R., Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localizes to cell surface projections, has sequence similarities to G protein β subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appleton B.A., Wu P., Wiesmann C. The crystal structure of murine coronin-1: A regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Humphries C.L., Balcer H.I., D'Agostino J.L., Winsor B., Drubin D.G. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodal A.A., Sokolova O., Robins D.B., Daugherty K.M., Hippenmeyer S. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 18.Gerisch G., Albrecht R., Heizer C., Hodgkinson S., Maniak M. Chemoattractant-controlled accumulation of coronin at the leading edge of Dictyostelium cells monitored using a green fluorescent protein-coronin fusion protein. Curr. Biol. 1995;5:1280–1285. doi: 10.1016/s0960-9822(95)00254-5. [DOI] [PubMed] [Google Scholar]
- 19.De Hostos E.L., Rehfuess C., Bradtke B., Waddell D.R., Albrecht R. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniak M., Rauchenberger R., Albrecht R., Murphy J., Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 21.Uetrecht A.C., Bear J.E. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Axelrodt D. Total internal-reflection fluorescence microscopy. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
- 23.Diez S., Gerisch G., Anderson K., Müller-Taubenberger A., Bretschneider T. Subsecond reorganization of the actin network in cell motility and chemotaxis. Proc. Natl. Acad. Sci. USA. 2005;102:7601–7606. doi: 10.1073/pnas.0408546102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manstein D.J., Titus M.A., De Lozanne A., Spudich J. Gene replacement in Dictyostelium: Generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung G., Wu X., Hammer J.A., III Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falk D.L., Wessels D., Jenkins L., Pham T., Kuhl S. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J. Cell Sci. 2003;116:3985–3999. doi: 10.1242/jcs.00696. [DOI] [PubMed] [Google Scholar]
- 27.Wu L., Valkema R., Van Haastert P.J.M., Devreotes P.N. The G protein β subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peracino B., Borleis J., Jin T., Westphal M., Wu J.-M. G protein β subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J. Cell Biol. 1998;141:1529–1537. doi: 10.1083/jcb.141.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machesky L.M., Insall R.H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 30.Bear J.E., Rawls J.F., Saxe C.L., III Scar, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blagg S.L., Stewart M., Sambles C., Insall R.H. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y.-L. Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J. Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mallavarapu A., Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai F.P.L., Szczodrak M., Block J., Faix J., Breitsprecher D. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 36.Fukui Y., Lynch T.J., Brzeska H., Korn E.D. Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature. 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- 37.Morita Y.S., Jung G., Hammer J.A., III, Fukui Y. Localization of Dictyostelium myoB and myoD to filopodia and cell-cell contact sites using isoform- specific antibodies. Eur. J. Cell Biol. 1996;71:371–379. [PubMed] [Google Scholar]
- 38.Miyata H., Bowers B., Korn E.D. Plasma membrane association of Acanthamoeba myosin I. J. Cell Biol. 1989;109:1519–1528. doi: 10.1083/jcb.109.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zot G., Doberstein S.K., Pollard T.D. Myosin-I moves actin filaments on a phospholipid substrate: Implications for membrane targeting. J. Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai L., Marshall T.W., Uetrecht A.C., Schafer D.A., Bear J.E. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senda S., Titus M.A. A potential mechanism for regulating myosin I binding to membranes in vivo. FEBS Lett. 2000;484:125–128. doi: 10.1016/s0014-5793(00)02137-2. [DOI] [PubMed] [Google Scholar]
- 42.Jontes J.D., Ostap E.M., Pollard T.D., Milligan R.A. Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIP) determined by cryoelectron microscopy of decorated actin filaments. J. Cell Biol. 1998;141:155–162. doi: 10.1083/jcb.141.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostap E.M., Pollard T.D. Biochemical kinetic characterization of the Acanthamoeba myosin-1 ATPase. J. Cell Biol. 1996;132:1053–1060. doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozlov M.M., Bershadsky A.D. Processive capping by formin suggests a force-driven mechanism of actin polymerization. J. Cell Biol. 2004;167:1011–1017. doi: 10.1083/jcb.200410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.le Guyader H., Hyver C. Periodic activity of the cortical cytoskeleton of the lymphoblast: modelling by a reaction-diffusion system. C. R. Acad. Sci. Paris. Life Sci. 1997;320:59–65. doi: 10.1016/s0764-4469(99)80087-0. [DOI] [PubMed] [Google Scholar]
- 46.Doubrovinski K., Kruse K. Cytoskeletal waves in the absence of molecular motors. Europhys. Lett. 2008;83 18003 p1–p6. [Google Scholar]
- 47.Bretschneider T., Jonkman J., Köhler J., Medalia O., Barisic K. Dynamic organization of the actin system in the motile cells of Dictyostelium. J. Muscle Res. Cell Motil. 2002;23:639–649. doi: 10.1023/a:1024455023518. [DOI] [PubMed] [Google Scholar]
- 48.Soldati T. Unconventional myosins, actin dynamics and endocytosis: A ménage à trois? Traffic. 2003;4:358–366. doi: 10.1034/j.1600-0854.2003.t01-1-00095.x. [DOI] [PubMed] [Google Scholar]
- 49.Clarke M., Maddera L. Phagocyte meets prey: Uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur. J. Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Clarke M., Müller-Taubenberger A., Anderson K.I., Engel U., Gerisch G. Mechanically induced actin-mediated rocketing of phagosomes. Mol. Biol. Cell. 2006;17:4866–4875. doi: 10.1091/mbc.E06-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinrich D., Youssef S., Schroth-Diez B., Engel U., Aydin D. Actin-cytoskeleton dynamics in non-monotonic cell spreading. Cell Adhes. Migrat. 2008;2:58–68. doi: 10.4161/cam.2.2.6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


