Abstract
Fluorescence microscopy provides a powerful method to directly observe single enzymes moving along a DNA held in an extended conformation. In this work, we present results from single EcoRV enzymes labeled with quantum dots which interact with DNA manipulated by double optical tweezers. The application of quantum dots facilitated accurate enzyme tracking without photobleaching whereas the tweezers allowed us to precisely control the DNA extension. The labeling did not affect the biochemical activity of EcoRV checked by directly observing DNA digestion on the single molecule level. We used this system to demonstrate that during sliding, the enzyme stays in close contact with the DNA. Additionally, slight overstretching of the DNA resulted in a significant decrease of the 1D diffusion constant, which suggests that the deformation changes the energy landscape of the sliding interaction. Together with the simplicity of the setup, these results demonstrate that the combination of optical tweezers with fluorescence tracking is a powerful tool for the study of enzyme translocation along DNA.
Main Text
Recent years have seen the development of single molecule fluorescence into a valuable tool for the quantitative study of diffusive or directed translocation of proteins along DNA (1,2). In these approaches, DNA molecules were extended either on a surface or using a flow and fluorescence was usually detected by labeling with organic dyes (3–9).
In this study, we take this idea one step further and present a system that combines single molecule fluorescence microscopy with dual optical tweezers. Using this system, we measured the diffusion constant of single EcoRV type II restriction enzymes (8) along a DNA molecule held in solution by tweezers-manipulated beads. As the fluorescent probe we used semiconductor nanocrystals (quantum dots, (QDots)), which combine brightness with high photostability (10). The dual optical tweezers not only circumvent the need of a flow application or a nearby surface for DNA extension, but also allow for the localization of DNA molecules without DNA-binding dyes that could potentially affect the protein translocation (11). Furthermore, DNA extension can be adjusted at will and overstretching of the DNA can be achieved (12), which can yield new information about the DNA-enzyme interaction. Although the combination of optical tweezers with single molecule fluorescence tracking has been realized using organic dyes (13–15) previously, the superior brightness of the QDots enable localization suitable for accurate tracking without photobleaching. QDot labeling has been previously reported for the in vitro study of enzyme-DNA interactions (16), however, without the benefit of tweezers-manipulated DNA. Moreover, we directly observed for the first time, to our knowledge, DNA cleavage by a single fluorescently-labeled restriction enzyme, verifying that EcoRV activity is retained under our experimental conditions.
Experiments were carried out on a standard setup for single molecule fluorescence microscopy, which was combined with a dual optical tweezers setup (see the Supporting Material). Briefly, a fiber-coupled solid state laser (5 W output, 1070 nm) was split via a polarizing beam splitter into two independently steerable pathways which were recombined by a second beam splitter before coupling into the microscope. The trapping beams were imaged onto the back aperture of an oil-immersion objective (PLAPON, 60x, NA 1.42, Olympus, Tokyo, Japan) by a hot beam splitter placed below the standard beam splitter mount for fluorescence imaging. The fluorescence emission of QDots and the transmitted light from bright field illumination were recorded on an ICCD camera (I-PentaMAX-512AEF, Roper Scientific, Trenton, NJ). Beam tilting of the excitation laser light allowed us to increase the fluorescence signal/noise ratio significantly compared to epifluorescence illumination (17). Tracking of fluorescently-labeled enzymes was achieved similar to previous experiments (8) whereas the DNA extension was controlled by bright-field imaging of the beads (Supporting Material).
DNA attachment between two beads was accomplished in a single flow channel by attaching one bead to the surface via a second DNA molecule (see Fig. 1 A and Movie S2 and Movie S3 in the Supporting Material). This facilitated an easy assembly of bead-DNA constructs and, in case of a rupture of the DNA extended between the trapped beads, experiments could be directly resumed by picking up a new construct from the surface without the necessity of buffer exchange. The surface attachment via a second DNA enabled us to move the bead-DNA construct molecule far enough into the solution (>3 μm) to prevent any interaction with the surface.
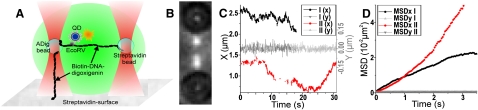
Figure 1.
Interaction of QDot-labeled EcoRV with tweezers-manipulated DNA. (A) Scheme of the setup: a DNA molecule is extended between two beads held by double optical traps, respectively. In addition, one of the beads is attached to the surface via a second DNA molecule. Enzymes sliding along the DNA are detected by the fluorescence emission of a QDot. (B) Overlaid bright field and fluorescence image of the two trapped beads and two successive sliding events of QDot-labeled enzymes on the extended DNA (129 nm/pixel). (C) Trajectories from the two interaction events recorded in B in X (direction of DNA extension) and Y direction. Gaps in the trajectories are caused by fluorescence intermittency. (D) MSD curves calculated from the interaction events in B depend linearly on time at short times in X direction, which demonstrates the sliding of the enzymes along the DNA. Nonlinear deviation at longer timescales can be explained by the stochastic nature of the linear diffusion (compare Fig. 2).
For fluorescence detection, a biotinylated EcoRV variant was labeled with commercially available semiconductor nanocrystals (Qdot 655, Invitrogen, Cergy, France), which were excited by the 488 nm line from an Argon laser. To ensure that QDot labeling did not alter the biological activity of the enzyme, DNA digestion was tested and directly visualized using a single-molecule assay based on surface-elongated DNA containing a single cognate site (Supporting Material).
Fig. 1 B–D (see also Movie S4) displays typical data obtained from single QDot-labeled enzymes sliding along a DNA molecule extended between two trapped beads. The use of QDots facilitates enzyme tracking with 40 ms time and 20 nm spatial resolution without restrictions by photobleaching. Although QDot fluorescence intermittency (“blinking”) occasionally leads to gaps in the enzyme trajectories, it does not affect the estimation of the enzyme diffusion constant by mean-square displacement (MSD) calculation (Supporting Material). Moreover, blinking can be used as a proof that the recorded interaction events involved a single QDot (18).
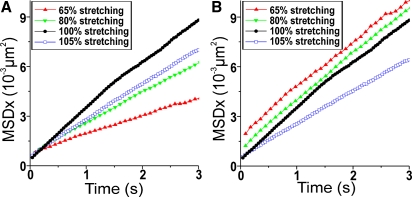
Apart from moving the DNA away from the surface, our setup allowed us to accurately adjust and determine the relative DNA extension sL (ratio of the end-to-end distance to the DNA contour length). We used this facility to demonstrate that during EcoRV interaction the enzyme follows the DNA contour, i.e., small jumps below the spatial resolution do not play a significant role in the observed translocation. Would small jumps be mainly responsible for the enzyme translocation, then the slope of the averaged MSD curves versus time should not depend on the relative extension sL of the DNA. In contrast, our data show that the slope increases significantly when sL is increased from 65% to 100% (Fig. 2 A). After correction for sL (8), however, we find identical values for the D1 within the experimental error (Fig. 2 B; D1 is 3.17 ± 0.23 × 10−3 μm2/s, 3.01 ± 0.2 × 10−3 μm2/s, and 3.15 ± 0.15 × 10−3 μm2/s for a DNA extension of 65%, 80%, and 100%, respectively), which confirms that sliding is the predominant mode of translocation of EcoRV along the DNA.
Figure 2.
Averaged MSD curves for different relative DNA extensions. Shown are the curves without (A) and with (B) correction for the relative extensions sL.
We also examined the translocation of QDot-labeled enzymes on surface extended DNA (Supporting Material) and obtained a similar value of D1 = 3.17 ± 0.38 × 10−3 μm2/s. This result shows that a nearby surface does not significantly alter the sliding of the enzyme, justifying previous experiments based on surface-attached DNA (8). Note that the experimental error is reduced in the tweezers-based setup, since the determination of sL, from which the main error contribution to D1 stems, is in this case more accurate. The reduced value of D1 for QDot-labeled EcoRV compared to dye-labeled EcoRV (D1(QDot) ≈ D1(dye)/3) can be explained by the larger hydrodynamic radius of the QDots compared to the enzyme, which leads to a decrease of the diffusion constant. Jumps, i.e., direct observation of fast 3D translocations larger than 200 nm (8), were also observed (Supporting Material, Movie S5); however, the limited number of jumps (15 jumps observed during 185 interaction events), made it difficult to deduce any quantitative information from the distribution of the jump lengths.
To confirm that QDot-labeling does not impede the biochemical activity of the enzyme, we used a single-molecule assay to directly visualize DNA digestion by a single fluorescently-labeled EcoRV enzyme (Supporting Material, Movie S6). Briefly, a DNA containing a single cognate site for EcoRV (GATATC) was immobilized on a surface in an extended conformation similar to previous experiments. After incubation with QDot-labeled EcoRV in a buffer containing Ca2+, site recognition by the labeled enzymes was observed as immobile bright spots on the extended DNA molecules. Rapid DNA cleavage was induced by flushing of the cell with a Mg2+ containing buffer, which ensures that the QDot-labeled enzymes are still biochemically active. To our knowledge, a direct observation of site recognition and subsequent DNA cleavage by a single restriction enzyme has never been reported before (2).
Although the extension-corrected D1 remains constant in the sL range from 65% to 100%, a different behavior was observed when the DNA was overstretched by only 5% with respect to its contour length. Corrected by sL, the measured D1 drops by more than 30% to 2.04 ± 0.15 μm2/s, a reduction that is much larger than can be accounted for by the experimental error. This indicates that the DNA deformation induced by a slight overstretching changes the energetic landscape of the sliding interaction. Although a more detailed discussion is beyond the scope of this work, this suggests that the DNA overstretching can yield useful information about the sliding mechanism. DNA force application is already a well established method to study the binding of immobile enzymes to the DNA (19).
In summary, we measured the linear diffusion of single QDot-labeled EcoRV on a DNA molecule extended by optical tweezers. The biochemical activity of labeled enzymes was validated by direct visualization of DNA cleavage with a single molecule assay. Using different DNA extensions, we could demonstrate that during sliding, the enzyme stays most of the time in close contact with the DNA, and that slight overstretching has a strong impact on DNA-EcoRV interaction. Together with the simplicity of our setup we are confident that our approach can be applied to a wide variety of different DNA-enzyme interactions.
Acknowledgements
We thank N. Porté for software development; I. Bonnet, E. Mercey, and F. Rusconi for DNA preparation; and K. Neuman, J.-F. Allemand, and M. Dahan for discussions and support.
This work was supported by funds from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Egide, Ministère de la Recherche (ACI Nanosciences NR069), Deutscher Akademischer Austauschdienst, Deutsche Forschungsgemeinschaft, and European Union. A.B. was supported by the Fondation pour la Recherche Médicale (20051206325) and CNano IdF-CNRS.
Supporting Material
References and Footnotes
- 1.van Mameren J., Peterman E.J.G., Wuite G.J.L. See me, feel me: methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Res. 2007;36:4381–4389. doi: 10.1093/nar/gkn412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorman J., Greene E.C. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 3.Handa N., Bianco P.R., Baskin R.J., Kowalczykowski S.C. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after Chi recognition. Mol. Cell. 2005;17:745–750. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Granéli A., Yeykal C.C., Robertson R.B., Greene E.C. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc. Natl. Acad. Sci. USA. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blainey P.C., van Oijen A.M., Banerjee A., Verdine G.L., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y.M., Austin R.H., Cox E.C. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 7.Kim J.H., Larson R.G. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet I., Biebricher A., Loverdo C., Voituriez R., Benichou O. Sliding and jumping of single EcoRV restriction enzymes on noncognate DNA. Nucleic Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tafvizi A., Huang F., Leith J.S., Fersht A.R., Mirny L.A., van Oijen A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alivisatos A.P. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996;271:933–937. [Google Scholar]
- 11.Crut A., Géron-Landre B., Bonnet I., Bonneau S., Desbiolles P. Detection of single DNA molecules by multicolor quantum-dot end-labeling. Nucleic Acids Res. 2005;33:e98. doi: 10.1093/nar/gni097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cluzel P., Lebrun A., Heller C., Lavery R., Viovy J.-L. DNA: An extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 13.Ishijima A., Kojima H., Funatsu T., Tokunaga M., Higuchi H. Simultaneous observation of individual ATPase and mechanical events by single myosin molecule during iteraction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 14.Harada Y., Funatsu T., Murakami K., Nonoyama Y., Ishijiama A. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Mameren J., Modesti M., Kanaar R., Wyman C., Wuite G.J.L. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophys. J. 2006;91:L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman J., Chowdhury A., Surtees J.A., Shimada J., Reichman D.R. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 18.Dahan M., Lévi S., Luccardini C., Rostaing P., Riveau B. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 19.Allemand J.-F., Bensimon D., Croquette V. Stretching DNA and RNA to probe their interactions with proteins. Curr. Opin. Struct. Biol. 2003;13:266–274. doi: 10.1016/s0959-440x(03)00067-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.