Abstract
The dysregulation of Ras-RAF signaling is associated with many types of human cancer. However, the kinetic and dynamic features of the mutual molecular recognition of Ras and RAF remain unknown. Here, we developed a technique for imaging single-pair fluorescence resonance energy transfer in living cells, and coupled this technique to single-molecule kinetic analysis to investigate how C-RAF (a subtype of RAF) molecules distinguish the active form of Ras (RasGTP) from the inactive form (RasGDP). Functional fragments of C-RAF containing the Ras-binding domains did not detect the switch in Ras activity in living cells as efficiently as did C-RAF. Single-molecule analysis showed that RasGDP associates with closed-conformation C-RAF, whereas the association of C-RAF with RasGTP immediately triggers the open RAF conformation, which induces an effective interaction between C-RAF and RasGTP. Spontaneous conformational changes from closed C-RAF to the open form rarely occur in quiescent cells. The conformational change in C-RAF is so important to Ras-RAF molecular recognition that C-RAF mutants lacking the conformational change cannot distinguish between RasGDP and RasGTP. The manipulation of the conformation of an effector molecule is a newly identified function of RasGTP.
Introduction
The Ras-RAF-MAPK system is a conserved intracellular reaction network that is involved in diverse biological functions, including growth, survival, and cell differentiation (1–3). In this network of protein reactions, the signaling between Ras and RAF is especially important because the dysregulation of this process is found in many types of human cancer (3–5). Ras is a member of the small GTPases and is inactive in the GDP-bound form (RasGDP). RasGDP is activated through GDP/GTP exchange (6), and the active GTP-bound form (RasGTP) interacts with the Ras-binding domains of downstream signaling proteins, called “effectors” (5,7). RAF, a cytoplasmic serine/threonine kinase, is one of the effectors of Ras (1–3). Irrespective of its nucleotide status, Ras is predominantly attached to the inner leaflet of the plasma membrane (8), and the association between RasGTP and RAF results in the translocation of cytoplasmic RAF to the plasma membrane, where RAF is activated by unknown kinases (3). Consequently, this translocation is the initiating event in RAF activation and signaling to the mitogen-activated protein kinase (MAPK) pathway (1,2).
Despite the biological importance of Ras-RAF signaling, the details of the molecular mechanism of Ras-RAF recognition remain unknown. The GDP/GTP exchange on Ras causes changes in its conformation (6), and it has long been thought that the accuracy of Ras-RAF signaling is maintained by an increase in the affinity between Ras and the Ras-binding domains of RAF with the conformational changes in Ras (6,9–13). Thus, RAF activation was considered to be regulated solely via the intracellular redistribution of RAF. However, recent reports have suggested a more complicated molecular recognition process. The N-terminal half of RAF contains two domains that contribute to Ras binding: the Ras-binding domain (RBD) and the cysteine-rich domain (CRD). Furthermore, two conformations have been detected in living cells, at least for a subtype of RAF (C-RAF), by means of fluorescence resonance energy transfer (FRET) measurements (14). In the inactive form of C-RAF, CRD is thought to form an intramolecular interaction with the C-terminal catalytic domain to suppress its kinase activity (15). FRET measurements suggest that C-RAF takes this “closed” conformation in quiescent cells, but in cells expressing a constitutively active mutant form of Ras (RasV12), RAF takes the elongated “open” conformation (14). It is thought that in the open conformation, CRD does not interact with the catalytic domain, and that in this conformation C-RAF is phosphorylated at several serine, threonine, and tyrosine residues, and is activated. Based on these recent findings, new models of Ras-RAF recognition have been proposed that include the functions of the two Ras-binding domains and conformational changes in RAF (3,16,17). However, these new models are based on biochemical ensemble measurements and low-resolution imaging studies. Thus, it remains unclear how the opening of the RAF conformation occurs and whether it is the cause or the result of RAF activation.
Here, we studied the molecular recognition between subtypes of Ras (H-Ras) and C-RAF in single molecules. In previous studies, we successfully used single-molecule kinetic analysis to reveal the details of intracellular molecular recognition (18–20), and we recently developed a technique to visualize intramolecular FRET signals from single molecules in living cells (21). In this study, we investigated the kinetics and dynamics of the molecular recognition between Ras and C-RAF using single-molecule kinetic analyses coupled with single-molecule imaging of the C-RAF conformation in living cells. We concentrated on the initial association state, which is essential for accurate molecular recognition. The kinetics of the later events in Ras-RAF recognition and the activation of RAF via phosphorylation will be reported elsewhere (K. Hibino, T. Shibata, T. Yanagida, and Y. Sako, unpublished results).
Materials and Methods
Plasmid construction and cell culture
pEGFP-C2, pEYFP-N1, pCMV-Ras, and pCMV-Raf-1 (c-raf) were purchased from Clontech (TaKaRa, Ohtsu, Japan). A monomeric mutation in green fluorescent protein (GFP; A206K) (22) was introduced into EG(Y)FP (mEG(Y)FP), and raf from pCMV-Raf-1 was cloned into pmEGFP-C2 (GFP-RAF). GFP-RBD (amino acids 51–131), RBDCRD (51–220)-GFP, and RAF-GFP were kindly provided by Dr. T. Balla (National Institute of Child Health and Human Development, National Institutes of Health; Bethesda, MD). GFP-RAF-yellow fluorescent protein (YFP) was made by reconstructing GFP-RAF, RAF-GFP, and pmEYFP-N1. The spacers between GFP and RAF, and between RAF and YFP were SGRTQISSSSFEF and RILQSTVPRARDPPVAT, respectively. Point mutations were introduced by site-directed mutagenesis, and all mutations were confirmed by DNA sequencing. HeLa cells were used for all experiments. The methods used for cell culture, transfection of plasmids, and stimulation with epidermal growth factor (EGF; Sigma-Aldrich, Tokyo, Japan) were described previously (18). For stimulation with EGF, the cells were incubated with 2–10 nM EGF and observed for 2–5 min after the addition of EGF to the culture medium at 25–27°C.
Microscopy and data processing
The intracellular distributions of the GFP-tagged proteins were observed in live HeLa cells using a scanning confocal microscope (TCS SP2; Leica, Welzlar, Germany) with a 100× oil-immersion objective (HCX PL APO CS ×100 1.4 oil; Leica), with excitation at 488 nm and detection at 510–600 nm. Image processing was performed with MetaMorph software (Molecular Devices, Sunnyvale, CA). The methods used for single-molecule imaging of GFP-tagged proteins were described previously (18). The images were acquired using an EB-CCD camera (C7190-20; Hamamatsu, Hamamatsu, Japan) equipped with an image intensifier (C8600-05; Hamamatsu) attached to a total internal reflection fluorescence (TIRF) microscope based on an IX-70 inverted microscope (Olympus, Tokyo, Japan), and recorded on digital videotape. Image processing was performed with MetaMorph and Image-Pro Plus (Media Cybanetics, Bethesda, MD). Single-molecule detection and tracking were performed with our custom-made software (21). Statistical and kinetic analyses were performed with Origin (OriginLab, Northampton, MA) and Mathematica (Wolfram, Champaign, IL).
Ensemble detection of FRET in GFP-RAF-YFP
The intramolecular FRET in GFP-RAF-YFP was visualized to detect conformational changes in RAF. This type of FRET probe was originally reported using cyan fluorescent protein (CFP) instead of GFP (14). We used GFP because the photostability of CFP is not sufficient for single-molecule imaging. The fluorescence emission spectra of GFP-RAF-YFP and its point mutants (C168S and S621A) were measured in live HeLa cells with a confocal microscope (TCS SP2). Specimens were illuminated with a 476 nm laser line, and 12 images at different emission wavelengths were acquired sequentially in the same field of view. The center of emission was shifted from 500 to 600 nm at regular intervals. The fluorescence emission spectra were measured in single cells coexpressing Ras and GFP-RAF-YFP. The emission spectra of GFP and YFP expressed individually in cells were used as references.
Intramolecular single-pair FRET imaging
Single molecules of GFP-RAF-YFP were visualized in live cells with the use of a TIRF microscope based on a TE-2000E inverted microscope (Nikon, Tokyo, Japan). The specimens were illuminated with a 488 nm laser (Sapphire 200; Coherent, Palo Alto, CA), and the fluorescence signal from the probe was directed through a 495 nm long-pass filter to block out laser scattering, and separated into GFP (500–525 nm) and YFP (525–540 nm) channels by means of custom-made dual-view optics (see Fig. S1 in the Supporting Material). To avoid the effects of photobleaching, we observed different cells before and after the addition of EGF. Image recording and data processing were performed as for the single-molecule GFP imaging. The GFP and YFP signals were separated after correction of the crosstalk between the GFP and YFP channels, using cells expressing only GFP or YFP as reference samples (Fig. S2).
Results
Intracellular distribution of RAF and its fragments containing Ras-binding domains
We prepared constructs to express GFP-tagged C-RAF (RAF) (GFP-RAF), RBD (GFP-RBD), or RBD-CRD (RBDCRD-GFP), and transfected the individual constructs into HeLa cells. The expression of all the GFP-tagged proteins in HeLa cells, at the correct molecular weights, was confirmed by immunoblot analysis (data not shown). Previous studies confirmed a direct interaction between Ras and these GFP-tagged proteins using intermolecular FRET imaging in cells (18) and immunoprecipitation (23). We monitored the localization of GFP fluorescence before and after the cells were stimulated with EGF, which induced the GDP/GTP exchange on Ras (Fig. 1 and Fig. S3). With the induced activation of Ras, GFP-RAF translocated from the cytoplasm to the plasma membrane, as previously reported (12,13,18). The time course of the translocation of RAF was negligibly affected by the GFP tag (18). Unlike RAF, GFP-RBD was mainly distributed in the cytoplasm, and even after Ras activation, only a small population of molecules moved to the plasma membrane. These results are similar to those of a previous study that used the same GFP-RBD construct (23). Conversely, RBDCRD-GFP was localized entirely at the plasma membrane, independently of Ras activation. The accumulation of RBDCRD-GFP was even observed in quiescent cells overexpressing a dominant negative mutant form of Ras (RasN17), which suppresses the basal activation of Ras, indicating that RBDCRD associates with inactive Ras molecules (Fig. S4). GFP alone was distributed in the cytoplasm and nucleus, independently of Ras activation. These results suggest that the dynamic interactions between the Ras-binding domains and the C-terminal catalytic domain of RAF are required for RAF to distinguish clearly between RasGDP and RasGTP, and consequently neither RBD nor RBDCRD, which lose these dynamic interactions, can fully distinguish between RasGDP and RasGTP in living cells. Confirmation of this suggestion requires a precise kinetic analysis of the interaction between Ras and RAF, and detection of the conformation of RAF. To achieve this, we used single-molecule detection in living cells (24,25).
Figure 1.
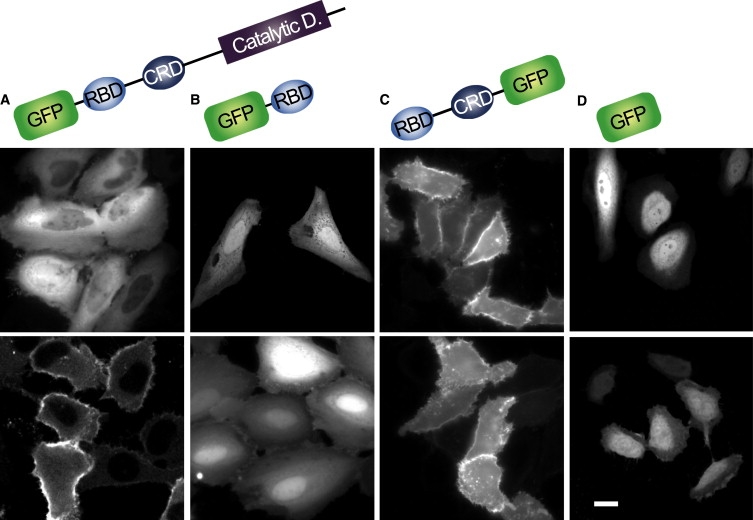
Intracellular distribution of RAF and its Ras-binding domains (RBD and CRD). GFP-RAF (A), GFP-RBD (B), RBDCRD-GFP (C), or GFP alone (D) was transiently coexpressed with Ras in HeLa cells. The intracellular distributions of the proteins were observed by scanning confocal microscopy before (upper panels) and after (lower panels) stimulation with EGF to induce Ras activation. GFP-RAF in quiescent cells (A, upper) and GFP-RBD (B) were predominantly distributed in the cytoplasm. GFP-RAF in cells after EGF stimulation (A, lower) and RBDCRD-GFP (C) were localized on the plasma membrane. GFP distributed in the cytoplasm and nucleus irrespective of the Ras activation (D). Scale bar: 10 μm. Catalytic D.: catalytic domain.
Single-molecule kinetic analysis of Ras-RAF recognition in living cells
Single molecules of GFP-RAF, GFP-RBD, and RBDCRD-GFP were observed in the basal membranes of living cells by TIRF microscopy (Figs. 2 A and Fig. S5, Movie S1, and Movie S2). Significant numbers of GFP fluorescent spots were observed in quiescent cells transfected with each GFP-tagged construct, indicating that single-molecule microscopy detected weak interactions between the RAF molecule and RasGDP (Fig. 2 A, left, and Movie S1) that could not be detected with confocal microscopy (Fig. 1). The GFP spots of these proteins in the quiescent cells associated specifically with RasGDP because the number of spots increased with the coexpression of Ras (Fig. S6). GFP alone associated only slightly with the plasma membrane (Fig. 2 B). The density of the GFP-RAF molecules increased with Ras activation, consistent with the RasGTP-dependent translocation of RAF from the cytoplasm to the plasma membrane (Fig. 2 A, right). The individual fluorescent GFP spots observed on the basal membranes of cells represented single molecules of GFP-tagged proteins. Single-molecule detection was confirmed by single-step photobleaching and the distribution of the fluorescence intensity of each spot (Fig. S5). Individual molecules of GFP-RAF (as well as GFP-RBD and RBDCRD-GFP) rapidly cycled between the cytoplasm and the plasma membrane, both before and after Ras activation (Movie S1 and Movie S2) (18).
Figure 2.
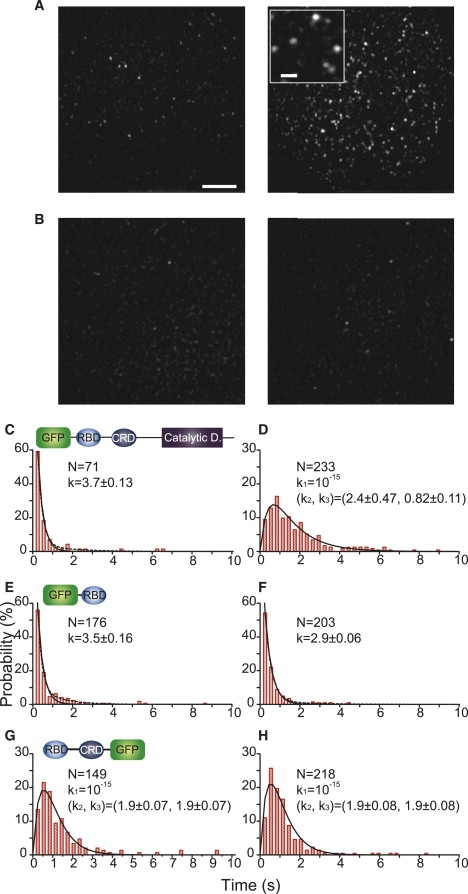
Single-molecule kinetics of dissociation between Ras and RAF. HeLa cells transiently expressing GFP-RAF (A) or GFP alone (B) were observed by TIRF microscopy before (left) and after (right) stimulation with EGF to induce Ras activation. Single molecules of GFP-RAF bound to the basal cell membrane were detected as fluorescent spots (18) (scale bar: 5 μm). The inset (A, right panel) is a magnified view of single GFP-RAF molecules (scale bar: 1 μm). Few single-molecule spots of GFP alone were observed on the basal cell membrane, indicating that the nonspecific binding of GFP to the plasma membrane is negligible. (C–H) On-time distributions of GFP-RAF (C and D), GFP-RBD (E and F), and RBDCRD-GFP (G and H) bound to Ras on the plasma membrane before (C, E, and G) and after (D, F, and H) stimulation with EGF. N indicates the number of spots examined. Solid lines were fitted to the data using kinetic models (Supporting Material). The best-fit values of the rate constants (s−1) obtained from these fittings are shown with the error ranges. Dotted lines in C, E, and F are the results of fitting with a two-component exponential function. The best-fit values and fractions of the fittings are 4.4 ± 0.27 s−1 (97%) and 0.66 ± 0.24 s−1 (3%) for C, 4.8 ± 0.49 s−1 (94%) and 0.85 ± 0.23 s−1 (6%) for E, and 3.2 ± 0.11 s−1 (98%) and 0.50 ± 0.19 s−1 (2%) for F.
The durations of the association of individual RAF molecules with the plasma membrane (on-times) were related kinetically to the interactions between Ras and RAF. Two patterns in the distribution of on-times were observed for GFP-RAF, GFP-RBD, and RBDCRD-GFP before and after Ras activation (Fig. 2, C–H), i.e., the on-times of GFP-RAF binding to RasGDP and GFP-RBD binding to RasGDP or RasGTP showed simple exponential distributions, whereas the on-time distributions of GFP-RAF binding to RasGTP and RBDCRD-GFP binding to RasGDP or RasGTP peaked, implying multiple rate-limiting steps in the dissociation process. These data indicate that the recruitment of RAF to the plasma membrane was not caused simply by an increase in the affinity between Ras and the Ras-binding domains of RAF. Instead, the qualitatively different reaction kinetics allowed RAF to distinguish between RasGDP and RasGTP.
The on-time distributions were analyzed with the use of mathematical models (Supporting Material). Molecules that did not accumulate or accumulated only weakly at the plasma membrane (RAF with RasGDP, and RBD) dissociated directly from the initial association state via a single stochastic rate-limiting step with a rate constant of 3–4 s−1. This result suggests that the weak accumulation of GFP-RBD on the plasma membrane of cells with RasGTP was probably caused by the higher association rate of RBD with RasGTP than with RasGDP. The similar rate constants for the dissociation of RAF from RasGDP and RBD from RasGDP (and RasGTP) suggest that RAF associates with RasGDP through the RBD of RAF. In contrast, for molecules that accumulated on the plasma membrane (RAF with RasGTP, and RBDCRD), the initial association passed through an intermediate state. Their direct dissociation from the initial association state was negligible, even though the rate of transition to the intermediate (0.8–2 s−1) was similar to or slower than the rates of the direct dissociation of RAF from RasGDP and RBD from RasGDP or RasGTP. Thus, the detected “initial association state” of RAF with RasGTP and RBDCRD with Ras differed from that of RAF with RasGDP and RBD with Ras.
Ensemble FRET measurements of the conformation of RAF molecules
The results of the kinetic analysis suggest the following hypothesis: RAF takes a closed conformation in quiescent cells and interacts with RasGDP only through the RBD, but when RAF is in contact with RasGTP, its closed conformation changes to an open conformation and it associates tightly with RasGTP via both the RBD and CRD. Regarding the interface between Ras and RAF, it is thought that RBDCRD is similar to the open state of RAF, which exposes both RBD and CRD to Ras, and that RBD is similar to the closed state of RAF, which exposes only RBD to Ras.
To confirm this hypothesis, we constructed a FRET-based probe (GFP-RAF-YFP) to detect the conformational changes in RAF (Fig. 3 A). The FRET signal from this probe was expected to decrease with the conformational change from closed to open. First, we evaluated whether this probe translocates with Ras activation. With Ras activation, GFP-RAF-YFP translocated from the cytoplasm to the plasma membrane, as do intrinsic RAF and GFP-RAF (18), with a normal time course after the stimulation of the cells (Fig. 3 B). The on-time distributions observed for single molecules of GFP-RAF-YFP were similar to those for GFP-RAF both before and after Ras activation; GFP-RAF-YFP dissociated from RasGDP in a single rate-limiting process, but dissociated from RasGTP in multiple rate-limiting processes (Fig. S7). Next, we confirmed the capacity of GFP-RAF-YFP to detect conformational changes in RAF in living cells (Fig. 4). The fluorescence emission spectrum of GFP-RAF-YFP (wild-type) in living cells under excitation at 476 nm was compared with those of two RAF mutants, GFP-RAF(C168S)-YFP and GFP-RAF(S621A)-YFP, which have been reported to be predominantly biased to the open conformation (14). For these measurements, the cells were not stimulated with EGF. S621A accumulated on the plasma membrane in quiescent cells (Fig. 5 C). No significant spectral differences were observed for the wild-type and C168S when they were in the cytoplasm or at the plasma membrane of quiescent cells. Compared with the spectrum for GFP-RAF-YFP (Fig. 4 A), the spectrum for each of the two mutants was depressed around the emission wavelength of YFP (Fig. 4, B and C). The emission spectrum of YFP or GFP alone was insensitive to stimulation with EGF (data not shown). These results indicate that this probe is useful for monitoring conformational changes in RAF molecules.
Figure 3.
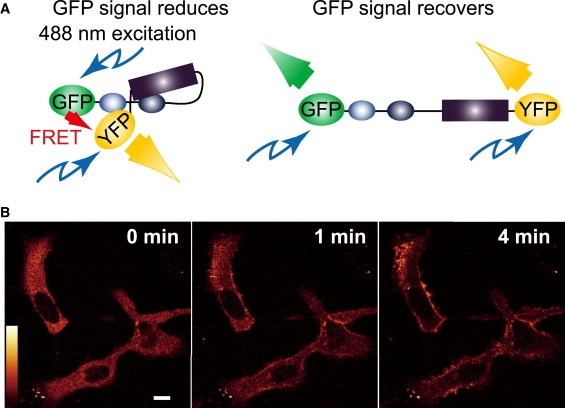
GFP-RAF-YFP probe. (A) Design of GFP-RAF-YFP probe to detect conformational changes in RAF. GFP signals were reduced by FRET from GFP to YFP in the closed conformation (left) and were recovered in the open conformation (right). (B) HeLa cells transiently coexpressing GFP-RAF-YFP and Ras were observed using a confocal microscope. Cells were stimulated with EGF to induce Ras activation, and successive images of the same cells were acquired at the indicated times after stimulation. These images show a summation of GFP and YFP fluorescence. Scale bar: 10 μm.
Figure 4.
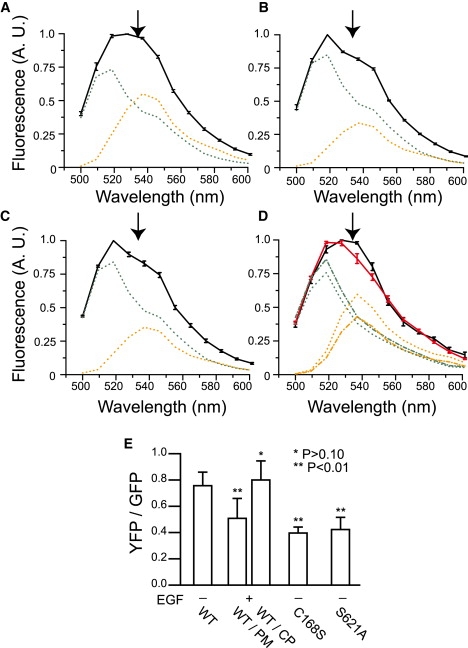
Ensemble detection of FRET in GFP-RAF-YFP. Fluorescence emission spectra were measured in single HeLa cells coexpressing Ras and GFP-RAF-YFP (A and D), Ras and GFP-RAF(C168S)-YFP (B), and Ras and GFP-RAF(S621A)-YFP (C). Only the cells in D were stimulated with EGF. The ensemble average for the spectra of six to seven cells is shown with the standard error. In D, the spectra in the cytoplasm (black) and at the plasma membrane (red) are shown separately. In the measurements shown in A and B, all regions of the cells had similar spectra. The spectra in C were obtained from the plasma membrane region. All spectra for the FRET probes are normalized to the peak intensity. The green and orange lines are the unmixed spectra for GFP and YFP, respectively, from the emission spectrum of the FRET probe observed under each condition. In D, dotted and dashed lines are unmixed spectra obtained from the cytoplasm and the plasma membrane, respectively. Depression of the spectrum around 530 nm (arrow) indicates low FRET efficiency, suggesting the open conformation of RAF. Each emission spectrum in A–D was separated into GFP and YFP signals, and the averages of the relative YFP signals normalized to the GFP signals are shown with their standard deviations (E). The YFP signals in B and C, and from the plasma membrane in D are significantly smaller than that in A.
Figure 5.
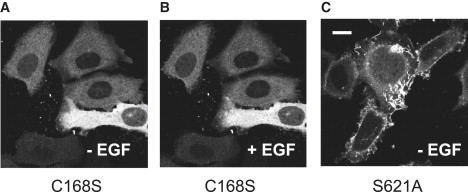
Intracellular distributions of RAF mutants. (A and B) C168S and (C) S621A mutants of RAF were coexpressed in HeLa cells with Ras. The cells were observed by confocal microscopy before (A and C) and after (B) stimulation with EGF to induce Ras activation. Scale bar: 10 μm.
The FRET signal (emission at YFP fluorescence) of GFP-RAF-YFP was reduced on the plasma membrane in cells <10 min after stimulation with EGF (Fig. 4, D and E). The spectrum of GFP-RAF-YFP that accumulated at the plasma membrane was similar to those of GFP-RAF(C168S)-YFP and GFP-RAF(S621A)-YFP, whereas the spectrum of GFP-RAF-YFP in the cytoplasm was similar to that in quiescent cells (Fig. 4). These results are consistent with the hypothesis that RAF adopts the closed conformation in the cytoplasm of quiescent cells and changes to the open conformation at the plasma membranes of stimulated cells, where RAF interacts with RasGTP.
Single-molecule FRET imaging of RAF molecules
The conformational change in RAF upon its association with RasGTP was confirmed by the detection of single-pair FRET in GFP-RAF-YFP molecules in living cells (Fig. 6 and Fig. S1, and Movie S3 and Movie S4). In quiescent cells, the fluorescence emission was suppressed in the GFP channel, even for the probe molecules that were retained at the cell surface by association with RasGDP (Fig. 6 A, upper, and Fig. S2, left), indicating a high FRET efficiency from GFP to YFP. In contrast, in cells stimulated with EGF to induce Ras activation, fluorescence emission was detected in both the GFP and YFP channels for the same individual probe molecules, indicating that FRET efficiency was reduced after Ras activation (Fig. 6 A, lower, and Fig. S2, right). Thus, like the results of the ensemble FRET measurements (Fig. 4), single-molecule FRET imaging suggests that RAF associates with RasGDP and RasGTP in its closed and open conformations, respectively. Fluorescence signals from GFP and YFP were separated by calculation at every fluorescent spot by removing the fluorescence leakage between the two channels (Fig. S2 B). In the single-pair FRET experiment, the GFP intensity was inversely proportional to the FRET efficiency. Binding to RasGTP rapidly induced conformational changes in RAF, as shown in the immediate (<0.1 s) reduction in FRET efficiency after the association of GFP-RAF-YFP with RasGTP on the plasma membrane (Fig. 6, B and C). This time course is consistent with our kinetic analysis, which indicated that the initial association state between RasGDP and RAF differed from that between RasGTP and RAF (Fig. 2).
Figure 6.
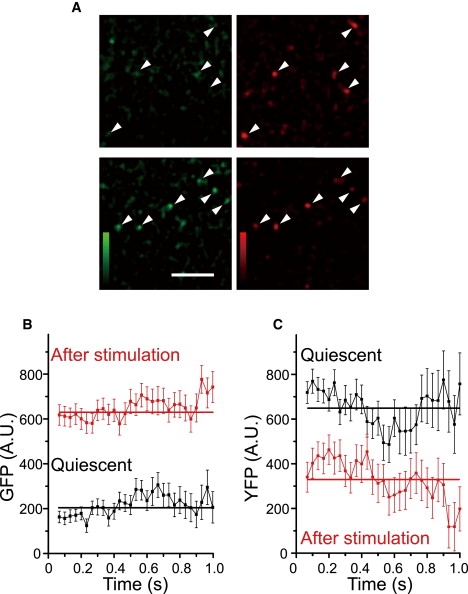
Single-pair FRET imaging in live cells. (A) Single molecules of GFP-RAF-YFP were observed in HeLa cells with a TIRF microscope before (upper) and after (lower) stimulation with EGF. Signals in the GFP (left, 500–525 nm) and YFP (right, 525–540 nm) channels were separated and detected simultaneously with dual-view optics (Fig. S1). Arrowheads indicate typical single molecules. Signals in the GFP channel increased upon stimulation with EGF, indicating conformational changes in GFP-RAF-YFP induced by RasGTP. Signals in the YFP channel at low FRET efficiency resulted from the direct excitation of YFP by the 488 nm laser beam and leakage of the GFP signals. Scale bar: 5 μm. (B and C) Changes in GFP (B) and YFP (C) fluorescence intensities are shown for GFP-RAF-YFP molecules after their association with Ras, with (red) or without (black) stimulation of the cells with EGF. The ensemble averages for the GFP signals from 242 (black) and 183 (red) molecules from two different cells under each condition are plotted with their standard error. Solid lines are time averages for the fluorescence signals. Because of the limited temporal resolution of the measurement, intensities within 0.1 s could not be determined (18).
Interaction between Ras and RAF mutants
Three point mutants of RAF (R89A, C168S, and S621A) were examined for their recognition of RasGTP. The point mutation of Arg89 is reported to disrupt the Ras-binding activity of RBD (26). Very little R89A mutant RAF was observed at the plasma membrane, similar to the case of GFP alone (unattached to the RAF molecule), both before and after the cells were stimulated with EGF (Figs. 2 B and 7, A and C). The C168S mutation disrupts both the Ras-binding activity of CRD and the intramolecular interaction between CRD and the catalytic domain (11,14,15,23,27,28). Because of its disruption of the intramolecular interaction, the C168S mutant RAF molecule adopts the open conformation (Fig. 4 B). However, in ensemble imaging, negligible accumulation of C168S at the membrane was observed (Fig. 5, A and B). In single molecules, an association between C168S RAF and RasGDP was observed (Fig. 7, B and C), with a frequency similar to that of the association between wild-type RAF and RasGDP (1.3 ± 0.18 s−1/100 μm2; Fig. 2 A, left). Compared with C168S, R89A RAF associated with Ras with lower frequency both before and after Ras activation. This result suggests that the RBD defines the association rate of RAF with Ras. The S621A mutation of RAF causes the loss of one of the phosphorylation sites, which act as the binding sites for the adaptor/scaffold protein 14-3-3 (3,16,17), and the S621A RAF molecule adopts the open conformation (Fig. 4 C). S621A RAF accumulated at the plasma membrane of quiescent cells (Fig. 5 C), like RBDCRD (Fig. 1 C).
Figure 7.
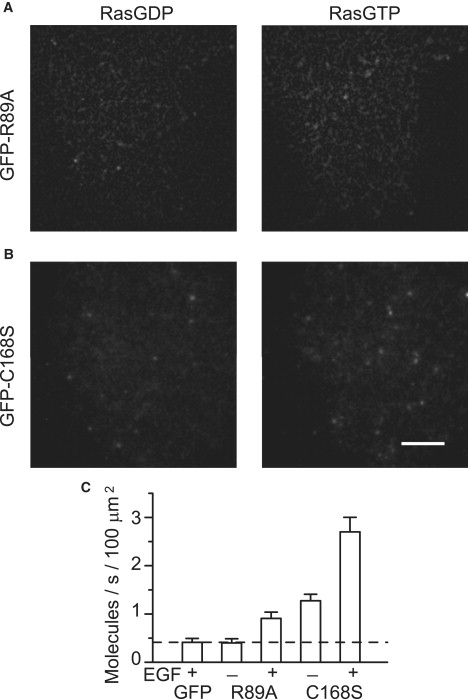
Single-molecule imaging of RAF mutants. (A) R89A and (B) C168S mutants of RAF were tagged with GFP and expressed in HeLa cells with Ras. The cells were observed by TIRF microscopy before (left) and after (right) stimulation with EGF to induce Ras activation (scale bar: 5 μm). (C) The averages of the molecules recruited to the basal cell membrane per second per 100 μm2 area of the membrane are shown with their standard error.
Discussion
Model of Ras-RAF recognition
The results of this study allow us to construct a model of how C-RAF distinguishes RasGTP from RasGDP (Fig. 8). C-RAF adopts a closed conformation in quiescent cells (Figs. 4 A and 6), in which the intramolecular interaction between CRD and the C-terminal domain of C-RAF suppresses the association between CRD and Ras and the catalytic activity of C-RAF. The closed conformation of C-RAF associates with RasGDP via RBD alone. This association is transient and C-RAF immediately dissociates from RasGDP (Figs. 2 and 7). In cells stimulated with EGF, in which Ras is activated through the GDP/GTP exchange on Ras, C-RAF in the cytoplasm still takes the closed conformation (Fig. 4 D), at least in the early phase of Ras activation, and interacts with RasGTP on the plasma membrane initially via RBD (Fig. 7). Because of the limited temporal resolution of our experiments, this earliest stage of the interaction between C-RAF and RasGTP was not observed. The conformation of C-RAF then rapidly changes to the open form (Figs. 4 D and 6) and C-RAF associates with RasGTP via both RBD and CRD (Fig. 2). This is the initial association state between C-RAF and RasGTP that we observed in this study. Consequently, the association of C-RAF with the plasma membrane is prolonged. This prolonged association must be essential for effective C-RAF activation.
Figure 8.
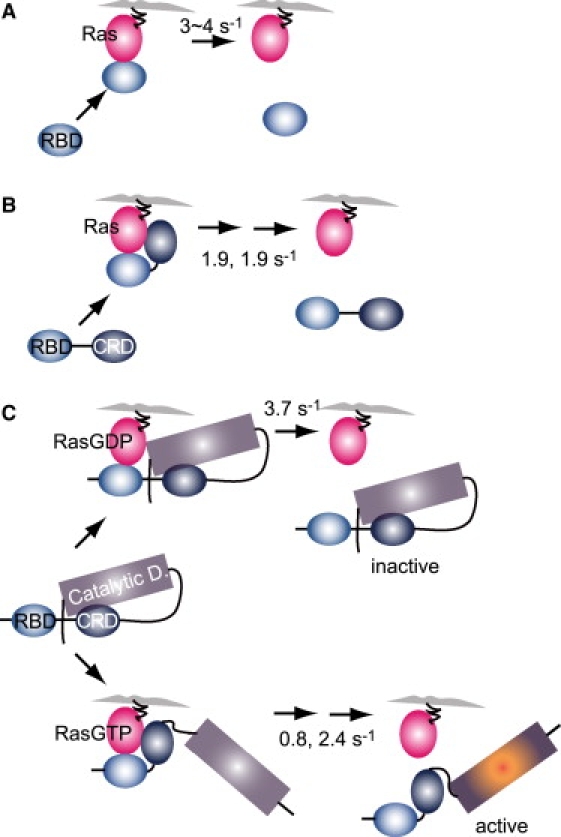
Models of the mutual molecular recognition of Ras and RAF. (A) Irrespective of the nucleotide status of Ras, RBD dissociates directly from the initial association state. (B) Irrespective of the nucleotide status of Ras, RBDCRD associates with Ras via both RBD and CRD in the initial association state, which changes to an intermediate state before its dissociation from Ras. (C) RAF takes a closed conformation in quiescent cells and interacts with RasGDP only through RBD. When RAF contacts RasGTP, RasGTP immediately changes the RAF structure to the open conformation (<100 ms) and RAF associates with RasGTP via both RBD and CRD. This process is essential for RAF to distinguish between RasGDP and RasGTP. Thereafter, like RBDCRD, RAF changes to an intermediate state before it dissociates from Ras.
Both the closed-to-open conformational change in C-RAF and its simultaneous association with Ras via its two Ras-binding domains are essential for C-RAF to distinguish RasGTP from RasGDP. The mutant molecules of C-RAF that are biased to the open conformation in the cytoplasm (RBDCRD and S621A) accumulate at the plasma membrane independently of Ras activation (Figs. 1 C and 5 C). Despite the open conformation of the C168S mutant in the cytoplasm (Fig. 4 B), the inhibition of the Ras-binding activity of CRD resulted in its loss of translocation to the plasma membrane after Ras activation (Fig. 5, A and B). The RBD mutant (R89A) also did not translocate to the plasma membrane after Ras activation (Fig. 7, A and C).
Interaction between RBD and Ras
In the single-molecule experiments, we detected small but significant numbers of GFP-RBD and GFP-RAF molecules on the plasma membrane of quiescent cells. These observations indicate a specific association between RBD of C-RAF and RasGDP. A small population of Ras could be in the GTP-bound form, even in quiescent cells. However, the on-time distributions of GFP-RAF were different in cells with RasGTP and quiescent cells (Fig. 2, C and D). The overexpression of Ras induced an increase in GFP-RAF molecules on the plasma membrane of quiescent cells (Fig. S6). This increase was not induced by an increase in spontaneously activated Ras molecules, because the on-time distribution was not altered by the overexpression of Ras (Fig. S6 D). It is plausible that RBD of C-RAF associates with RasGDP with a certain affinity.
Although many previous biochemical studies used the RBD fragment of C-RAF as a probe to detect RasGTP, we observed only a weak association between GFP-RBD and RasGTP in living cells. This discrepancy can be explained when we consider the effective concentrations of RasGTP and RBD in each experiment. In the biochemical studies, microbeads with a high density of protein on their surfaces were used for pull-down assays. Under such conditions, the GFP-RBD used in this study was able to pull down the active form of Ras (23). Similarly, in an intramolecular FRET probe to detect GEF activity for Ras, the distance between Ras and RBD is short (29–31). In recent imaging studies, the translocation of the GFP-tagged single RBD domain to the plasma membrane was observed only in cells overexpressing Ras (23,32,33). Therefore, a GFP-tagged trimeric RBD probe was used to detect Ras activation in normal cells (33). Moreover, in our experience with conventional microscopy, the coexpression of Ras is required to detect the translocation of GFP-RAF to the plasma membrane after Ras activation. However, in single-molecule imaging, the coexpression of Ras is not essential (Fig. 2 A). With other Ras effectors (phosphoinositide 3-kinase and phospholipase Cɛ), the GTP dependence of the association of RBD with Ras was observed in pull-down assays (34–36). With the Ras association (RA) domain of Ral guanine nucleotide exchange factor, GFP-tagged single RA detected the activation of endogenous Rap1 (a subtype of Ras) in COS-1 cells (37). However, in general, it is thought that GFP-RBD of Ras effectors is not sensitive enough to report the activation of Ras by simple recruitment in normal cells without the overexpression of Ras (38).
In a previous study (18), we measured the on-time distribution of GFP-RAF at a later stage (>30 min) after stimulation with EGF. In the later stage, GFP-RAF formed micrometer-scale patches that accumulated in the membrane. The on-time distribution of GFP-RAF in the bulk membrane, which was outside the patches, was similar to that of GFP-RAF in quiescent cells in this study. This result suggests that RasGTP rarely exists in the bulk membrane, where GFP-RAF interacts with RasGDP. However, the on-time was prolonged in the patches. Based on the results of this study, it is highly likely that both RasGDP and RasGTP are present in the patches, and that the on-time distribution is a mixture of RasGDP and RasGTP.
Spontaneous conformational changes in C-RAF in the cytoplasm
Our results suggest that the structure of C-RAF in quiescent cells is biased toward the closed conformation, and that the spontaneous fluctuation of the structure toward the open conformation is small. If this were not the case, C-RAF in the open conformation would bind firmly to both RasGDP and RasGTP, and C-RAF could not accurately recognize the activation of Ras, as with the RBDCRD fragment and the S621A mutant (Figs. 1 C, 2, and 5 C, and Fig. S4 B). As mentioned above, the phosphorylated serine 621 residue of C-RAF acts as the binding site for the adaptor/scaffold protein 14-3-3. It is thought that 14-3-3 crosslinks the phosphorylated S259 and S621 residues of C-RAF (16,17,39). S621 is constitutively phosphorylated, even in quiescent cells (40,41), and the mutation S621A induces a shift in the equilibrium of the C-RAF conformation to the open form (Fig. 4 C). Thus, 14-3-3 seems to function as a stabilizer of the closed conformation of C-RAF in quiescent cells, to avoid missignaling from RasGDP to C-RAF. However, its association with 14-3-3 may not completely suppress the conformational fluctuations of C-RAF. Another mutation, C168S, also induces a shift in the equilibrium of the C-RAF conformation to open (Fig. 4 B). The C168S mutation causes the loss of the intramolecular interaction between CRD and the C-terminal catalytic domain. Therefore, the cooperative effect of its association with 14-3-3 and its intramolecular interaction maintains C-RAF in the closed conformation in quiescent cells.
Activity of RasGTP in opening the conformation of C-RAF
Single-molecule analysis of the interactions between Ras and C-RAF revealed that RasGTP is not merely a high-affinity site for the recruitment of C-RAF to the plasma membrane; it also induces a change in the C-RAF conformation. RasGTP, but not RasGDP, actively opens the conformation of C-RAF from the closed state. This RasGTP activity has not been previously demonstrated, and seems to be triggered by an interaction between RBD of C-RAF and RasGTP. The R89A mutant of C-RAF, in which the ability of RBD to associate with Ras is lost, did not accumulate at the plasma membrane (Fig. 7, A and C), and another loss-of-function mutant of RBD, R89L, is reported to adopt the closed conformation, even in cells expressing RasV12 (14). After its association with RBD, RasGTP probably induces a dissociation of the intramolecular interaction between CRD and the C-terminus of C-RAF and/or the intermolecular interaction between C-RAF and 14-3-3, and RasGTP associates with the CRD of C-RAF in addition to RBD. Considering the structural similarity among the members of the RAF family, it is possible that the molecular recognition between RasGTP and other subtypes of RAF (A-RAF and B-RAF) is governed by a mechanism similar to that proposed between RasGTP and C-RAF in this study.
Activation of C-RAF through phosphorylation on the plasma membrane
After C-RAF is recruited by RasGTP to the plasma membrane, it is activated by its phosphorylation by unknown kinases on the plasma membrane (3). The fact that C-RAF associates with RasGDP and RasGTP in different conformations should be important to the high fidelity of the regulation of the kinase activity of C-RAF in association with RasGTP. Because intermolecular interactions are inherently stochastic, a subset of C-RAF molecules associated with RasGDP would encounter the kinase responsible for C-RAF activation, even though their affinity for RasGDP is low. Therefore, missignaling is inevitable, with a certain probability, if molecular switching depends on simple changes in affinity. The conformational change in C-RAF upon binding to RasGTP probably functions as a checkpoint for accurate signal transduction. It is possible that the open conformation of C-RAF is required for the presentation of C-RAF to the kinase. Clarification of the activation process of C-RAF after its association with RasGTP is the next challenge.
Acknowledgments
We thank Dr. Tamas Balla (National Institute of Child Health and Human Development, National Institutes of Health Bethesda, MD) for the GFP-RBD, RBDCRD-GFP, and RAF-GFP constructs, and members of our laboratories for valuable discussions and encouragement.
Supporting Material
References
- 1.Avruch J., Zhang X., Kryiakis J.M. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem. Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 2.Daum G., Eisenmann-Tappe I., Fries H.-W., Troppmair J., Rapp U.R. The ins and outs of Raf kinases. Trends Biochem. Sci. 1994;19:474–480. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 3.Wellbrock C., Karasarides M., Marais R. The Raf proteins take center stage. Nat. Rev. Mol. Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 4.Downward J. Targeting Ras signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 5.Repasky G.A., Chenette E.J., Der C.J. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Milburn M.V., Tong L., deVos A.M., Brunger A., Yamaizumi Z. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 7.Wittinghofer A., Nassar N. How Ras-related proteins talk to their effectors. Trends Biochem. Sci. 1996;21:488–491. doi: 10.1016/s0968-0004(96)10064-5. [DOI] [PubMed] [Google Scholar]
- 8.Hancock J.F. Ras proteins: different signals from different localizations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 9.Vojtek A.B., Hollenberg S.M., Cooper J.A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 10.Warne P.H., Viciana P.R., Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., Settleman J., Kyriakis J.M., Takeuchi-Suzuki E., Elledge S.J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 12.Leevers S.J., Paterson H.F., Marshall C.J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 13.Stokoe D., Macdonald S.G., Cadwallader K., Symons M., Hancock J.F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 14.Terai K., Matsuda M. Ras binding opens c-Raf to expose the docking site for mitogen-activated protein kinase kinase. EMBO Rep. 2005;6:251–255. doi: 10.1038/sj.embor.7400349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler R.E., Stephens R.M., Saracino M.R., Morrison D.K. Autoregulation of the Raf-1 serine/threonine kinase. Proc. Natl. Acad. Sci. USA. 1998;95:9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 17.Avruch J., Khokhlatchev A., Kyriakis J.M., Luo Z., Tzivion G. Ras activation of the Raf kinase: Tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 18.Hibino K., Watanabe T.M., Kozuka J., Iwane A.H., Okada T. Single- and multiple-molecule dynamics of the signaling from H-Ras to cRaf-1 visualized on the plasma membrane of living cells. ChemPhysChem. 2003;4:748–753. doi: 10.1002/cphc.200300731. [DOI] [PubMed] [Google Scholar]
- 19.Teramura Y., Ichinose J., Takagi H., Nishida K., Yanagida T. Single-molecule analysis of epidermal growth factor binding on the surface of living cells. EMBO J. 2006;25:4215–4222. doi: 10.1038/sj.emboj.7601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morimatsu M., Takagi H., Ota K.G., Iwamoto R., Yanagida T. Multiple-state reactions between the epidermal growth factor receptor and Grb2 as observed using single-molecule analysis. Proc. Natl. Acad. Sci. USA. 2007;104:18013–18018. doi: 10.1073/pnas.0701330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibino K., Hiroshima M., Takahashi M., Sako Y. Single-molecule imaging of fluorescent proteins expressed in living cells. Methods Mol. Biol. 2009;544:451–460. doi: 10.1007/978-1-59745-483-4_30. [DOI] [PubMed] [Google Scholar]
- 22.Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 23.Bondeva T., Balla A., Várnai P., Balla T. Structural determinants of Ras–Raf interaction analyzed in live cells. Mol. Biol. Cell. 2002;13:2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sako Y., Yanagida T. Single-molecule visualization in cell biology. Nat. Rev. Mol. Cell Biol. 2003;4:SS1–SS5. [PubMed] [Google Scholar]
- 25.Sako Y. Imaging single molecules in living cells for systems biology. Mol. Syst. Biol. 2006;56:1–6. doi: 10.1038/msb4100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian J.R., Vojtek A.B., Cooper J.A., Morrison D.K. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc. Natl. Acad. Sci. USA. 1994;91:5982–5986. doi: 10.1073/pnas.91.13.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruder J.T., Heidecker G., Rapp U. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 28.Luo Z., Diaz B., Marshall M.S., Avruch J. An intact Zinc finger is required for optimal binding to processed Ras and for Ras-dependent Raf activation in situ. Mol. Cell. Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 30.Sawano A., Takayama S., Matsuda M., Miyawaki A. Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev. Cell. 2002;3:245–257. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 31.Yokono T., Kotaniguchi H., Fukui Y. Clear imaging of Förster resonance energy transfer (FRET) signals of Ras activation by a time-lapse three-dimensional deconvolution system. J. Microsc. 2006;223:9–14. doi: 10.1111/j.1365-2818.2006.01593.x. [DOI] [PubMed] [Google Scholar]
- 32.Chiu V.K., Bivona T., Hach A., Sajous J.B., Silletti J. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 33.Augsten M., Pusch R., Biskup C., Rennert K., Wittig U. Live-cell imaging of endogenous Ras-GTP illustrates predominant Ras activation at the plasma membrane. EMBO Rep. 2006;7:46–51. doi: 10.1038/sj.embor.7400560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Viciana P., Warne P.H., Dhand R., Vanhaesebroeck B., Gout I. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Viciana P., Warne P.H., Vanhaesebroeck B., Waterfield M.D., Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 36.Song C., Hu C.-D., Masago M., Kariyai K., Yamawaki-Kataoka Y. Regulation of a novel human phospholipase C, PLCɛ, through membrane targeting by Ras. J. Biol. Chem. 2001;276:2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- 37.Bivona T.G., Wiener H.H., Ahearn I.M., Silletti J., Chiu V.K. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bivona T.G., Quatela S., Philips M.R. Analysis of Ras activation in living cells with GFP–RBD. Methods Enzymol. 2006;407:128–143. doi: 10.1016/S0076-6879(05)07012-6. [DOI] [PubMed] [Google Scholar]
- 39.Tzivion G., Luo Z., Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 40.Morrison D.K., Heidecker G., Rapp U.R., Copeland T.D. Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 41.Zhu J., Balan V., Bronisz A., Balan K., Sun H. Identification of Raf-1 S471 as a novel phosphorylation site critical Raf-1 and B-Raf kinase activities and for MEK binding. Mol. Biol. Cell. 2005;16:4733–4744. doi: 10.1091/mbc.E05-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


