Abstract
AmrZ, a member of the Ribbon-Helix-Helix family of DNA binding proteins, functions as both a transcriptional activator and repressor of multiple genes encoding Pseudomonas aeruginosa virulence factors. The expression of these virulence factors leads to chronic and sustained infections associated with worsening prognosis. In this study, we present the X-ray crystal structure of AmrZ in complex with DNA containing the repressor site, amrZ1. Binding of AmrZ to this site leads to auto-repression. AmrZ binds this DNA sequence as a dimer-of-dimers, and makes specific base contacts to two half sites, separated by a five base pair linker region. Analysis of the linker region shows a narrowing of the minor groove, causing significant distortions. AmrZ binding assays utilizing sequences containing variations in this linker region reveals that secondary structure of the DNA, conferred by the sequence of this region, is an important determinant in binding affinity. The results from these experiments allow for the creation of a model where both intrinsic structure of the DNA and specific nucleotide recognition are absolutely necessary for binding of the protein. We also examined AmrZ binding to the algD promoter, which results in activation of the alginate exopolysaccharide biosynthetic operon, and found the protein utilizes different interactions with this site. Finally, we tested the in vivo effects of this differential binding by switching the AmrZ binding site at algD, where it acts as an activator, for a repressor binding sequence and show that differences in binding alone do not affect transcriptional regulation.
Author Summary
The bacterium Pseudomonas aeruginosa causes a variety of human infections and is the leading cause of death in patients with cystic fibrosis. The main reason for the severity of these infections arises from the ability of P. aeruginosa to express virulence factors that protect it from the host immune system. Several of these processes are controlled by a transcription factor called AmrZ, a potential target for anti-microbial therapeutics. AmrZ is unusual in that it has the ability to both activate some genes, such as for alginate biofilm, and repress others, as with flagellum and itself. Here we determine the three dimensional structure of AmrZ bound to DNA containing a repressor sequence. Our structure shows the specific interactions the protein makes with the DNA for binding and repression. It also reveals that both the sequence and shape of the DNA are important for tight association. We next examined the binding of the protein to DNA containing an activator sequence and found that it has different interactions. However, by switching the AmrZ binding site at algD, where it acts as an activator, for a repressor binding sequence in P. aeruginosa, we show that differences in binding alone do not account for transcriptional regulation.
Introduction
Pseudomonas aeruginosa is an opportunistic, Gram negative bacterium that causes a variety of infections, mainly in immune-challenged patients [1]–[3]. More notably, chronic lung infection by P. aeruginosa is the leading cause of death in patients with the autosomal recessive disorder cystic fibrosis (CF) [4]. The underlying cause of the severity of these infections is due in part to the arsenal of virulence factors P. aeruginosa has at its disposal, including type III secretion systems, production of biofilms, phospholipase, exotoxin A, motility, and lipopolysaccharide. In alginate producing strains isolated from CF patients, the transcription factor AmrZ (Alginate and Motility Regulator Z, formerly AlgZ) is highly expressed [5]. Our previous work has shown AmrZ functions as both a transcriptional activator and repressor of several virulence factors. AmrZ is necessary for alginate production, via the activation of algD, which is the first gene in the alginate biosynthetic operon [6]. Reciprocal to this, AmrZ represses fleQ, which encodes an activator of flagellum expression [7]. AmrZ is also required for the regulation of genes responsible for type IV pili localization and twitching motility, through the interaction with a currently unknown gene target [8]. Finally, AmrZ also represses its own transcription by binding to two sites on the amrZ promoter, amrZ1 and amrZ2 [9].
The 108 amino acid, 12.3 kD AmrZ protein is a member of the ribbon-helix-helix (RHH) family of DNA binding proteins, sharing highest sequence similarity to the Arc and Mnt repressors from bacteriophage P22 [10]. Sequence analyses predict that there are over 2300 proteins containing RHH domains found in bacteria, Archaea, and bacteriophages; however, less than twenty of these proteins have been studied with structural or biochemical techniques [11]. Structural information from RHH proteins both in the presence [12]–[19] and absence [20]–[27] of operator DNA, show that they exist as dimers, formed by a hydrophobic core created by the two α-helices. The majority of RHH proteins are transcriptional repressors. AmrZ and Helicobacter pylori NikR are currently the only characterized RHH proteins known to function as both transcriptional activators and repressors [23]. DNA binding by RHH proteins occurs by the insertion of the anti-parallel β-sheet formed by one β-strand from each monomer into the major groove of DNA. The interactions between the protein and the recognition site are very specific, and mutations to either the DNA binding β-sheet, or the operator site often have a negative effect on DNA binding [27], [28]. In addition to binding DNA as a dimer, RHH proteins also assemble as tetramers, which are stabilized by other domains in the protein, such as occurs with the C-terminal domain of Mnt [29].
Information from sequence alignments and structural predictions define three regions of the AmrZ protein, an extended N-terminus spanning residues 1–16, the RHH domain, located from residues 13–66, and a C-terminal domain from residues 67–108 [30]. Both the extended N-terminus and the C-terminal domain do not share any sequence similarity to other proteins, and their exact function has remained an open question. The extended N-terminus has been hypothesized to play a role in DNA binding, and is conserved in other AmrZ orthologs of P. putida and P. syringae [31]. Extended N-termini of other RHH proteins have been examined, although their functions vary between cofactor binding, oligomerization and protein-protein interactions, ATP hydrolysis, in addition to having roles in DNA recognition. The C-terminal domain of AmrZ is proposed to be involved in protein oligomerization, which is supported by glutaraldehyde cross linking assays that show AmrZ forms oligomeric species consistent with the molecular weight of dimers and tetramers in solution [30].
Of the genes that are regulated by AmrZ, the specific locations of the binding sites are only known for two of them. AmrZ functions as a transcriptional activator at the algD promoter and binds 282 base pairs upstream of the transcriptional start site. Additionally, AmrZ acts as a transcriptional repressor of its own gene and recognizes two sites on the amrZ promoter (amrZ1 and amrZ2) at positions −93 and −161. Interestingly, DNA foot-printing has been performed at each of these three sites, and little sequence consensus is shared among them [6], [9]. Both the algD operon and amrZ are under the control of the alternative sigma factor AlgT (AlgU/σ22) [32]. Expression of the algD operon requires additional factors, including the response regulators AlgB and AlgR, the nucleosome proteins IHF and AlgP, and the AlgQ protein, each being necessary, but not sufficient to activate transcription on their own [5]. To date, only the AmrZ and AlgT proteins are known to interact with the amrZ promoter region.
Open questions have remained as to the exact strategies employed by AmrZ to function as both a transcriptional activator and repressor. It is unclear what specific interactions the protein makes with both activator and repressor sequences within DNA in order to carry out these functions. To answer these questions, we determined the crystal structure of an AmrZ C-terminal truncation mutant, Δ42 AmrZ, in complex with an 18 bp oligonucleotide containing the amrZ1-binding site. This structure defines the specific recognition site as two half sites separated by a linker region, and provides evidence that the extended N-terminus of AmrZ interacts with the DNA in a sequence independent manner. Site directed mutagenesis experiments of the amrZ1 DNA reveal that recognition is not only based on the direct readout of the nucleotide sequences, but also relies on recognition of the intrinsic shape of the DNA. These data allow for the creation of a model for transcriptional repression by AmrZ, where a combination of specific base recognition at two half sites and recognition of intrinsic DNA structure allow for binding. We also examined the interaction of AmrZ with the algD-binding site, where AmrZ binding functions as an activator of alginate biosynthesis. The results from these assays only identify one AmrZ binding site on algD and also suggest that the protein may utilize an additional residue in DNA binding. Finally, we demonstrate that while there are different protein interactions at the activator and repressor sequences, these differences alone do not account for the activator and repressor activity of AmrZ.
Results/Discussion
Structural overview of the Δ42 AmrZ: 18 bp amrZ1 complex
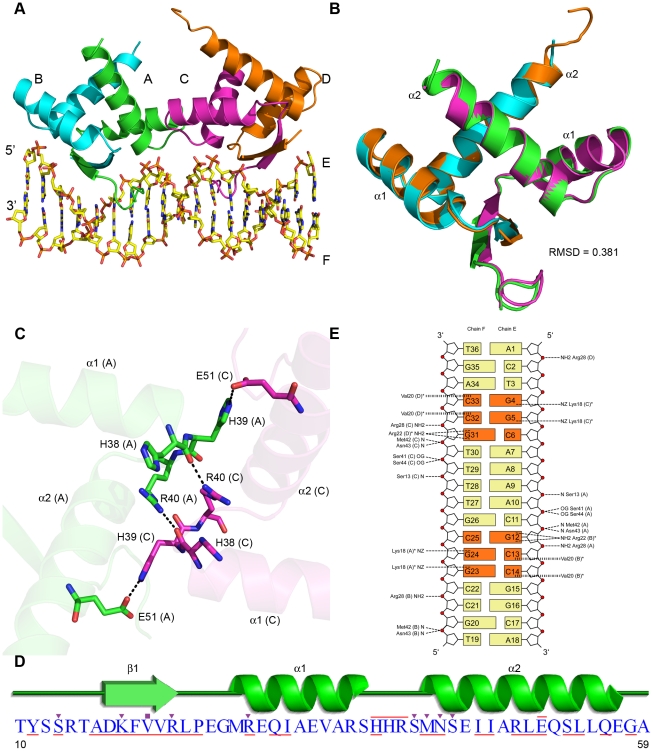
We determined the structure of a C-terminal truncation mutant of AmrZ, Δ42, in complex with an 18 bp oligonucleotide containing the amrZ1 site to 3.1 Å resolution (Table 1 and Figure 1A). The Δ42 variant of AmrZ (residues 1–66) contains the extended N-terminus and the RHH DNA binding domain, but has a truncated C-terminal domain. Many C-terminal truncation variants of AmrZ were used in crystallization experiments, and Δ42 was the only AmrZ construct tested that crystallized either in the presence or absence of DNA. The Δ42 AmrZ protein was tested for DNA binding affinity, and compared with the wild type protein, no reduction in affinity to any of the three known AmrZ binding sequences (amrZ1/amrZ2/algD) was observed (data not shown). The structure reveals AmrZ binds the amrZ1 site as a dimer-of-dimers, and DNA recognition occurs by the interaction with two half sites on the DNA, separated by five base pairs. There are no major structural differences between each AmrZ dimer (Cα RMSD = 0.381 Å) (Figure 1B). The dimer-dimer interface, which occludes approximately 290 Å2 of surface area on each dimer, is formed by a series of interactions between specific residues located on the loop connecting α-helix 1 and α-helix 2 on chains B and C (Figure 1C). The interactions in this region are symmetric, with the backbone carbonyl of His38 of one protomer forming a hydrogen bond to Arg40 of the opposing protomer, and the side chain of His39 forming a salt bridge to Glu51, also across the interface. The relatively small interface between each dimer, in combination with evidence that AmrZ forms higher order oligomers in solution [30], suggests there are likely additional dimer-dimer interactions mediated by the C-terminal domain of the protein.
Table 1. Crystallographic data collection and refinement statistics.
| Data Collection Statistics | |
| Wavelength (nm) | 0.9793 |
| Spacegroup | I422 |
| Unit Cell Parameters (a, b, c)(Å) | 129.5, 129.5, 152.6 |
| (α,β,γ)(°) | 90, 90, 90 |
| # Complexes/Asymmetric Unit | 1 |
| Unique Reflections | 22435 |
| Resolution Range (Å) | 40.00 – 3.1 (3.15 – 3.1) |
| Mean Redundancy | 5.7 (5.8) |
| Overall Completeness (%) | 99.7 (100.0) |
| Rmerge (%)(a) | 6.9 (54.3) |
| Mean I/σ | 29.0 (2.98) |
| # Se Atoms Found | 8 |
| FOM initial | 0.41 |
| FOM after density modification | 0.65 |
| Model Refinement Statistics | |
| Rwork (%) | 26.1 |
| Rfree (%)(b) | 29.5 |
| # Protein Atoms | 1522 |
| # DNA Atoms | 731 |
| # Water Molecules | 7 |
| RMSD Bond Angles (°) | 0.006 |
| RMSD Bond Lengths (Å) | 1.180 |
| Ramachandran Statisticsc | |
| Most Favored Regions | 153 (89.0%) |
| Additionally Allowed Regions | 19 (11.0%) |
| Generously Allowed Regions | 0 (0.0%) |
| Disallowed Regions | 0 (0.0%) |
Rmerge = (Σ|I−<I>|)/ΣI, where I is the observed intensity and <I> is the average intensity.
Rfactor = Σ∥Fo|−|Fc∥/Σ|Fo|. Rfree is calculated with the same equation, but with 5% of reflections not used in the refinement.
Ramachandran statistics are given as the number of amino acids that lie within each region, and the percentage is given in parenthesis.
Values in parenthesis are for the outermost resolution shell (3.15 Å – 3.1 Å).
Figure 1. Structural overview of AmrZ - amrZ1 complex.
(A) The Δ42 AmrZ protein binds to the 18 bp amrZ1 binding site as a dimer of dimers. One dimer is composed of chains A and B (green/cyan), while the other dimer is composed of chains C and D (magenta/orange). (B) The superposition of AmrZ dimers show no major structural differences between them (Cα RMSD = 0.381 Å). (C) The dimer - dimer interface is created by a network of hydrogen bonds between the residues in the loop region between α-helix 1 and α-helix 2 of chains A and C. (D) Secondary structure representation of one AmrZ ribbon-helix-helix monomer. Residues forming hydrogen bonds to DNA are indicated by the purple triangles, while residues forming hydrophobic interactions to DNA are indicated by purple squares. Residues forming the dimer interface between each monomer are underlined in red, and the residues which form the dimer-dimer interface are overlined in red. (E) Schematic of both the sequence dependent and sequence independent interactions between AmrZ and amrZ1. Hydrogen bonding interactions to the DNA are illustrated with a short dashed line, while hydrophobic interactions are illustrated with a vertical dashed line. Nucleotides involved in sequence specific interactions are represented in orange. The peptide chain for each residue is labeled in parentheses, and residues that make contacts to more than one nucleotide are notated with an asterisk.
The interface between AmrZ monomers to form a dimer is primarily composed of α-helix 1 and α-helix 2 of each monomer that come together to form a hydrophobic core. The dimer interface is quite extensive, composed of 25 residues (Figure 1D, underlined residues) and buries approximately 1600 Å2 of each monomer. Each AmrZ dimer interacts with the amrZ1 binding site through sequence dependent interactions (see below), mediated by the insertion of the anti-parallel β-sheet, formed by dimerization, into the major groove of DNA. Additionally, a number of sequence independent interactions to the phosphate backbone are formed, further supporting the protein-DNA complex (Figure 1E). The protein - DNA interactions exclude a total surface area of 1469 Å2 and are symmetric on both halves of the DNA. The one exception to this is the α-helix 2 N-terminus of chain D, which is not positioned to interact with the phosphate backbone; however, this is most likely due to the lack of a 5′ phosphate group on the nucleotide A1, an artifact of chemical DNA synthesis.
Sequence dependent binding by AmrZ
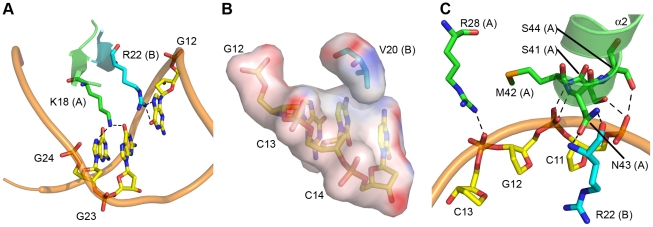
The structure allows us to determine the specific nucleotide sequence recognized by AmrZ, as well as other factors that contribute to DNA recognition. The insertion of the anti-parallel β-sheet from each AmrZ dimer into the major groove of the amrZ1 site provides for the recognition of two half sites, each with the sequence 5′-GGC (Figure 1E, orange bases). Sequence dependent binding by AmrZ occurs via the interaction of three residues, Lys18, Val20, and Arg22, with the nucleotides. Lys18 from one AmrZ monomer is positioned where it can form hydrogen bonding interactions to the O6 and N7 atoms of the two guanine nucleotides, G23 and G24 (G4 and G5 on the other half site) (Figure 2A). The DNA binding β-sheet also orients the residue Arg22, from the other monomer of the dimer, to form a bidentate hydrogen bond to both the O6 and N7 atoms of the nucleotide G12 (G31 on the other half site). This is on the opposite strand of the two bases with which Lys18 interacts. Bidentate hydrogen bonding, specifically between arginine residues and guanine nucleotides, are a major determinant in the selectivity of DNA bases [33]. Another relevant residue located on the DNA binding β-sheet is Val20. Interestingly, among other RHH proteins, this position in the DNA binding β-sheet is generally conserved as a neutral hydrophilic residue. One other exception to this is the Neisseria gonorrhoeae FitAB protein in which there is also a valine at this position [18]. The structure of FitAB in complex with DNA shows the valine forms a van der Waals interaction with the C5 methyl group of a thymine base. In the AmrZ structure, it appears that residue Val20 is poised to select for cytosine bases C13 and C14 (C32 and C33 on other half site) via a hydrophobic interaction (Figure 2B). Purine nucleotides would not be favorable in these locations since the N7 atom of the purine base would interfere with the hydrophobic pocket that is formed by Val20, while a thymine nucleotide in this position would sterically clash with the isopropyl side chain of the valine residue.
Figure 2. AmrZ - amrZ1 interactions.
(A) Sequence dependent binding by AmrZ is mediated by the insertion of the DNA binding β-sheet into the major groove of DNA. AmrZ recognizes two half sites on the DNA, each with the sequence 5′-GGC. Lys18 is positioned to form hydrogen bonds to multiple O6 and N7 atoms on the adjacent guanine nucleotides, G23 and G24 (G4 and G5 on other half site), while Arg22 forms hydrogen bonds to the O6 and N7 atoms on G12 (G31 on other half site). (B) Electrostatic surface representation of Val20, which is positioned to form hydrophobic interactions to the two nucleotides C13 and C14 (C32 and C33 on other half site). Electropositive surface potential is denoted in blue, while an electronegative surface potential is denoted in red. (C) Sequence independent binding by AmrZ is mediated by the N-terminus of α-helix 2, as well as Arg28 from α-helix 1, which form hydrogen bonding interactions to phosphodiester backbone of multiple nucleotides.
To confirm the requirement for each half site in amrZ1 recognition by AmrZ, a series of mutations were created to the amrZ1 DNA binding site. Both 1 and 2 nucleotides in each amrZ1 half site were mutated, and the affinity of WT AmrZ to each of these mutant sequences was measured using fluorescence anisotropy (Table 2). Mutating one nucleotide in each 5′-GGC AmrZ recognition half site to 5′-GTC caused a 9.8 fold reduction in affinity, while mutating two of the nucleotides in each half site to 5′-TTC caused a 12.9 fold reduction in affinity compared to binding to the native amrZ1 sequence. These results confirm the observations from the structure that sequence dependent recognition occurs through the interactions with two half sites, each with the sequence 5′-GGC.
Table 2. AmrZ affinity for amrZ1 binding site mutants.
| Binding Site | Sequencea | Kd b (nM) ± SE | Fold over WT amrZ1 c |
| WT amrZ1 | GTACTGGCAAAACGCCGGCACG CATGACCGTTTTGCGGCCGTGC | 8.41±0.8 | 1.0 |
| GTC amrZ1 | GTACTGTCAAAACGACGGCACG CATGACAGTTTTGCTGCCGTGC | 82.7±8.6 | 9.8 |
| TTC amrZ1 | GTACTTTCAAAACGAAGGCACG CATGAAAGTTTTGCTTCCGTGC | 109±13 | 12.9 |
| CGCG amrZ1 | GTACTGGCCGCGCGCCGGCACG CATGACCGGCGCGCGGCCGTGC | 63.4±7.3 | 7.5 |
| TTC/GC amrZ1 | GTACTTTCCGCGCGAAGGCACG CATGAAAGGCGCGCTTCCGTGC | 2050±180 | 244 |
| TTTT amrZ1 | GTACTGGCTTTTCGCCGGCACG CATGACCGAAAAGCGGCCGTGC | 41.7±5.1 | 5.0 |
| AATT amrZ1 | GTACTGGCAATTCGCCGGCACG CATGACCGTTAAGCGGCCGTGC | 58.6±7.5 | 7.0 |
| ATAT amrZ1 | GTACTGGCATATCGCCGGCACG CATGACCGTATAGCGGCCGTGC | 39.1±4.6 | 4.7 |
The two AmrZ binding sites on the wild type amrZ1 sequence are represented by the underlined nucleotides. Mutations to the wild type amrZ1 binding site are notated by the bolded nucleotides in each mutant sequence.
The Kd was calculated by fitting the hyperbolic equation for a single ligand binding model with saturation (eq 2) to the data in Figure S2, which were averaged from four independent experiments.
Fold over (wild type) WT amrZ1 is defined by (Kd of sample)/(Kd of wild type) for each sample.
We previously evaluated the contribution of Lys18 and Arg22 to AmrZ activity using in vitro DNA binding assays at amrZ1 and transcriptional reporter assays to measure amrZ repression [30]. The mutation of Lys18 to an alanine (K18A) resulted in a drastic reduction in the DNA binding activity, causing a 274-fold increase in the dissociation constant (Kd), compared to WT AmrZ. When amrZ was replaced in the P. aeruginosa chromosome with a gene encoding K18A AmrZ, amrZ derepression was observed, which was similar in magnitude to that observed in strains harboring null amrZ alleles. Similar results were obtained for the R22A mutant of AmrZ, which had a 44-fold increase in Kd compared to WT AmrZ in vitro, and comparable effects of amrZ transcription in vivo. When the effects of mutating the valine at position 20 to an alanine (V20A) were tested in vitro, a 10-fold increase in Kd was observed. Even with a smaller reduction in DNA binding ability compared to the K18A and R22A mutants, V20A AmrZ was unable to repress amrZ transcription in vivo.
The extended N-terminus of AmrZ
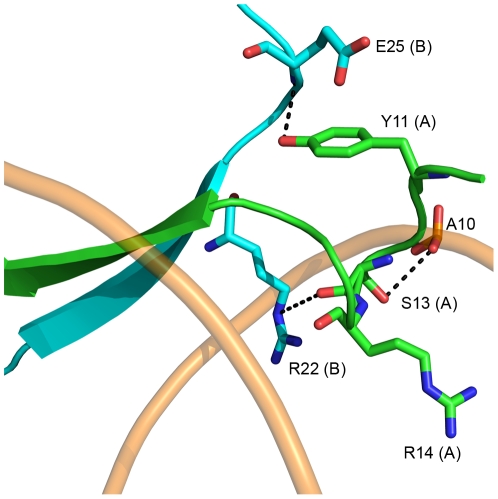
We observe electron density for the extended N-terminus starting at residue 10 on chains A and C. This density is only observed on the side of the AmrZ dimer that makes the specific contacts to the amrZ1 binding site. The lack of electron density of the extended N-terminus on the side of the AmrZ dimer that does not contact the DNA suggests that the N-terminus is disordered in solution, and becomes structured upon DNA binding. Residues 10–17 of the N-terminus form a looped structure, allowing the amino acids Ser13 and Arg14 in the major groove to interact with the DNA (Figure 3). This looped structure is supported by the residue Tyr11, which forms a hydrogen bond to the backbone of Glu25 from the other monomer in the dimer, and by the head-on orientation of the carboxyl side chain of Glu25 perpendicular to the aromatic ring of Tyr11. The side chain of Ser13 forms a hydrogen bond to a phosphate in the DNA backbone, and also positions the residue Arg14 into the major groove of the DNA; however, no contacts between Arg14 and the DNA bases are observed in the structure. This is consistent with previous studies of an AmrZ R14A mutant, which has no change in binding affinity for the amrZ1 DNA when compared to WT protein in vitro, as well as no effect on amrZ repression in vivo [30]. Other RHH proteins contain extended N-termini that contribute to DNA binding. The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein has a 16 residue N-terminal domain that is necessary for recognition of at least one of the binding sites of the protein [19]. Additionally, the seven residue extended N-terminus of the Arc repressor is disordered in solution, but adopts a tandem-turn structure upon binding DNA [13], and mutations to the N-terminus result in decreased binding to operator sites [34]. Although mutations to the extended N-terminus in AmrZ do not reduce affinity to the amrZ1 repressor site, AmrZ may act in a manner similar to ArtA, where the extended N-terminus may provide specificity for DNA binding at other sites.
Figure 3. The extended N-terminus of AmrZ.
The N-terminus of AmrZ is ordered only when in contact with the DNA (shown as wheat colored tubes). It forms a looped structure which is stabilized by Glu25 from chain B and Tyr11 from chain A. This region positions the side chains of two residues, Ser13 and Arg14 in the major groove of DNA. Ser13 forms hydrogen bonding contacts to the phosphate backbone, while Arg14 is not positioned to contact any nucleotides. This loop also interacts with the DNA binding residue Arg22 via the hydrogen bond between the backbone carbonyl of Ser13 and the εN of the arginine side chain.
Sequence independent binding
There are a number of interactions between AmrZ and the phosphodiester backbone of the DNA that act to position the DNA binding β-sheet in the major groove. The majority of these sequence independent interactions occur with residues located in the N-terminus of α-helix 2, which points down towards the DNA backbone (Figure 2C). The positioning of this helix allows the formation of hydrogen bonding interactions between the side chains of Ser41 and Ser44, and the backbone amide nitrogens of Met42 and Asn43 to the phosphate groups of the DNA. This interaction is further bolstered by the positive dipole of the N-terminal end of α-helix 2 and the negatively charged phosphate backbone of the DNA. Another sequence independent interaction between AmrZ and the DNA occurs via the side chain of Arg28, from α-helix 1, to the backbone of the DNA. Interestingly, the location of α-helix 2 also allows the side chain of Asn43 to form two hydrogen bonding interactions to the backbone amide nitrogen and carbonyl oxygen of the DNA binding residue, Arg22, in the opposite monomer. This also helps position Arg22 for interaction with the DNA bases. Sequence independent interactions formed by the N-terminus of α-helix 2 are one of the main structural features of RHH proteins [11]. These contacts are often observed to anchor the protein onto the DNA; however, in the case of AmrZ, this electrostatic interaction may play an additional role in recognition of the intrinsic shape of the DNA, particularly in the linker region.
The linker region between amrZ1 half sites confers specificity
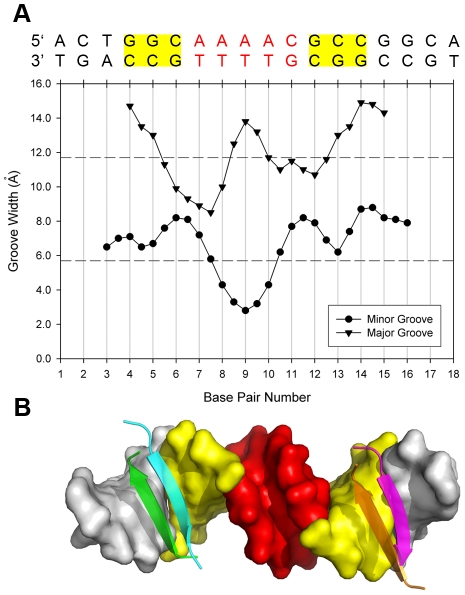
Analysis of the amrZ1 DNA in the structure reveals a significant narrowing of the minor groove to 2.8 Å in the A/T rich region between the two amrZ1 half sites (Figure 4A). In addition to the narrow minor groove, there is an increase in the width of the major grooves where AmrZ interacts with each half site, most likely to accommodate the width of the anti-parallel β-sheet in this region (Figure 4B). A-tract DNA, as in the amrZ1 site, has specific properties in that each ApA base pair step exhibits a negative roll, and bifurcated hydrogen bonds between each adenine and two thymine nucleotides on the opposite strand lead to propeller twisting and minor groove narrowing; A-tract DNA is also thought to be less flexible due to the extra stabilization provided by the additional bifurcated hydrogen bonds [35]. Based on this we investigated the region between the two amrZ1 half sites for any role in AmrZ binding, and whether the binding of AmrZ causes distortions to the amrZ1 DNA, or if the amrZ1 DNA is intrinsically distorted, allowing for AmrZ recognition.
Figure 4. The DNA linker region between the two AmrZ binding half sites has a distorted structure.
(A) Major and minor groove widths of the amrZ1 binding site are plotted on the graph with the corresponding nucleotide sequence above. Values are of inter-phosphate distance minus their van der Waals surface. Also indicated are the average major and minor groove widths of 11.7 Å, and 5.7 Å, respectively, of B-form DNA (horizontal dashed lines) [68]. The minor groove in the region between each AmrZ binding half site is narrowed to 2.8 Å (B) Surface representation of the amrZ1 DNA. The two AmrZ binding half sites are colored in yellow, while the distorted minor groove is colored in red. Also shown is the position of the DNA binding β-sheets on each binding half site.
To test if the linker region between each AmrZ binding half site contributes to AmrZ affinity at amrZ1, the native A/T rich linker sequence 5′-AAAAC was mutated to a G/C rich linker region with the sequence 5′-CGCGC, which resulted in a 7.5 fold reduction in binding (Table 2). It is important to note that this reduction in binding is not caused by the removal of specific protein - nucleotide interactions, since there are no contacts between the AmrZ protein and amrZ1 DNA in this region. Combining the mutations in the AmrZ binding half site with the mutations to the linker region (TTC/GC amrZ1) caused a severe aberration in binding affinity (244-fold reduction). The results show that binding affinity is regulated by both the sequence dependent interactions between AmrZ and amrZ1 and the linker region separating these binding sites.
Additional binding experiments were performed to determine if the intrinsic structure of the A/T rich linker contributes to binding affinity. The five base pair linker region on the native amrZ1 binding site was mutated to three sequences, each having their own unique properties. A sequence with the linker region mutated to 5′-TTTTC resulted in a 5.0-fold reduction in AmrZ binding, when compared to the WT amrZ1 sequence (Table 2). TpT base pair steps have the same properties of ApA base pair steps, including a narrow minor groove and less flexibility [35]. Mutating the amrZ1 sequence to 5′-AATTC caused a 7.0-fold reduction in affinity when compared to AmrZ binding to the WT amrZ1 sequence (Table 2). Molecular dynamics simulations of the interactions between the papillomavirus E2 transcription factors and their binding sites have shown that the 4 nucleotide sequence AATT has similar minor groove and propeller twist properties to A-tract DNA [36]. The last amrZ1 mutant binding site tested had a linker region containing the sequence 5′-ATATC, and AmrZ binding to this site was also altered compared to the WT amrZ1 sequence, causing a 4.7-fold reduction in affinity (Table 2). This site was designed to test if flexibility in the linker region allowed AmrZ to distort the DNA and form a complex. The TpA step in this sequence permits variations in roll, twist and slide due to poor stacking between these base pairs, and DNA containing these steps contain wider minor grooves, caused by the steric clashing of cross strand adenines [37]. It should be noted that the properties described for these sequences are average parameters derived from structures and that individual structures show a range of properties, specifically minor groove width [38]. Although AmrZ had reduced affinity for each of these three sequences, the most dramatic effect was mutation of the linker region to 5′-CGCGC. Binding of AmrZ to the sequence 5′-ATATC was reduced suggesting that A/T content of the sequence, which is usually thought to impart flexibility to DNA, was not the main contributor to AmrZ specificity. AmrZ binding to the two sequences harboring mutant linker regions with similar properties to the A-tract sequence (5′-TTTTC and 5′-AATTC) was decreased, suggesting that there are properties unique to the 5′-AAAAC linker sequence in the native amrZ1 binding site that allow for binding specificity. Taken together, these data allow us to propose that binding specificity is directed by intrinsic distortions to the DNA, rather than the flexibility conferred by the A/T rich sequence composition. Recognizing a physical feature of the DNA rather than a specific sequence introduces degeneracy in the recognition sequence that would influence the number of potential recognition sites for AmrZ. We queried the P. aeruginosa PAO1 genome [31] for the number of binding sites with the exact amrZ1 repressor sequence and found 5 sites. If we allow the A/T linker region to be degenerate, the number of potential binding sites increases to 77. Further biological studies will be required to determine how many of these sites function as actual regulators.
Narrowed minor grooves of DNA have a strong correlation between the width and increased electronegative potential of the minor groove [38]. There are many examples of transcription factors that recognize local distortions of the minor groove in addition to sequence specific recognition in both prokaryotic and eukaryotic organisms. The Listeria monocytogenes helix-turn-helix (HTH) transcription factor MogR recognizes two half sites on the flaA operator site [39]. The minor groove between the two half sites is distorted, and contributes to MogR specificity for this site. Another example is the myocyte enhancer factor-2 (MEF2), a member of the MADS-box superfamily, which recognizes a narrowed minor groove on the consensus sequence to bind and activate transcription [40]. These two examples, in addition to others, use positively charged residues, specifically arginine, to recognize and form contacts with the enhanced electronegative potential of the narrow minor groove [38]. However, there are examples of proteins similar to AmrZ that recognize minor groove shape, but do not make any contacts to the minor groove. The classical example is the bacteriophage 434 repressor recognition of six binding sites on the two operator regions, OR and OL, which is greatly modulated by the sequence composition of the central region of these sites [41]. These variations in binding affinities have been shown to be biologically important in directing the lysogenic or lytic fate of bacteriophage 434 [42]. Although the 434 repressor positions an arginine residue near the minor groove, there are no specific contacts by the protein to this region, and mutational analysis shows that this arginine does not contribute to binding affinity [41]. Recently, recognition of the intrinsic structure of narrowed minor grooves has been studied with the DNA bending protein Fis, which is responsible for the compaction of bacterial DNA [43]. An A/T rich (5′-AATTT) narrowed minor groove, located between two Fis binding sites is compressed, allowing for the insertion of two HTH domains into the adjacent major grooves of DNA. Mutations to this narrow minor groove sequence cause changes in binding, with the biggest change occurring by mutating the sequence to a G/C rich sequence (5′-GGCGC). Narrowed minor grooves between binding half sites have been observed in other RHH protein - DNA structures. In the structure of Arc in complex with DNA, the minor groove between half sites is narrowed to 1.2 Å, and the sequence in this region has the sequence 5′-GTGCT [13]. Likewise, in the Streptococcus sp. CopG-DNA structure the minor groove is narrowed to 1.9 Å and has the sequence 5′-TTGAG [14]. The DNA in complex with the inc18 plasmid encoded omega protein has a minor groove width of 2.7 Å, and the A/T rich sequence 5′-AAAT. Also, due to a fortuitous packing arrangement in the crystal, both bound and free DNA were observed, with the free DNA having similar secondary structure as the omega bound DNA [16]. For each of these proteins, the contributions of the linker region between the two half sites to binding have not been determined.
Alterations in the sequence specific half sites, the linker region, or both can modulate affinity for AmrZ to amrZ1; however, the exact mechanism by which AmrZ recognizes the distorted structure of the minor groove remains enigmatic. In the Δ42AmrZ-amrZ1 structure (Figure 1A), there are no positively charged amino acids that contact the minor groove. The extended N-terminus of AmrZ contains an arginine at position 2 which might make these contacts; however, in vitro DNA binding assays performed with various N-terminal truncation mutants of AmrZ showed no decrease in binding to amrZ1 [30]. The phosphate backbone on either side of the narrow minor groove of amrZ1 is contacted on each side by the N-terminus of α-helix 2 from chains A and C (Figure 1A, 2C). This attraction is most likely enhanced due to the positive dipole formed by the N-terminus of the α-helix and the increased electronegative potential of the narrowed minor groove.
AmrZ binding to the activator site, algD
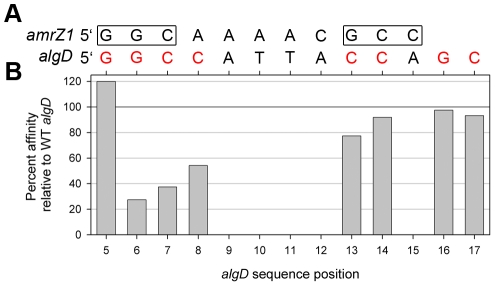
In addition to functioning as a repressor when bound to amrZ1, AmrZ binding to the algD site is necessary for the activation of genes responsible for alginate biosynthesis. Interestingly, there are significant divergences between the activator and repressor sequences, and AmrZ affinity to the algD binding site is approximately 24 fold reduced compared to the amrZ1 binding site (Tables 2 & 3). Using the information from the AmrZ interaction with the repressor amrZ1 binding site, we asked if we could predict how AmrZ interacts with the binding site on the algD promoter (Figure S1). We set out to determine the features of the algD sequence necessary for AmrZ recognition and activation. By aligning the left half AmrZ binding site on amrZ1 (5′-GGC) to the algD sequence (positions 5–7), it became apparent that there is no similar right half binding site on algD, and additionally, the sequence of the linker region is also different (Figure 5A). In order to probe the interaction between AmrZ and algD, multiple single nucleotide mutations of the algD site were created, and binding affinity of AmrZ to each of the mutant algD sequences was measured with fluorescence anisotropy.
Table 3. AmrZ affinity for algD binding site mutants.
| Binding Site | Sequencea | Kd b (nM) ± SE | Fold over WT algD c |
| WT algD | CATTGGCCATTACCAGCCTCCC GTAACCGGTAATGGTCGGAGGG | 198±14 | 1.0 |
| m5 algD | CATTTGCCATTACCAGCCTCCC GTAAACGGTAATGGTCGGAGGG | 164±17 | 0.8 |
| m6 algD | CATTGTCCATTACCAGCCTCCC GTAACAGGTAATGGTCGGAGGG | 723±37 | 3.7 |
| m7 algD | CATTGGACATTACCAGCCTCCC GTAACCTGTAATGGTCGGAGGG | 530±52 | 2.7 |
| m8 algD | CATTGGCAATTACCAGCCTCCC GTAACCGTTAATGGTCGGAGGG | 365±30 | 1.8 |
| m13 algD | CATTGGCCATTAACAGCCTCCC GTAACCGGTAATTGTCGGAGGG | 256±20 | 1.3 |
| m14 algD | CATTGGCCATTACAAGCCTCCC GTAACCGGTAATGTTCGGAGGG | 216±17 | 1.1 |
| m16 algD | CATTGGCCATTACCATCCTCCC GTAACCGGTAATGGTAGGAGGG | 203±17 | 1.0 |
| m17 algD | CATTGGCCATTACCAGACTCCC GTAACCGGTAATGGTCTGAGGG | 212±19 | 1.1 |
Mutations to the wild type algD binding site are notated by the bolded nucleotides in each mutant sequence.
The Kd was calculated by fitting the hyperbolic equation for a single ligand binding model with saturation (eq 2) to the data in Figure S3, which were averaged from four independent experiments.
Fold over (wild type) WT algD is defined by (Kd of sample)/(Kd of wild type) for each sample.
Figure 5. AmrZ binding to the algD activator site.
(A) Alignment of the amrZ1 and algD DNA sequences that have been derived from previous footprinting experiments [6], [9]. The two binding half sites on the amrZ1 sequence are boxed, while nucleotides that were mutated in the algD sequence are shown in red. (B) Results from the scanning mutagenesis of the algD site (Table 3). Only mutations of guanine nucleotides at positions 6 (forward strand), and 7 and 8 (reverse strand) resulted in a noticeable decrease in binding affinity of AmrZ compared to the WT algD sequence. Mutations to the right side binding half site resulted in no major decrease in binding affinity of AmrZ. Percent affinity was calculated by dividing the Kd for the WT algD binding site by the Kd for each mutant binding site.
Through mutagenesis of nucleotides in the proposed left half binding site in algD, we show that AmrZ recognizes the sequence 5′-GGC at this site. The guanine nucleotides at positions 5 and 6 on one strand of the algD binding site and positions 7 and 8 on the other strand (Figure 5A) were mutated to thymine bases, and the binding affinity of AmrZ to each of these mutant sequences was measured (Table 3, Figure 5B). The mutation to position 5 resulted in a slight increase to AmrZ affinity, while mutations to positions 6, 7, and 8 each resulted in significant reductions in affinity. To determine the nucleotides AmrZ interacts with on the right half of the algD binding site, guanine bases at position 16 on one strand, and positions 13, 14, and 17 on the opposite strand of algD were mutated to thymine residues. No significant differences in binding affinity to these sequences are observed (Table 3, Figure 5B), suggesting that AmrZ interacts with this half site in a different manner than what is observed at amrZ1.
Our previous binding experiments show the same residues, Lys18, Val20, and Arg22 are involved in the sequence dependent interactions with algD [30]. In addition, we found that Arg14 is also necessary for binding, with the R14A mutant of AmrZ exhibiting a 5 fold reduction in binding affinity at algD. This arginine residue is also required for transcriptional activation of algD, where R14A AmrZ only retains 3% of WT activity in vivo. From the AmrZ - amrZ1 structure, the extended N-terminus forms a looped structure which positions Arg14 into the major groove of DNA (Figure 3); however, no specific contacts between this residue and the amrZ1 DNA are observed, and mutations of this residue have no effects on in vitro and in vivo activity at the repressor site.
algD promoter swapping
The differences we observe in interactions of AmrZ with the activator algD binding sequence versus the repressor amrZ1 binding sequence led us to ask if these different binding modes alone could account for activation or repression activity. To test this hypothesis we switched the AmrZ binding site in the algD promoter (activator) with the amrZ1 binding site (repressor) and introduced this variant into an algD::lacZ transcriptional fusion, which was stably integrated into the genome of the mucoid P. aeruginosa strain FRD1 (FRD1 palgDamrZ1-lacZ). The position and length of the switched binding site were the same as in the native algD promoter. With this construct we measured relative activation at algD with a β-galactosidase activity assay compared to an algD::lacZ transcriptional fusion containing the wild type algD AmrZ binding site (FRD1 palgD-lacZ). The results of this experiment (Table 4) reveal activation of algD remains unchanged when the amrZ1 binding site replaced the native site. Cell lysates from the FRD1 palgDamrZ1-lacZ strain had 527.7 units of β-galactosidase activity compared to 536.1 units for FRD1 palgD-lacZ. Expression of both palgD-lacZ and palgDamrZ1-lacZ were significantly reduced in amrZ mutant P. aeruginosa strains (Table 4), indicating that the reporter fusion faithfully reproduced what has been observed previously regarding AmrZ activation of algD [5], [10], [30]. The activation of algD with the amrZ1 repressor site at its promoter supports a model in which AmrZ binding alone does not regulate activation or repression of transcription, but rather interactions of AmrZ with other regulators at the amrZ and algD promoters likely contribute to repression or activation, respectively. This is consistent with the previous evidence that the AlgB, AlgR, IHF, and CysB regulators are known to bind on the algD promoter and are necessary for activation [44]–[48], suggesting a possible interaction of AmrZ with one of these proteins. To date, no other regulators have been identified to bind the amrZ promoter; however, it is possible one of these same regulators may also interact with AmrZ there as well. An additional determinant likely dictating activation versus repression is the position of the AmrZ binding site relative to the start of transcription, which differs for algD (−282) and amrZ1 (−93).
Table 4. Transcriptional activation of algD containing amrZ1-repressor binding sequence.
| P. aeruginosa Genotype | Units β-galactosidase |
| FRD1 palgD-lacZ | 536.1+/−20.5 |
| FRD1 lacZ | 73.9+/−41.3 |
| FRD1 palgDamrZ-lacZ | 527.7+/−11.6 |
| FRD1 ΔamrZ palgD-lacZ | 114.7+/−15.3 |
| FRD1 ΔamrZ lacZ | 72.2+/−34.3 |
| FRD1 ΔamrZ palgDamrZ-lacZ | 151.3+/−24.4 |
Conclusions
AmrZ functions as both a transcriptional activator and repressor of P. aeruginosa virulence genes. We have determined the structure of Δ42 AmrZ in complex with an 18 base pair oligonucleotide containing the amrZ1 binding site. AmrZ binding to this site results in the repression of amrZ transcription. By combining structural and biochemical data, we developed a model for AmrZ recognition at amrZ1. Using the suggested terminology from the recent review by Rohs et al. [49], the protein-DNA specificity of AmrZ can be classified by major groove base readout through protein residues in the β-sheet with two GGC half sites in the DNA. This is combined with local shape readout utilizing minor groove distortions in the linker region between the half sites. We also probed the interaction of AmrZ with another biologically important binding site algD, which leads to the activation of alginate biosynthesis. In contrast, we observed stark differences in the physical interactions that AmrZ makes with the algD sequence that suggest the protein likely utilizes a different mode of recognition at this site. AmrZ binds the algD sequence with lower affinity, and mutagenesis of the algD sequence shows that only one half site contributes to AmrZ binding. However, these differences in protein binding at the promoter sequences are alone not sufficient to account for the activator or repressor activity of AmrZ, and likely the position of AmrZ binding at the promoter and/or protein interactions with other regulators are also necessary for biological function.
Materials and Methods
Molecular cloning, expression and purification of WT and Δ42 P. aeruginosa AmrZ
The gene encoding WT AmrZ was PCR amplified from the P. aeruginosa strain PAO1 with the primers amrZ_F (5′-CGCCATCACATATGCGCCCACTGAAACAGGC) and amrZ_wt_R (5′-CGCCATCAGGATCCTCAGGCCTGGGCCAGCTC). The resulting gene product was then inserted into a modified pET19 expression vector (Novagen) which encodes an N-terminal poly-Histidine tag, followed by a Rhinovirus 3C protease cleavage site, which permits the removal of the affinity tag (PreScission Protease, GE Healthcare). The pET19-amrZ vector was transformed into E. coli C41(DE3) cells for expression. One liter of LB-Broth (Luria-Bertani) supplemented with 50 µg/ml of ampicillin was inoculated with 10 ml of an overnight culture of the C41 cells containing the pET19-amrZ vector. The cells were grown at 37°C to an OD600 = 0.5, and induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 16°C for 20 hours. Prior to induction with IPTG, cells were rapidly cooled on ice to 20°C to bring the temperature of the culture close to the induction temperature. Induction of the cells at low temperature was necessary for protein solubility during overexpression. Cells were harvested by centrifugation, resuspended in lysis buffer (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 4 M urea), and lysed using an EmulsiFlex C-5 cell homogenizer (Avestin). Cell debris was removed at 30,000× g and the supernatant was passed over a 10 ml Ni-NTA (Qiagen) column equilibrated with lysis buffer. This column was washed with 20 column volumes of wash buffer 1 (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 35 mM imidazole, 3 M urea), followed by 10 column volumes of wash buffer 2 (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 50 mM imidazole, 2 M urea). Bound AmrZ was eluted with elution buffer (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 500 mM imidazole, 1 M urea), treated with PreScission Protease according to the manufacturer's directions, and dialyzed over night at 4°C against 100 mM Bis-Tris pH 5.5, 100 mM NaCl, 5% glycerol, 2 mM dithiothreitol (DTT), and 0.5 mM EDTA. The partial denaturing conditions introduced by the 4 M urea were necessary for protein solubility and affinity to the Ni-NTA column, and no change in secondary structure or DNA binding affinity was observed compared to protein purified without urea present. AmrZ was then passed over a MonoS cation exchange column, and eluted with a 0.1 M–1 M gradient of NaCl. Purity of the peak fractions was verified by SDS-PAGE, and fractions containing pure WT AmrZ were pooled. For crystallization experiments, AmrZ was dialyzed against 100 mM Bis-Tris pH 5.5, 100 mM NaCl, 2% glycerol, while for DNA binding assays, AmrZ was dialyzed against a buffer containing 100 mM Bis-Tris pH 6.5, 150 mM NaCl, 5% glycerol. WT AmrZ was then concentrated to 20 mg/ml for crystallization experiments, or 1 mg/ml for DNA binding assays, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. Concentration of WT AmrZ was measured using the BCA assay (Thermo Scientific) using a standard curve of lysozyme as a reference.
The Δ42 C-terminal truncation mutant of AmrZ was amplified from the P. aeruginosa strain PAO1 using the primers amrZ_F and amrZ_Δ42_R (5′-CGCCATCAGGATCCTCAAACACCGAGATTGTCTTG). Expression and purification of this protein was carried out using the procedures outlined for WT AmrZ.
Crystallization of Δ42 AmrZ - amrZ1 complex
Crystallization trials of AmrZ were carried out by screening multiple AmrZ C-terminal deletion constructs against a library of double stranded DNA oligonucleotides containing the amrZ1 binding site (Integrated DNA Technologies). Initial crystals were obtained only with the Δ42 AmrZ C-terminal truncation and an 18 bp oligonucleotide in a condition containing 6% PEG 8 K, 0.1 M MES pH 6.0, 0.1 M CaCl2, 0.1 M NaCl. For experimental phasing, selenomethionine (Se-Met) derivatized Δ42 AmrZ was prepared using published methods [50]. Purification of this protein was performed using the methods described above, with the only exception being the addition of 5 mM DTT in the final dialysis buffer. Crystals of the Se-Met Δ42 AmrZ - 18 bp amrZ1 complex were obtained by mixing the protein and DNA in a 1∶1.5 molar ratio (810 µM AmrZ: 607.5 µM amrZ1) in the presence of 50 mM MgSO4. This complex was crystallized by the hanging drop vapor diffusion method at 25°C at a 1∶1 ratio with reservoir solution containing 3% PEG8K, 0.1 M MES pH 6.0, 0.15 M NaCl, and 2 mM TCEP pH 8.0. Crystals grew within 2–3 weeks and were soaked in a solution containing 20% 2-methyl-1,3 propanediol for cryo-protection before being frozen in liquid nitrogen for data collection.
Data collection and refinement of the Δ42 AmrZ - amrZ1 structure
Diffraction data for crystals containing the Δ42 AmrZ: 18 bp amrZ1 complex were collected on beamline X25 at the National Synchrotron Light Source (NSLS), Brookhaven National Labs. The dataset was collected at the selenium peak, with an X-ray wavelength of 0.9793 nm. Indexing, integration and scaling of the data were performed using HKL2000 program suite [51]. Phasing of the structure was performed using SAD methods with the program SOLVE [52], and density modification was performed using RESOLVE [53]. Manual model building was performed in Coot [54], and refinement was carried out using the programs REFMAC5 [55] within the CCP4 program suite [56], and CNS [57]. Data collection and refinement statistics are found in Table 1. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the PDB id 3QOQ.
DNA binding measurements
Binding affinity of the various amrZ1 and algD binding site mutants were performed using fluorescence anisotropy as previously described [30], [58]. In brief, increasing concentrations of WT AmrZ were incubated in a reaction (25 µl) containing 1 nM 22-mer 5′-6-carboxy-fluorescein (6-FAM) labeled DNA oligonucleotide (IDT) containing either the amrZ1 or the algD sequences, 100 nM nonspecific DNA of random sequence, 100 µg/ml bovine serum albumin (BSA), 100 mM Bis-Tris pH 6.5, 150 mM NaCl, and 5% glycerol. DNA concentrations were kept 1 nM (<<Kd) to ensure equilibrium measurements of binding constants. Anisotropy measurements were recorded at 25°C on a Safire2 microplate reader with a fluorescence polarization module (Tecan Group, Ltd.), using an excitation wavelength of 470 nm and an emission wavelength of 525 nm. Anisotropy data were scaled and normalized using Equation 1 below:
| (1) |
In this equation, Aobs is the measured anisotropy value for each AmrZ concentration, A0 is the anisotropy of the unbound DNA, and Amax is the maximum anisotropy observed in each experiment. The dissociation constant (Kd) was calculated by fitting the data to the equation for a single state binding model (Equation 2).
| (2) |
Fitting of the data to Equation 2 was performed using SigmaPlot. Raw data and fits for AmrZ binding to each DNA sequence can be found in Figures S2 and S3, with the results from these experiments being presented in Tables 2 and 3. Results presented are the averages of four independent experiments.
Calculation of protein and DNA parameters
The program CURVES+ [59] was used to measure the major and minor groove widths of the amrZ1 DNA. The buried surface areas formed by protein-DNA interactions and by protein-protein interactions were measured using the programs AREAIMOL in the CCP4 suite [56] and PDBsum [60], respectively. Ramachandran statistics found in Table 1 were calculated with PDBsum [60]. Electrostatic surface representations of the protein and DNA were created by first generating a PQR file, which contains charge and radius information for each atom, with the program PDB2PQR [61], followed by visualization of the electrostatic surface using the APBS program [62]. All figures of structural representations were prepared using the program PyMol [63].
amrZ promoter switching
The AmrZ binding site (ABS) (CCATTGGCCATTACCAGCCTCCC) in the algD promoter was replaced by the same-length amrZ1 ABS (GTACTGGCAAAACGCCGGCACGC) from the amrZ promoter by site-directed mutagenesis [64]. Mutagenesis was achieved by primers algD74 (GCGTGCCGGCGTTTTGCCAGTACATTACGCCGGAGATGGCATTTC) and algD75 (GTACTGGCAAAACGCCGGCACGCGCCATTACATGCAAATTACGATTGC), together with flanking primers algD65 (CCCCAAGCTTCTCTTTCGGCACGCCGAC) and algD66 (CCGGGATCCCCGACATAGCCCAAACCAAAG). PCR products of algD65/algD74 and algD66/algD75 were denatured and hybridized. The products were used as the template for the second PCR, with primers algD65 and algD66. With HindIII and BamHI sticky ends, the final PCR product was cloned into HindIII and BamHI double digested mini-CTX-lacZ transcriptional fusion vector [65], resulting in a new plasmid pBX8, which harbors modified algD::lacZ transcriptional fusion (palgDamrZ1-lacZ). The sequence of the palgDamrZ1-lacZ promoter was verified by PCR and sequencing.
Construction of transcriptional fusion in P. aeruginosa chromosome
The plasmid pBX8 was transferred into P. aeruginosa FRD1 using E. coli strain SM10. The modified palgDamrZ1-lacZ transcriptional fusion was integrated at the attB site within the chromosome of FRD1 and FRD1 ΔamrZ [30], and the unnecessary portion of the fragment was removed by pFLP2 [66], resulting in FRD1 palgDamrZ1-lacZ or FRD1 ΔamrZ palgDamrZ1-lacZ, respectively.
β-galactosidase assays
P. aeruginosa in mid-log phase were pelleted and washed with Z-buffer (110 mM Na2HPO4, 45 mM NaH2PO4, 10 mM KCl, 2 mM MgSO4, pH7.0). Cells were lysed through three rounds of fast freezing at −80°C then thawing at 37°C, followed by mild sonication. Samples were centrifuged at 18 k× g and 4°C for 10 min at 21,000× g. The supernatants were analyzed for β-galactosidase activity by mixing 10 µl of a sample supernatant with 80 µl of Z-buffer. To start the reaction 20 µl of 4 mg/ml orthonitrophenol was added. The color change in the reaction was monitored with time and reactions were stopped by addition of 40 µl 1 M Na2CO3 for reading. Wild type FRD1 palgD::lacZ transcriptional fusion was the positive control, and FRD1 lacZ with no promoter acted as the negative control. The absorbance at both 420 nm and 550 nm of each reaction solution was read in a Molecular Devices M5 microplate reader. Miller Units were calculated from different strains as outlined [67].
Supporting Information
Sequences of known AmrZ binding sites. The two known AmrZ binding sites on the amrZ promoter leading to amrZ repression (amrZ1 and amrZ2), and the one known binding site on the algD promoter, leading to activation of the alginate biosynthetic pathway are shown here. These sites have been determined experimentally through DNA footprinting experiments [6], [9], and share little consensus. The sequences are aligned based on the region of highest similarity. In the consensus above the sequences, uppercase nucleotides represent bases that are present in all three sequences, while lowercase nucleotides represent bases that are present in only two of the sequences.
(TIF)
AmrZ - amrZ1 binding data. Fluorescence anisotropy was utilized to calculate the binding affinity for AmrZ to multiple sequences harboring mutations in the amrZ1 binding site. Each data point is an average from four independent experiments, and the error bars are calculated from the standard deviation. Data was processed as described in the Materials and Methods section, and results from these data are shown in Table 2. (A) AmrZ: WTamrZ1 (B) AmrZ: GTCamrZ1 (C) AmrZ: TTCamrZ1 (D) AmrZ: CGCGamrZ1 (E) AmrZ: TTC/GCamrZ1 (F) AmrZ: ATATamrZ1 (G) AmrZ: TTTTamrZ1 (H) AmrZ: AATTamrZ1.
(TIF)
AmrZ - algD binding data. To determine the AmrZ binding site on the algD promoter, multiple mutations to the algD DNA sequence were created and binding affinities between AmrZ and these sequences were determined using fluorescence anisotropy. Each data point is an average from four independent experiments, and the error bars are calculated from the standard deviation. Data from these experiments were processed as described in the Materials and Methods section, and results are shown in Table 3. (A) AmrZ: WTalgD (B) AmrZ: m5algD (C) AmrZ: m6algD (D) AmrZ: m7algD (E) AmrZ: m8algD (F) AmrZ: m13algD (G) AmrZ: m14algD (H) AmrZ: m16algD (I) AmrZ: m17algD.
(TIF)
Acknowledgments
The authors would like to thank Annie Heroux and the staff of Beamline X-25 at the National Synchrotron Light Source (NSLS) in Brookhaven, NY for assistance in data collection. We are also grateful to Dr. Paul J. Holland for the critical reading of this manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a Cystic Fibrosis Foundation Student Traineeship PRYOR10H0 (EEP), an American Heart Association predoctoral fellowship, 0815249E (EAW) and Public Health Service grants AI061396 and HL58334 (DJW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Regules JA, Glasser JS, Wolf SE, Hospenthal DR, Murray CK. Endocarditis in burn patients: clinical and diagnostic considerations. Burns. 2008;34:610–616. doi: 10.1016/j.burns.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Carruthers MM, Kanokvechayant R. Pseudomonas aeruginosa endocarditis. Report of a case, with review of the literature. Am J Med. 1973;55:811–818. doi: 10.1016/0002-9343(73)90262-3. [DOI] [PubMed] [Google Scholar]
- 3.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 4.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 6.Baynham PJ, Wozniak DJ. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 7.Tart AH, Blanks MJ, Wozniak DJ. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol. 2006;188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J Bacteriol. 2006;188:132–140. doi: 10.1128/JB.188.1.132-140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsey DM, Baynham PJ, Wozniak DJ. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J Bacteriol. 2005;187:4430–4443. doi: 10.1128/JB.187.13.4430-4443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baynham PJ, Brown AL, Hall LL, Wozniak DJ. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol Microbiol. 1999;33:1069–1080. doi: 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- 11.Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 12.He YY, McNally T, Manfield I, Navratil O, Old IG, et al. Probing met repressor-operator recognition in solution. Nature. 1992;359:431–433. doi: 10.1038/359431a0. [DOI] [PubMed] [Google Scholar]
- 13.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 14.Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, et al. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. Embo J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, et al. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J Mol Biol. 2006;364:170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 16.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiter ER, Wang SC, Zamble DB, Drennan CL. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc Natl Acad Sci U S A. 2006;103:13676–13681. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J Biol Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 19.Ni L, Jensen SO, Ky Tonthat N, Berg T, Kwong SM, et al. The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes. Nucleic Acids Res. 2009;37:6970–6983. doi: 10.1093/nar/gkp756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgering MJ, Boelens R, Gilbert DE, Breg JN, Knight KL, et al. Solution structure of dimeric Mnt repressor (1–76). Biochemistry. 1994;33:15036–15045. doi: 10.1021/bi00254a012. [DOI] [PubMed] [Google Scholar]
- 21.Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution. J Mol Biol. 2001;314:789–796. doi: 10.1006/jmbi.2001.5157. [DOI] [PubMed] [Google Scholar]
- 22.Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 23.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, et al. Crystal structure of the nickel-responsive transcription factor NikR. Nat Struct Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 24.Popescu A, Karpay A, Israel DA, Peek RM, Jr, Krezel AM. Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators. Proteins. 2005;59:303–311. doi: 10.1002/prot.20406. [DOI] [PubMed] [Google Scholar]
- 25.Larson JD, Jenkins JL, Schuermann JP, Zhou Y, Becker DF, et al. Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition. Protein Sci. 2006;15:2630–2641. doi: 10.1110/ps.062425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallo M, Ferrari E, Eliseo T, Amata I, Pertinhez TA, et al. A new member of the ribbon-helix-helix transcription factor superfamily from the plant pathogen Xanthomonas axonopodis pv.citri. J Struct Biol. 2010;170:21–31. doi: 10.1016/j.jsb.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Huang L, Yin P, Zhu X, Zhang Y, Ye K. Crystal structure and centromere binding of the plasmid segregation protein ParB from pCXC100. Nucleic Acids Res. 2010;39:2954–2968. doi: 10.1093/nar/gkq915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight KL, Sauer RT. DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal residues. Proc Natl Acad Sci U S A. 1989;86:797–801. doi: 10.1073/pnas.86.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldburger CD, Sauer RT. Domains of Mnt repressor: roles in tetramer formation, protein stability, and operator DNA binding. Biochemistry. 1995;34:13109–13116. doi: 10.1021/bi00040a023. [DOI] [PubMed] [Google Scholar]
- 30.Waligora EA, Ramsey DM, Pryor EE, Jr, Lu H, Hollis T, et al. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J Bacteriol. 2010;192:5390–5401. doi: 10.1128/JB.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, et al. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coulocheri SA, Pigis DG, Papavassiliou KA, Papavassiliou AG. Hydrogen bonds in protein-DNA complexes: where geometry meets plasticity. Biochimie. 2007;89:1291–1303. doi: 10.1016/j.biochi.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Brown BM, Milla ME, Smith TL, Sauer RT. Scanning mutagenesis of the Arc repressor as a functional probe of operator recognition. Nat Struct Biol. 1994;1:164–168. doi: 10.1038/nsb0394-164. [DOI] [PubMed] [Google Scholar]
- 35.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 36.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen A, Higgins DE, Panne D. Recognition of AT-rich DNA binding sites by the MogR repressor. Structure. 2009;17:769–777. doi: 10.1016/j.str.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J Mol Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 41.Koudelka GB, Harrison SC, Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. Nature. 1987;326:886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- 42.Koudelka GB. Recognition of DNA structure by 434 repressor. Nucleic Acids Res. 1998;26:669–675. doi: 10.1093/nar/26.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delic-Attree I, Toussaint B, Garin J, Vignais PM. Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol Microbiol. 1997;24:1275–1284. doi: 10.1046/j.1365-2958.1997.4121799.x. [DOI] [PubMed] [Google Scholar]
- 45.Mohr CD, Deretic V. In vitro interactions of the histone-like protein IHF with the algD promoter, a critical site for control of mucoidy in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1992;189:837–844. doi: 10.1016/0006-291x(92)92279-7. [DOI] [PubMed] [Google Scholar]
- 46.Mohr CD, Leveau JH, Krieg DP, Hibler NS, Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toussaint B, Delic-Attree I, Vignais PM. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun. 1993;196:416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- 48.Wozniak DJ. Integration host factor and sequences downstream of the Pseudomonas aeruginosa algD transcription start site are required for expression. J Bacteriol. 1994;176:5068–5076. doi: 10.1128/jb.176.16.5068-5076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohs R, Jin X, West SM, Joshi R, Honig B, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 52.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 57.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 58.Holland PJ, Hollis T. Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS One. 2010;5:e8680. doi: 10.1371/journal.pone.0008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. Schrödinger, LLC.
- 64.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 65.Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–950, 952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- 66.Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 68.Saenger W. Principles of Nucleic Acid Structure. 1984. 556 Springer-Verlag, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of known AmrZ binding sites. The two known AmrZ binding sites on the amrZ promoter leading to amrZ repression (amrZ1 and amrZ2), and the one known binding site on the algD promoter, leading to activation of the alginate biosynthetic pathway are shown here. These sites have been determined experimentally through DNA footprinting experiments [6], [9], and share little consensus. The sequences are aligned based on the region of highest similarity. In the consensus above the sequences, uppercase nucleotides represent bases that are present in all three sequences, while lowercase nucleotides represent bases that are present in only two of the sequences.
(TIF)
AmrZ - amrZ1 binding data. Fluorescence anisotropy was utilized to calculate the binding affinity for AmrZ to multiple sequences harboring mutations in the amrZ1 binding site. Each data point is an average from four independent experiments, and the error bars are calculated from the standard deviation. Data was processed as described in the Materials and Methods section, and results from these data are shown in Table 2. (A) AmrZ: WTamrZ1 (B) AmrZ: GTCamrZ1 (C) AmrZ: TTCamrZ1 (D) AmrZ: CGCGamrZ1 (E) AmrZ: TTC/GCamrZ1 (F) AmrZ: ATATamrZ1 (G) AmrZ: TTTTamrZ1 (H) AmrZ: AATTamrZ1.
(TIF)
AmrZ - algD binding data. To determine the AmrZ binding site on the algD promoter, multiple mutations to the algD DNA sequence were created and binding affinities between AmrZ and these sequences were determined using fluorescence anisotropy. Each data point is an average from four independent experiments, and the error bars are calculated from the standard deviation. Data from these experiments were processed as described in the Materials and Methods section, and results are shown in Table 3. (A) AmrZ: WTalgD (B) AmrZ: m5algD (C) AmrZ: m6algD (D) AmrZ: m7algD (E) AmrZ: m8algD (F) AmrZ: m13algD (G) AmrZ: m14algD (H) AmrZ: m16algD (I) AmrZ: m17algD.
(TIF)