Abstract
Mitochondrial genomes have maintained some bacterial features despite their residence within eukaryotic cells for approximately two billion years. One of these features is the frequent presence of polycistronic operons. In land plants, however, it has been shown that all sequenced vascular plant chondromes lack large polycistronic operons while bryophyte chondromes have many of them. In this study, we provide the completely sequenced mitochondrial genome of a lycophyte, from Huperzia squarrosa, which is a member of the sister group to all other vascular plants. The genome, at a size of 413,530 base pairs, contains 66 genes and 32 group II introns. In addition, it has 69 pseudogene fragments for 24 of the 40 protein- and rRNA-coding genes. It represents the most archaic form of mitochondrial genomes of all vascular plants. In particular, it has one large conserved gene cluster containing up to 10 ribosomal protein genes, which likely represents a polycistronic operon but has been disrupted and greatly reduced in the chondromes of other vascular plants. It also has the least rearranged gene order in comparison to the chondromes of other vascular plants. The genome is ancestral in vascular plants in several other aspects: the gene content resembling those of charophytes and most bryophytes, all introns being cis-spliced, a low level of RNA editing, and lack of foreign DNA of chloroplast or nuclear origin.
Introduction
Mitochondria are the cellular power houses of nearly all eukaryotes [1]. Extensive sequencing of their genomes over the last three decades reveals that this organellar genome has maintained one of its ancestral bacterial features in most protists, fungi, animals, and early land plants: genes being organized into large syntenic blocks, many of which represent polycistronic operons [1], [2], [3], [4], [5], [6], [7]. A major exception, however, is found in flowering plants, whose chondromes contain mostly free-standing genes with their own transcriptional regulatory elements [7], [8], [9], [10], [11], [12]. Recent sequencing of a chondrome from the gymnosperm Cycas taitungensis [13] shows that this type of derived mitochondrial genome is likely shared by all seed plants. When this type of mitochondrial genome with a unique gene organization and transcription system arose in plant evolution has been a long-standing question in mitochondrial research. Sequencing of chondromes from representatives of major lineages of charophytic algae [14], [15], [16], [17] and land plants [13], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27] suggests that early vascular plants are likely the groups where the genome experienced a major change. In this study we report the completely sequenced mitochondrial genome of a lycophyte, from Huperzia squarrosa of Lycopodiaceae, which bridges the gap between the ancestral type of mitochondrial genomes found in bryophytes and the derived type in seed plants.
Lycophytes are sister to all other vascular plants [28], [29], and hence are an appropriate group for investigating the ancestral condition of mitochondrial genome in vascular plants. There are three lineages within lycophytes: Lycopodiaceae, Isoetaceae, and Selaginellaceae [30], [31], [32]. Lycopodiaceae represent the basalmost clade of lycophytes [29]; a species in the family becomes a natural choice to look for the most archaic chondrome of all vascular plants. Recent reports of nearly complete chondromes from Isoetes and Selaginella show that mitochondrial genomes in these two lineages have independently acquired some features found in angiosperm chondromes, e.g., rapid rearrangement of gene order, loss of many ribosomal protein and tRNA genes, trans-splicing of introns, heavy RNA editing, and invasion of foreign DNAs of chloroplast and nuclear origins [24], [25]. These studies make it more urgent to sequence a chondrome from Lycopodiaceae so that the accurate state of mitochondrial genome in the basalmost vascular plants can be determined.
Results and Discussion
General Features of the Huperzia Mitochondrial Genome
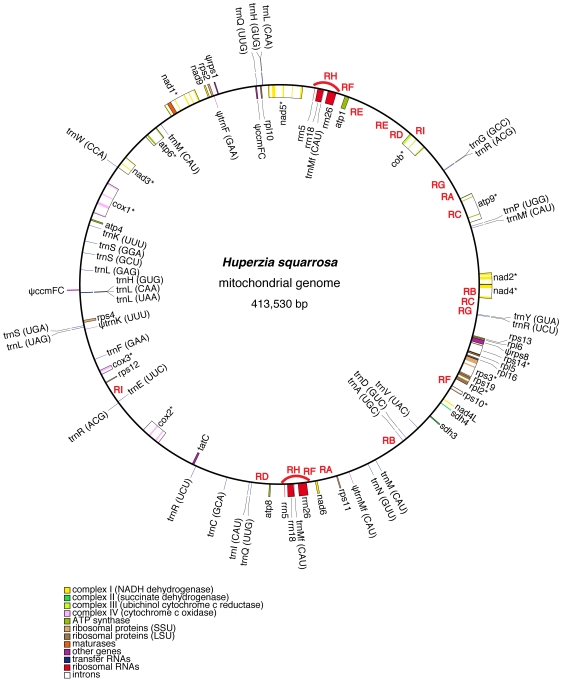
The mitochondrial genome of Huperzia squarrosa is assembled as a single circular molecule (Fig. 1, deposited in GenBank under the accession JQ002659). Its size is 413,530 base pairs (bp), with AT content of 55.8%. The genes account for 27% of the genome, 10% and 17% of which are exons and introns respectively (Table 1).
Figure 1. The gene map of Huperzia squarrosa mitochondrial genome.
Genes (exons indicated as closed boxes) shown on the outside of the circle are transcribed counter-clockwise, whereas those on the inside are transcribed clockwise. Genes with group II introns (open boxes) are labeled with asterisks. Pseudogenes are indicated with the prefix “ψ”. Repeats are marked with bold-face upper case letters (RA – RI) in regions where they are located. The two red arcs indicate the duplicated rRNA gene clusters.
Table 1. Genome sizes and proportions of the various types of sequence in the mitochondrial genomes of Chara vulgaris and seven land plants1.
| Species | Genome size (bp) | AT (%) | Genes (%) | Exons (%) | Introns (%) | Intergenic spacers (%) |
| Chara vulgaris | 67,737 | 59.1 | 91 | 52 | 39 | 9 |
| Marchantia polymorpha | 186,609 | 57.6 | 51 | 23 | 28 | 49 |
| Physcomitrella patens | 105,340 | 59.4 | 65 | 37 | 28 | 35 |
| Megaceros aenigmaticus | 184,908 | 54.0 | 50 | 16 | 34 | 50 |
| Huperzia squarrosa | 413,530 | 55.8 | 27 | 10 | 17 | 73 |
| Cycas taitungensis | 414,903 | 53.1 | 20 | 9 | 11 | 80 |
| Oryza sativa | 490,520 | 56.1 | 14 | 8 | 6 | 86 |
| Brassica napus | 221,853 | 54.8 | 29 | 16 | 13 | 71 |
Intron-encoded genes such as matR in vascular plants were excluded from calculation of genes and exons.
From our fosmid library screening experiments, we believe that the Huperzia chondrome sequence reported here represents a completely sequenced mitochondrial genome of an early vascular plant. With seven bryophyte chondromes and over two dozens of seed plant chondromes sequenced (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=33090&opt=organelle), this genome provides an important piece of data for comparison to identify the phylogenetic point at which the organellar genome experienced dramatic changes, particularly in genome size. The bryophyte chondromes are 100–200 kb in size (Table 1) [18], whereas the seed plant chondromes show a much broader size range, from slightly over 200 kb in Brassica [33], [34] to 11.3 mb (million base pairs) in Silene [35]. The over 400 kb mitochondrial genome in the lycophyte Huperzia is approximately twice the size of the largest bryophyte chondrome, that of the hornwort Phaeoceros laevis [23]. This size increase is mostly caused by expansion of intergenic spacers, whose percentage in the whole genome jumps from 35–50% in bryophytes to 73% in the lycophyte (Table 1). This expansion does not seem to be caused by transposons, as the percentages of transposon fragment sequences in the genomes have remained largely unchanged from bryophytes to the lycophyte based on a preliminary analysis (data not shown). Instead, insertion of a large number of pseudogene pieces has partially resulted in expansion of the spacers. Sixty-nine pseudogene pieces longer than 50 bp were detected. They added to 18,026 bp and account for 4.4% of the genome (Table S1). Previously, a moderate number of pseudogene pieces were found in intergenic spacers in the chondromes of the three liverworts, but only a few in the two hornworts and none in the two mosses, likely due to presence/absence of reverse transcriptase and different constraints on genome sizes in different species [18]. In the Marchantia chondrome, which has the most pseudogene pieces among bryophyte chondromes, their total length was only 5,402 bp, accounting for 2.9% of the genome [18]. During the same evolutionary transition, contribution to the genome size by exons and introns decreases significantly, from 50–65% to 27%, and the proportions of decrease for exons and introns are similar (Table 1).
RNA editing likely occurs in the Huperzia mitochondrial genome, as annotation of all protein-coding genes using the standard genetic code requires introduction of 19 editing events to reconstitute start or stop codons and to remove internal stop codons (Table 2). The in silico analyses with the software PREPACT [36] using sequences of genes from Marchantia or cDNAs from Isoetes and Selaginella as reference templates suggest that there are 334, 576, and 364 edited sites respectively (Table 3). Despite some uncertainty associated with these analyses, it is reasonable to say that the level of editing in the Huperzia chondrome is less than what have been reported in the chondromes of two other lycophytes, Isoetes and Selaginella, where 1,782 and 2,152 editing events are required to make entire transcript populations functional [25], [37].
Table 2. A list of start and stop codons created or removed through putative RNA editing events in coding sequences within Huperzia squarrosa mitochondrial genome1.
| Gene name | Start codon created | Stop codon removed | Stop codon created |
| sdh4 | ACG→AUG | CAA→UAA | |
| atp9 | CAA→UAA | ||
| rps13 | ACG→AUG | CGA→UGA | |
| nad9 | ACG→AUG | CAA→UAA | |
| nad1 | ACG→AUG | UAA→CAA | CGA→UGA |
| cox1 | ACG→AUG | ||
| nad5 | ACG→AUG | UAA→CAA | CAA→UAA |
| rps12 | UAA→CAA | ||
| rps2 | CAA→UAA | ||
| rpl6 | CAA→UAA | ||
| cob | UAG→CAG | ||
| cox3 | CAA→UAA |
GTG is the start codon for rpl16.
Table 3. Predicted RNA editing sites in the mitochondrial genome of Huperzia squarrosa using sequences of genes from Marchantia or cDNAs from Isoetes and Selaginella with the software PREPACT.
| gene | Marchantia polymorpha | Isoetes engelmannii | Selaginella moellendorffii | ||||||
| C>U | U>C | total | C>U | U>C | total | C>U | U>C | total | |
| atp1 | 3 | 0 | 3 | 8 | 26 | 34 | 6 | 5 | 11 |
| atp4 | 7 | 1 | 8 | 6 | 5 | 11 | |||
| atp6 | 9 | 0 | 9 | 14 | 19 | 33 | |||
| atp8 | 5 | 2 | 7 | 4 | 6 | 10 | 4 | 7 | 11 |
| atp9 | 7 | 0 | 7 | 8 | 0 | 8 | |||
| cob | 8 | 3 | 11 | 10 | 6 | 16 | |||
| cox1 | 19 | 0 | 19 | 30 | 38 | 68 | |||
| cox2 | 4 | 0 | 4 | 2 | 12 | 14 | 12 | 12 | 24 |
| cox3 | 6 | 0 | 6 | 13 | 19 | 32 | 13 | 12 | 25 |
| nad1 | 16 | 4 | 20 | 19 | 5 | 24 | 14 | 15 | 39 |
| nad2 | 16 | 0 | 16 | 23 | 29 | 52 | 18 | 26 | 44 |
| nad3 | 12 | 0 | 12 | 10 | 4 | 14 | 7 | 5 | 12 |
| nad4 | 21 | 2 | 23 | 23 | 3 | 26 | 17 | 25 | 42 |
| nad4L | 3 | 0 | 3 | 3 | 2 | 5 | 4 | 5 | 9 |
| nad5 | 15 | 7 | 22 | 41 | 42 | 83 | 26 | 40 | 66 |
| nad6 | 10 | 1 | 11 | 13 | 7 | 20 | 5 | 14 | 19 |
| nad9 | 4 | 1 | 5 | 8 | 14 | 22 | 6 | 10 | 16 |
| rpl2 | 15 | 24 | 39 | ||||||
| rpl5 | 6 | 0 | 6 | 4 | 8 | 12 | |||
| rpl6 | 4 | 0 | 4 | ||||||
| rpl10 | 4 | 14 | 18 | ||||||
| rpl16 | 2 | 0 | 2 | ||||||
| rps2 | 3 | 1 | 4 | 5 | 9 | 14 | |||
| rps3 | 5 | 7 | 12 | 16 | 12 | 28 | |||
| rps4 | 5 | 2 | 7 | 5 | 4 | 9 | |||
| rps10 | 1 | 2 | 3 | ||||||
| rps11 | 4 | 2 | 6 | ||||||
| rps12 | 1 | 0 | 1 | ||||||
| rps13 | 8 | 0 | 8 | ||||||
| rps14 | 0 | 0 | 0 | ||||||
| rps19 | 1 | 1 | 2 | ||||||
| sdh3 | 6 | 3 | 9 | 2 | 2 | 4 | |||
| sdh4 | 7 | 2 | 9 | ||||||
| tatC | 13 | 5 | 18 | 12 | 25 | 37 | 14 | 32 | 46 |
| sum | 250 | 84 | 334 | 279 | 297 | 576 | 146 | 208 | 364 |
No foreign DNA of chloroplast or nuclear origin was detected in the Huperzia mitochondrial genome. This result is the same as what was found in the Selaginella chondrome [25]. In the Isoetes chondrome, however, three short pieces of chloroplast and nuclear DNAs were detected despite the fact that the genome seemed to be relatively compact [24].
Gene Content
The Huperzia mitochondrial genome contains 66 genes, with 37 coding for proteins, 3 for ribosomal RNAs, and 26 for transfer RNAs (Fig. 1, Table S2). The 37 protein-coding genes include 8 genes for NADH:ubiquinone oxidoreductase (complex I of the respiratory chain, as designated in [1]; nad1-6, 4L, 9), 2 genes for succinate:ubiquinone oxidoreductase (complex II; sdh3, 4), 1 gene for ubiquinol:cytochrome c oxidoreductase (complex III; cob), 3 genes for cytochrome c oxidase (complex IV; cox1-3), 5 genes for adenosine triphosphate synthase (complex V; atp1, 4, 6, 8, 9), 1 gene for cytochrome c biogenesis (ccmFC), 16 genes for ribosomal proteins, and 1 gene for other functions (tatC).
There is a duplicated set of rRNA genes. Several tRNA genes also have duplicated copies, some up to 3 copies (Table S2). There is no chloroplast-originated tRNA gene in the Huperzia chondrome.
Among the total of 80 genes (66 unique ones plus 14 duplicated copies), six are pseudogenes. One of them is ccmFC, which is the only remaining member of the gene complex coding for cytochrome c biogenesis function. The pseudogene argument is supported by two lines of evidence: about 500 nucleotides are missing in the first exon and there are several indels that disrupt the reading frame (Fig. S1). One interesting aspect about this gene is that it is split into two pieces located on two different strands far apart in the genome, with 80 nucleotides of well conserved sequence of the 3′-end of the intron attached to the second exon (Figs. 1 and S1). The gene nad7, present in other two lycophytes (Isoetes [24] and Selaginella [25]) and other vascular plants, but absent or present as a pseudogene in some bryophytes, is absent in the Huperzia chondrome (Table S2). The repeated efforts to find this gene in the fosmid library screening experiments did not yield any positive clone. For ribosomal protein genes, many of which have been lost from the completely sequenced mitochondrial genomes of two hornworts [22], [23] and apparently also from the chondromes of Isoetes and Selaginella [24], [25], there are still 16 genes in the Huperzia mitochondrial genome and 14 of them are functional. In land plants, only liverworts have more ribosomal protein genes in their chondromes [18]. Likewise, the Huperzia mitochondrial genome is among the most tRNA gene-rich land plant chondromes, and this condition is in stark contrast to the other two lycophytes, which seem to have lost most or all tRNA genes from their chondromes [24], [25].
Three recent studies reported pseudogene pieces in intergenic spacers [18], [38], [39]. One of them performed a systematic survey of pseudogene pieces in all seven sequenced bryophyte mitochondrial genomes and found that the three liverworts had a few dozens of such fragments in the spacers, whereas the two hornworts had only a few pieces and the two mosses had none [18]. In the Huperzia chondrome, 69 pseudogene pieces were found in 32 spacers (Table S1), and they were derived from 24 of the 40 protein- and rRNA-coding genes. For all genes encoding functions involved in respiration (excluding the dysfunctional ccmFC), only nad2 and sdh4 had no pseudogene piece in the spacers. In addition, tatC and rrn18 lacked any piece in the spacers. In contrast, only six of the 14 functional ribosomal protein genes had pseudogenes, rpl2, rps2, rps3, rps4, rps10, and rps12. Pseudogenes were also detected for seven tRNA genes: trnFgaa, trnKuuu, trnLuaa, trnMfcau, trnPugg, trnWcaa, and trnYgua. Because tRNA genes in general are very short and show extreme sequence conservation, they could not be subject to the same kind of analyses as were done to the protein- and rRNA-coding genes for investigation of the sources and mechanisms of origins of the pseudogenes. Hence, they will not be discussed any further.
One question to ask is where these pseudogene pieces came from. We examined alignment of the functional copy and pseudogene pieces of the gene as well as its functional ortholog from other sequenced plant mitochondrial genomes (Fig. S2). In addition, we performed phylogenetic analysis using the alignment (phylogenetic trees not shown). Among the 24 genes of this kind, 17 genes have their pseudogene pieces grouped with the functional copy from the Huperzia chondrome. For the other seven genes (atp8, cob, nad4L, nad5, rps2, rps3, and rps12), one or a few pseudogene pieces were either short or somewhat divergent, and thus grouped with the functional ortholog from other species. Finally, the same kind of analyses were performed for five fragments of three group II introns in such pseudogene pieces: cox2i691, cox3i171, and rps10i235 (Fig. S3), and the results showed that all intron fragments in the pseudogene pieces were more closely related to the introns in the functional genes of the Huperzia chondrome than to those from other plant chondromes. Therefore, these data suggest that the most pseudogene pieces came from their corresponding functional genes in the Huperzia chondrome. For those that did not group with the functional copy from the Huperzia chondrome, one explanation may be that they have accumulated aberrant mutations after pseudogenization, and our examination of the alignment seems to support such an interpretation. The intron cox3i171 provides extra information to support that the pseudogene pieces in the Huperzia chondrome originated within the genome, not from outside, because this intron has only been found in liverworts and Lycopodiaceae so far [18], [19], [20], [40], and three copies of this intron from the Huperzia pseudogenes are all much more similar to the intron in the functional copy of the Huperzia chondrome than those from the three liverworts (Fig. S3).
A further question to ask is how these pseudogene pieces arose. One possible mechanism is retroposition: reverse transcription of the gene transcript and insertion of the cDNA back into the genome. A piece of evidence supporting this scenario is that several intron-containing genes have intron-less fragments in the spacers (Table S1, Fig. S4). However, some pseudogene pieces contain intron fragments. This situation can be explained by the use of intron-containing pre-mRNAs as templates for reverse transcription or by other mechanisms of sequence duplication that do not involve RNA intermediates. Our examination of alignment between the functional gene and pseudogene piece(s) in all cases (Fig. S4) showed that a majority of the pseudogene pieces that lack introns were resulted from cDNAs with precise splicing removal of introns and connection of exons. In cases where introns were still present, regions of the exon/intron juncture were well aligned; the introns also aligned well between the functional gene and the pseudogene piece(s) (Figs. S3 & S4). Ideally, RNA-edited sites can also be compared between the pseudogene pieces and the functional gene to test whether reverse transcription was involved, but lack of cDNA sequences data prevents this analysis from being done. The results of in silico analyses of RNA editing are just not accurate enough to permit such secondary analysis. Finally, we emphasize that despite the relatively strong evidence uncovered in this study that supports a retroposition mechanism for the origin of the pseudogene pieces in intergenic spacers, other mechanisms cannot be completely excluded, particularly for those pieces that did not group with the functional copy of the same species.
Regardless of mechanisms responsible for origins of these pseudogene pieces, their presence in such abundance from so many genes in the Huperzia mitochondrial genome poses an interesting question on why they exist. Recently, it has been reported that thousands of or even more pseudogenes are present in sequenced nuclear genomes of plants and animals and that retroposition seems to be the mechanism of their origin [41], [42], [43], [44]. Some of these pseudogenes produce antisense small RNAs with features similar to small interfering RNAs [43]. It will be desirable to investigate whether pseudogenes in plant mitochondrial genomes have similar functions.
Gene Order and Repeat Sequences
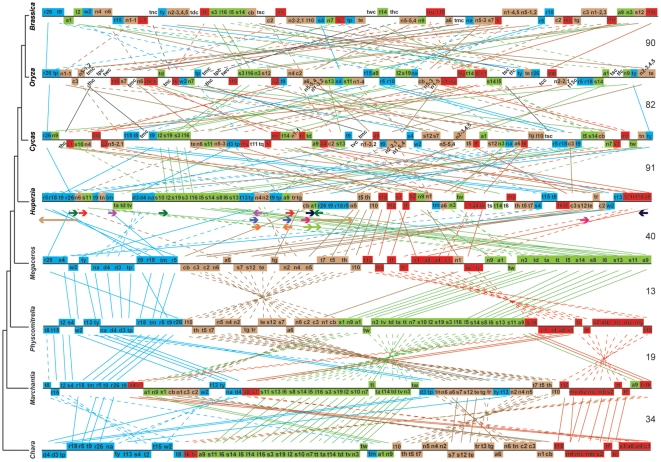
The gene order in the Huperzia mitochondrial genome can be described as half bryophyte-like and half seed plant-like. This genome exhibits the most dramatic rearrangement since the origin of land plants; 40 events of deletion, duplication, inversion, and translocation are required to bring the chondromes of Huperzia and Megaceros into complete synteny (Fig. 2). The level of rearrangement during the origin of vascular plants surpasses what the mitochondrial genome experienced when plants colonized land (34 events). Ten gene clusters conserved in the Chara and bryophyte chondromes are present in this early vascular plant chondrome: s10-l2-s19-s3-l16-l5-s14-s8-l6-s13, r5-r18-t9-r26, tv-td-ta, d3-d4, t13-ty, n2-n4, tr-tg, t7-t5-th, a4-c1, and te-s12 (see Table S2 for abbreviated and full gene names). The ribosomal protein gene cluster, a putative polycistronic operon that can be traced back to the mitochondrial genome of Reclinomonas americana, an early eukaryote [5], is still intact and comprises 10 genes in Huperzia. It is also interesting to note that the gene cluster of n5-t5-th-l10-t10-my-tf-s1-s2, formed through juxtaposition by parts of two gene clusters likely in the common ancestor of hornworts and vascular plants, survived genome shuffling during the bryophyte-vascular plant transition (Fig. 2). On the other hand, several blocks of genes in the chondromes of charophytes and bryophytes no longer stay together in the Huperzia chondrome, e.g., (n6)-c2-c3-(n1)-cb, n2-n4-n5, c1-a4-(a8-s1). In Figure 2, most genes shown in blue, brown, and red color and even some genes in green, which largely stayed together in the Chara and bryophyte chondromes, are dispersed all over the genome in Huperzia.
Figure 2. Gene order comparison among mitochondrial genomes of Chara vulgaris, Marchantia polymorpha, Physcomitrella patens, Megaceros aenigmaticus, Huperzia squarrosa, Cycas taitungensis, Oryza sativa, and Brassica napus.
Species are arranged according to the organismal phylogeny of land plants and the outgroup [29]. Solid lines connect orthologous genes between species with the same orientation, and dashed lines connect those with the reversed orientation. Repeat sequences (shown in colored arrows) in Huperzia are color-coded: RepA – red, RepB – purple, RepC – blue, RepD – black, RepE – light green, RepF – green, RepG – orange, RepH – brown, and RepI – pink. The inferred number of events of deletion, duplication, inversion, and translocation required to bring the two adjacent chondromes into complete synteny is shown on the right between the two genomes.
Nine classes of repeat sequences longer than 100 bp were detected in the Huperzia chondrome (Table 4). All of them have two copies except one, RepF, which has three copies. Some of them are direct repeats whereas others are inversely oriented. Six of the repeat classes (RepB, C, D, E, F and I) show homology to genes or introns in the genome, and three of them (RepB, C, and I) in fact involve introns as the repeats per se. These sequence homologies suggest that the repeats arose from duplication of pre-existing sequences within the genome, perhaps mediated initially by transposons. A preliminary examination of transposon fragment distribution shows that most of the nine repeat classes have such fragments located within 2 kb on at least one side (data not shown).
Table 4. Repeat sequences in the mitochondrial genome of Huperzia squarrosa 1.
| Name | Location | Length (bp) | Orientation | Origin of the repeat |
| RepA_Hs | atp9-trnRacg | 124 | direct | – |
| nad6-rps11 | ||||
| RepB_Hs | trnMcau-trnAugc | 1,241 | inverted | nad4i461g2 |
| part nad4i461g2 | ||||
| RepC_Hs | part atp9i87g2 | 681 | direct | atp9i87g2 |
| trnYgua-nad4 | ||||
| RepD_Hs | cob-atp1 | 362 | inverted | rrn26 |
| trnQuug-atp8 | ||||
| RepE_Hs | cob-atp1 | 143 | direct | atp8 |
| cob-atp1 | ||||
| RepF_Hs | rrn26-nad6 | 168 | direct | cox3i171g2 |
| rps10-rpl2 | ||||
| atp1-rrn26 | (inverted) | |||
| RepG_Hs | atp9-trnRacg | 100 | inverted | – |
| trnYgua-nad4 | ||||
| RepH_Hs2 | atp1-nad5 | 14,637 | inverted | rrn26, trnMfcau, |
| atp8-nad6 | rrn18, rrn5 | |||
| RepI_Hs | part cobi693g2 | 764 | direct | cobi693g2 |
| rps12-trnEuuc |
Only identical repeats are listed, except for RepH_Hs, which has one indel and one substitution, and RepI_Hs, which has two indels and one substitution between the two copies.
RepH_Hs includes the complete genes of rrn5, rrn18, rrn26, and trnMfcau and part of nad5 gene.
Fifteen microsatellite sequences of di-, tri-, and tetra-nucleotides were found in the Huperzia chondrome, with the tri-nucleotide type being the most abundant (9 sequences), the di-nucleotide type less so (4 sequences), and the tetra-nucleotide type the least so (2 sequences). None of them was located in any of the repeat sequences identified above, in a stark contrast to what was found in the Selaginella chondrome, where a much larger number of microsatellites were detected and 82 out of the total of 98 microsatellites occurred in five repeats [25].
A model was proposed more than twenty years ago on how repeat sequences were responsible for plant mitochondrial genome rearrangement and large repeats were generated via short direct repeats-mediated recombination [45], [46]. This model has recently been substantiated by data from the completely sequenced cucumber chondrome [47]. It also seems to explain the distribution of repeats and genes that have changed locations in the Huperzia chondrome (relative to the bryophyte chondromes). First, three classes of repeats were involved in disruption of some gene clusters in bryophytes and resulting in the current gene distribution pattern in the Huperzia: RepA for rps11-atp9 (which was linked in Chara, Physcomitrella, and Megaceros); RepD for cob-trnQuug(t10) (linked in Chara and Chaetosphaeridium (NC_004118)); RepG for nad4-nad2-trnGgcc(tg)-trnRacg(tr) (linked in Physcomitrella and Megaceros (tr is lost in the latter)); RepG for trnGgcc(tg)-trnRacg(tr)-trnRucu(t13)-trnYgua(ty) (linked in Marchantia) (Figs. 1 and 2). Second, two of the three copies of RepF are located near the two copies of RepH, which is the sole long (14 kb) repeat class in the Huperzia chondrome. Third, the RepB, C, and I are all duplicated intron portions and rearrangement involving them would disrupt genes. Given that there is lack of trans-splicing capability in the genome (no trans-splicing intron (see below)), it is understandable that these three repeat classes were not involved in genome rearrangement. Finally, for RepE, which has both copies located in the same long spacer between atp1 and cob, any rearrangement facilitated by them would not be detected.
One unexplained observation is that all repeats except one copy of RepI are located in half of the genome (Fig. 1). Nevertheless, in both halves of the genome that contain or lack repeats, there are regions that show many rearrangements or gene order conservation (Fig. 2). Thus, there are probably many repeats under 100 bp that escaped detection because of the search criterion of 100 bp.
Finally, we want to add that in the process of isolating mitochondrial DNA fragments for sequencing and assembling the genome, we did not detect existence of multipartite subgenomic circles as found in some angiosperms [47], [48].
Intron Content
The Huperzia mitochondrial genome contains 32 group II introns and no group I intron, according to the definitions of these mobile genetic elements [49]. They are located in 15 genes: atp6, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad5, rpl2, rps3, rps10, and rps14. All of them are cis-spliced (Table S3). The intron complement in the Huperzia chondrome is a mixed result of intron gains and losses at different stages of land plant evolution.
While intron content has been shown to be highly stable in the mitochondrial genomes of each of the three bryophyte lineages [18], it is not in the chondromes of lycophytes. Among a total of 48 intron positions in the chondromes of three lycophytes (Huperzia, Isoetes, and Selaginella), 32 positions show variability in intron content: atp6i80g2, atp9i95g2, cox1i227g2, cox1i266g2, cox1i323g2, cox1i395g1, cox1i511g2, cox1i876g1, cox1i1149g2, cox1i1305g1, cox2i373g2, cox2i691g2, cox3i171g2, nad1i477g2, nad1i669g288, nad1i728g2, nad2i542g2, nad2i709g2, nad4i976g2, nad4i1399g2, nad5i392g2, nad7i140g2, nad7i209g2, nad7i676g2, nad7i917g2, nad7i1113g2, rpl2i917g2, rps3i74g2, rps3i257g2, rps10i235g2, rps14i114g2, and rrn18i839g1 (Table S3). This level of intron distribution variation within a major lineage is unprecedented in land plants. It may be partly due to the fact that two of the three lycophytes, Isoetes [24] and Selaginella [25], have extremely unusual mitochondrial genomes while Huperzia has a rather conventional plant chondrome.
The Most Archaic Mitochondrial Genome of Vascular Plants in Huperzia
Lycophytes are the sister lineage to all other vascular plants [28], [29], and hence are likely to capture many ancestral features of vascular plants. The Huperzia chondrome represents the most archaic form of vascular plant mitochondrial genomes when compared with those of other vascular plants and the outgroup bryophytes. Its ancestral nature is primarily reflected in the gene order. Among more than two dozens of vascular plant chondromes sequenced to date (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=33090&opt=organelle), the Huperzia mitochondrial genome has the least rearranged gene order relative to the seven bryophyte chondromes [18], [19], [20], [21], [22], [23] (Fig. 2). First, it has a large conserved gene cluster (containing 10 genes) that has been well conserved since the origin of mitochondria – the ribosomal protein gene cluster [5]. In contrast, this gene cluster is broken into much smaller ones containing no more than four genes in the chondromes of seed plants and two other lycophytes, Isoetes and Selaginella [24], [25] (Fig. 2). Second, the low level of genome rearrangement in the Huperzia mitochondrial genome is reflected by the fact that all of its 32 introns are cis-spliced. In the highly rearranged chondromes of seed plants, several group II introns in nad1, nad2, and nad5 are trans-spliced [50] (Table S3), and one of them, nad1i728g2, has undergone cis- to trans-splicing transition many times independently [51], [52]. Not surprisingly, the highly rearranged chondromes of Isoetes and Selaginella contain their own sets of trans-splicing introns, and in fact a first ever trans-splicing group I intron has been discovered in Isoetes [24], [25] (Table S3). A third indicator of the archaic gene order in the Huperzia chondrome is that only 40 events of deletion, duplication, inversion, and translocation are required to bring this genome and that of Megaceros into complete synteny, whereas more than twice as many events are required to bring the chondromes of Huperzia, Cycas, Oryza, and Brassica into complete synteny (Fig. 2). It should be added that this indicator does not reflect accurately the level of genomic rearrangement that happened during evolution because of the following two facts. One is that evolutionary gaps between Huperzia and Cycas, between Cycas and Oryza, and between Oryza and Brassica are smaller than that between Megaceros and Huperzia [53]. The other is that in seed plants the number of events inferred to bring two chondromes into complete synteny is almost certainly underestimated because these genomes are so recombinogenic that the rate is likely saturated. For example, two cytotypes of one maize species differ by as many as 16 rearrangement events [54].
Presence of the large conserved ribosomal protein gene cluster and several small gene clusters in the Huperzia chondrome suggests that this genome still uses an ancestral type of gene expression system, presumably with a relatively small number of promoter sequences in the genome. In contrast, the mitochondrial genomes of seed plants probably have a derived type of gene expression system, with one or multiple promoters for each of their most genes, because of the high frequency of genome rearrangement among species and the genome structure of having mostly free-standing genes (or gene pieces in cases of trans-splicing intron-connected exons) [8], [9], [10], [11], [12].
Several other aspects of the Huperzia chondrome reinforces its archaic status among all vascular plant mitochondrial genomes. One is its gene content, with nearly the full set of genes found in the chondromes of Chara, Marchantia, and Physcomitrella still present in this genome. The only major categories of genes that are missing or have become pseudogenes are ccm genes and nad7. Ribosomal protein genes and tRNA genes, which have been lost in Isoetes, Selaginella and some angiosperms [24], [25], [55], are almost all present in Huperzia. Second, the level of RNA editing is quite low in the Huperzia mitochondrial genome when compared with that in the Isoetes and Selaginella chondromes [25], [37], but is comparable with the editing levels in several angiosperm mitochondrial genomes [34], [56], [57], [58]. Third, there is lack of foreign DNAs of chloroplast or nuclear origin in the Huperzia chondrome, unlike what was observed in Isoetes, Cycas, and some angiosperms, where chloroplast tRNA genes and other fragments, or nuclear DNAs have invaded the mitochondrial genome, sometimes on a massive scale [13], [24], [47], [56], [59]. Finally, even though the Huperzia chondrome is 2–4 times the sizes of bryophyte chondromes, it is in no position to compete with some monstrous angiosperm mitochondrial genomes [35], [47], [56], [59]. The genome size increase in the Huperzia mitochondria seems to be related to the overall tolerance of large genomes in cells of vascular plants when the diploid phase becomes dominant in the life cycle of a plant [60], [61]. It is perhaps caused mostly by retroposition of pseudogenes into intergenic spacers, not as a result of massive invasion of foreign DNAs from the chloroplast and nucleus as seen in some angiosperm chondromes [47], [56], [59].
Materials and Methods
Approximately 10 g of fresh tissue of Huperzia squarrosa (G. Forster) Trevis was collected in Matthaei Botanical Gardens at the University of Michigan. The material was brought to the lab for cleaning under a dissecting scope. A voucher specimen numbered Qiu 05001 was deposited at the University Herbarium.
Total cellular DNA was extracted with the CTAB method [62], and purified with phenol extraction to remove proteins. A fosmid library was constructed using the CopyControl™ kit (EPICENTRE Biotechnologies, Madison, Wisconsin, USA) from the total cellular DNA fragments of 35–45 kb size-selected by agarose gel electrophoresis. No restriction enzyme digestion or mechanical shearing was used before electrophoresis. Clones containing mitochondrial DNA fragments were identified through Southern hybridizations using the HRP chemiluminescent blotting kit (KPL, Inc., Gaithersburg, Maryland, USA), with major mitochondrial genes as probes. The probes were made by amplification from total cellular DNAs of Marchantia polymorpha and Arabidopsis thaliana.
The inserts were sequenced with two methods. First, fosmid DNA was sheared into 2–3 kb segments and then the DNA segments were purified by agarose gel and cloned in pUC-18 vector for shotgun-sequencing library construction. Thermo-cycling sequencing reaction was performed in a final volume of 24 µL containing 16-µL DYEnamic ET Terminator sequencing kit premix, 10 pM universal sequencing primers, and 500 ng plasmid DNA. The reaction conditions were 95°C for 2 min, followed by 35 cycles of 95°C denaturation for 15 s, 50°C annealing for 15 s, and 60°C extension for 90 s. The amplified DNA fragments were sequenced on an ABI-3730 DNA sequencer (Applied Biosystems, Foster City, California, USA). DNA sequences were assembled by using the software package phred/phrap/consed/ [63], [64] on a PC/UNIX platform. Approximately 270 kb was obtained with this method. Second, more inserts, which connected the entire genome circle, were sequenced using primer-walking on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, California, USA). Sequences were assembled using Sequencher (Gene Codes Corp., Ann Arbor, Michigan, USA).
The mitochondrial genomes were annotated in seven steps. First, genes for known mitochondrial proteins and rRNAs were identified by Basic Local Alignment Search Tool (BLAST) searches [65] (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) of the non-redundant database at the National Center for Biotechnology Information (NCBI). The exact gene and exon/intron boundaries were predicted by alignment of orthologous genes from annotated plant mitochondrial genomes available at the organelle genomic biology website at NCBI (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/organelles.html). Occurrence of RNA editing was inferred through creation of proper start and stop codons as well as removal of internal stop codons. Further, RNA editing sites were predicted by in silico analyses using the recently developed software PREPACT (www.prepact.de) and following the default settings [36]. Sequences of mitochondrial genes from Marchantia [19] or cDNAs from Isoetes (GenBank accessions HQ616410–HQ616434) [37] and Selaginella (GenBank accessions JF276233–JF276250) [25] were used as reference templates in three separate analyses to minimize the effect of sequence divergence among species. The Marchantia gene sequences could be used for such analyses because no RNA editing has been detected in this chondrome. Second, genes for hypothetical proteins were identified using the web-based tool - Open Reading Frames Finder (ORF-finder; http://www.ncbi.nlm.nih.gov/gorf/gorf.html) with the standard genetic code. Third, genes for tRNAs were found using tRNAscan-SE [66] (http://lowelab.ucsc.edu/tRNAscan-SE/). Fourth, repeated sequences were searched using REPuter [67] (http://bibiserv.techfak.uni-bielefeld.de/reputer/) or BLAST. Fifth, microsatellite sequences were screened using msatcommander 0.8.2 with the following settings: accepting di-nucleotide (di-) repeats of six or more, and tri-, tetra-, penta- and hexa-nucleotide repeats of four or more [68] (five was used for all five categories in the Selaginella study [25]). Finally, pseudogene pieces in intergenic spacers were identified by BLAST genes against spacers, and those longer than 50 bp were recorded in this study.
To detect DNAs of chloroplast and nuclear origin in the Huperzia mitochondrial genome, we compared the Huperzia chondrome with the chloroplast genome of Huperzia lucidula [69] and the nuclear genome of Selaginella moellendorffii [70] using the program blastn at NCBI. In both analyses, default settings were used.
The annotated GenBank file of the Huperzia mitochondrial genome was used to draw a gene map by using OrganellarGenomeDRAW tool (OGDRAW) [71]. The map was then examined for further comparison of gene order and content. When sequence homology in some parts of certain genes or intergenic spacers was uncertain, the sequences were aligned using CLUSTAL_X [72], with visual examination followed.
Supporting Information
Alignment of ccmFC Sequences from Physcomitrella , Cycas , and Huperzia .
(DOCX)
Alignment of 24 genes and their pseudogene piece(s) from the Huperzia mitochondrial genome and the functional ortholog from other plants. Most of these plants have their mitochondrial genomes sequenced, which are available at NCBI Organelle Genome Resources (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759&hopt=html). A small number of sequences are from GenBank and have their accession numbers listed after the taxon names. Coordinate numbers indicating location of a pseudogene piece within the Huperzia mitochondrial genome are listed in the sequence name. If desired, each matrix can be copied in “word” to make a “.txt” file and opened in PAUP to run a phylogenetic analysis to determine evolutionary relationships of the pseudogene pieces.
(DOCX)
Alignment of introns that are attached to pseudogene piece(s) or located in the functional gene in the Huperzia mitochondrial genome and the ortholog intron from other plants. Most of these plants have their mitochondrial genomes sequenced, which are available at NCBI Organelle Genome Resources (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759&hopt=html). Coordinate numbers indicating location of a pseudogene intron piece within the Huperzia mitochondrial genome are listed in the sequence name. If desired, each matrix can be copied in “word” to make a “.txt” file and opened in PAUP to run a phylogenetic analysis to determine evolutionary relationships of the introns attached to the pseudogene pieces.
(DOCX)
Alignment of functional genes and their pseudogene pieces in the Huperzia mitochondrial genome. All queries are functional genes whereas subjects are pseudogene pieces.
(DOCX)
Pseudogene pieces in intergenic spacers of Huperzia squarrosa mitochondrial genome.
(DOC)
Gene contents in mitochondrial genomes of selected charophyte and land plants.
(DOC)
Intron contents in mitochondrial genomes of selected charophyte and land plants.
(DOC)
Acknowledgments
We thank Richard W. Jobson, Jeffrey D. Palmer and Yasuo Sugiyama for helpful discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Science Foundation (NSF) grants DEB 0531689 and 0332298 to YQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 3.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paquin B, Laforest MJ, Forget L, Roewer I, Wang Z, et al. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- 5.Lang BF, Burger G, Okelly CJ, Cedergren R, Golding GB, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 6.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 7.Schuster W, Brennicke A. The plant mitochondrial genome - physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78. [Google Scholar]
- 8.Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenothera berteriana. J Biol Chem. 1993;268:7849–7855. [PubMed] [Google Scholar]
- 9.Binder S, Brennicke A. Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos Trans R Soc London Ser B. 2003;358:181–189. doi: 10.1098/rstb.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulligan RM, Maloney AP, Walbot V. RNA processing and multiple transcription initiation sites result in transcript size heterogeneity in maize mitochondria. Mol Gen Genet. 1988;211:373–380. doi: 10.1007/BF00425688. [DOI] [PubMed] [Google Scholar]
- 11.Fey J, Marechal-Drouard L. Compilation and analysis of plant mitochondrial promoter sequences: an illustration of a divergent evolution between monocot and dicot mitochondria. Biochem Biophys Res Commun. 1999;256:409–414. doi: 10.1006/bbrc.1999.0349. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn K, Weihe A, Borner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaw SM, Shih ACC, Wang D, Wu YW, Liu SM, et al. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 14.Turmel M, Otis C, Lemieux C. An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics. 2007;8:137. doi: 10.1186/1471-2164-8-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol Biol Evol. 2002;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- 16.Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci, USA. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turmel M, Otis C, Lemieux C. The mitochondrial genome of Chara vulgaris: Insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell. 2003;15:1888–1903. doi: 10.1105/tpc.013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Xue J-Y, Wang B, Li L, Qiu Y-L. The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution. PLoS One. 2011;6:e25836. doi: 10.1371/journal.pone.0025836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA - A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Xue J-Y, Li L, Liu L, Qiu Y-L. The complete mitochondrial genome sequence of the liverwort Pleurozia purpurea reveals extremely conservative mitochondrial genome evolution in liverworts. Curr Genet. 2009;55:601–609. doi: 10.1007/s00294-009-0273-7. [DOI] [PubMed] [Google Scholar]
- 21.Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, et al. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol. 2007;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Wang B, Liu Y, Qiu Y-L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol. 2009;68:665–678. doi: 10.1007/s00239-009-9240-7. [DOI] [PubMed] [Google Scholar]
- 23.Xue J-Y, Liu Y, Li L, Wang B, Qiu Y-L. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr Genet. 2010;56:53–61. doi: 10.1007/s00294-009-0279-1. [DOI] [PubMed] [Google Scholar]
- 24.Grewe F, Viehoever P, Weisshaar B, Knoop V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009;15:5093–5104. doi: 10.1093/nar/gkp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 27.Tian XJ, Zheng J, Hu SN, Yu J. The rice mitochondrial genomes and their variations. Plant Physiol. 2006;140:401–410. doi: 10.1104/pp.105.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255:1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- 29.Qiu Y-L, Li LB, Wang B, Chen ZD, Knoop V, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci, USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jermy AC. Isoetaceae. In: Kramer KU, Green PS, editors. The Families and Genera of Vascular Plants, Vol I, Pteridophytes and Gymnosperms. Berlin: Springer-Verlag; 1990. pp. 26–31. [Google Scholar]
- 31.Jermy AC. Selaginellaceae. In: Kramer KU, Green PS, editors. The Families and Genera of Vascular Plants, Vol I, Pteridophytes and Gymnosperms. Berlin: Springer-Verlag; 1990. pp. 39–45. [Google Scholar]
- 32.Ollgaard B. Lycopodiaceae. In: Kramer KU, Green PS, editors. The Families and Genera of Vascular Plants, Vol I, Pteridophytes and Gymnosperms. Berlin: Springer-Verlag; 1990. pp. 31–39. [Google Scholar]
- 33.Chang S, Yang T, Du T, Huang Y, Chen J, et al. Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC Genomics. 2011;12:497. doi: 10.1186/1471-2164-12-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenz H, Rüdinger M, Volkmar U, FIscher S, Herres S, et al. Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Curr Genet. 2009;56:189–201. doi: 10.1007/s00294-009-0283-5. [DOI] [PubMed] [Google Scholar]
- 37.Grewe F, Herres S, Viehoever P, Polsakiewicz M, Weisshaar B, et al. A unique transcriptome: 1728 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011;39:2890–2902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groth-Malonek M, Rein T, Wilson R, Groth H, Heinrichs J, et al. Different fates of two mitochondrial gene spacers in early land plant evolution. Int J Plant Sci. 2007;168:709–717. [Google Scholar]
- 39.Wahrmund U, Groth-Malonek M, Knoop V. Tracing plant mitochondrial DNA evolution: rearrangements of the ancient mitochondrial gene cluster trnA- trnT- nad7 in liverwort phylogeny. J Mol Evol. 2008;66:621–629. doi: 10.1007/s00239-008-9114-4. [DOI] [PubMed] [Google Scholar]
- 40.Hiesel R, von Haeseler A, Brennicke A. Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proc Natl Acad Sci, USA. 1994;91:634–638. doi: 10.1073/pnas.91.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Zheng H, Fan C, Li J, Shi J, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Zhang Z, Gerstein MB, Zheng D. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comp Biol. 2009;5:e1000449. doi: 10.1371/journal.pcbi.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podlaha O, Zhang JZ. Pseudogenes and their evolution. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, Ltd; 2010. pp. 1–8. [Google Scholar]
- 45.Small I, Suffolk R, Leaver CJ. Evolution of plant mitochondrial genomes via substoichiometric intermediate. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 46.André C, Levy A, Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- 47.Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011;23:2499–2513. doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 49.Michel F, Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2:33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malek O, Knoop V. Trans-splicing group II introns in plant mitochondria: The complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA. 1998;4:1599–1609. doi: 10.1017/s1355838298981262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dombrovska O, Qiu Y-L. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogen Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Qiu Y-L, Palmer JD. Many independent origins of trans splicing of a plant mitochondrial group 2 intron. J Mol Evol. 2004;59:80–89. doi: 10.1007/s00239-004-2606-y. [DOI] [PubMed] [Google Scholar]
- 53.Qiu Y-L. Phylogeny and evolution of charophytic algae and land plants. J Syst Evol. 2008;46:287–306. [Google Scholar]
- 54.Allen JO, Fauron CM, Minx P, Roark L, Oddiraju S, et al. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics. 2007;177:1173–1192. doi: 10.1534/genetics.107.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci, USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alverson AJ, Wei XX, Rice DW, Stern DB, Barry K, et al. Insights into the Evolution of Mitochondrial Genome Size from Complete Sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mower JP, Palmer JD. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genomics. 2006;276:285–293. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- 58.Giege P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci, USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez-Moreno L, González VM, Benjak A, Martí MC, Puigdomènech P, et al. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 2011;12:424. doi: 10.1186/1471-2164-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu Y-L, Taylor AB, McManus HA. Evolution of the life cycle in land plants. J Syst Evol. 2012 in press. [Google Scholar]
- 61.Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta). Ann Bot. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doyle JJ, Doyle JS. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bulletin. 1987;19:11–15. [Google Scholar]
- 63.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 64.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faircloth BC. MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour. 2008;8:92–94. doi: 10.1111/j.1471-8286.2007.01884.x. [DOI] [PubMed] [Google Scholar]
- 69.Wolf PG, Karol KG, Mandoli DF, Kuehl J, Arumuganathan K, et al. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene. 2005;350:117–128. doi: 10.1016/j.gene.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW) - a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 72.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of ccmFC Sequences from Physcomitrella , Cycas , and Huperzia .
(DOCX)
Alignment of 24 genes and their pseudogene piece(s) from the Huperzia mitochondrial genome and the functional ortholog from other plants. Most of these plants have their mitochondrial genomes sequenced, which are available at NCBI Organelle Genome Resources (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759&hopt=html). A small number of sequences are from GenBank and have their accession numbers listed after the taxon names. Coordinate numbers indicating location of a pseudogene piece within the Huperzia mitochondrial genome are listed in the sequence name. If desired, each matrix can be copied in “word” to make a “.txt” file and opened in PAUP to run a phylogenetic analysis to determine evolutionary relationships of the pseudogene pieces.
(DOCX)
Alignment of introns that are attached to pseudogene piece(s) or located in the functional gene in the Huperzia mitochondrial genome and the ortholog intron from other plants. Most of these plants have their mitochondrial genomes sequenced, which are available at NCBI Organelle Genome Resources (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759&hopt=html). Coordinate numbers indicating location of a pseudogene intron piece within the Huperzia mitochondrial genome are listed in the sequence name. If desired, each matrix can be copied in “word” to make a “.txt” file and opened in PAUP to run a phylogenetic analysis to determine evolutionary relationships of the introns attached to the pseudogene pieces.
(DOCX)
Alignment of functional genes and their pseudogene pieces in the Huperzia mitochondrial genome. All queries are functional genes whereas subjects are pseudogene pieces.
(DOCX)
Pseudogene pieces in intergenic spacers of Huperzia squarrosa mitochondrial genome.
(DOC)
Gene contents in mitochondrial genomes of selected charophyte and land plants.
(DOC)
Intron contents in mitochondrial genomes of selected charophyte and land plants.
(DOC)