Abstract
Liver-specific and nonliver-specific methionine adenosyltransferases (MATs) are products of two genes, MAT1A and MAT2A, respectively, that catalyze the formation of S-adenosylmethionine (AdoMet), the principal biological methyl donor. Mature liver expresses MAT1A, whereas MAT2A is expressed in extrahepatic tissues and is induced during liver growth and dedifferentiation. To examine the influence of MAT1A on hepatic growth, we studied the effects of a targeted disruption of the murine MAT1A gene. MAT1A mRNA and protein levels were absent in homozygous knockout mice. At 3 months, plasma methionine level increased 776% in knockouts. Hepatic AdoMet and glutathione levels were reduced by 74 and 40%, respectively, whereas S-adenosylhomocysteine, methylthioadenosine, and global DNA methylation were unchanged. The body weight of 3-month-old knockout mice was unchanged from wild-type littermates, but the liver weight was increased 40%. The Affymetrix genechip system and Northern and Western blot analyses were used to analyze differential expression of genes. The expression of many acute phase-response and inflammatory markers, including orosomucoid, amyloid, metallothionein, Fas antigen, and growth-related genes, including early growth response 1 and proliferating cell nuclear antigen, is increased in the knockout animal. At 3 months, knockout mice are more susceptible to choline-deficient diet-induced fatty liver. At 8 months, knockout mice developed spontaneous macrovesicular steatosis and predominantly periportal mononuclear cell infiltration. Thus, absence of MAT1A resulted in a liver that is more susceptible to injury, expresses markers of an acute phase response, and displays increased proliferation.
When Kinsell et al. (1) first observed, in 1947, a marked impairment of methionine metabolism in patients with liver cirrhosis, they established the crucial importance of the liver in the regulation of blood methionine concentration. A few years later, Cantoni (2) demonstrated that the first step in methionine metabolism is its conversion to S-adenosylmethionine (AdoMet), a reaction that is catalyzed by the enzyme now known as methionine adenosyltransferase (MAT). The MAT gene is one of 482 genes required for survival of an organism (3). It is important because AdoMet is the principal biological methyl donor and the ultimate source of the propylamine moiety used in polyamine biosynthesis (3, 4). In mammals, two genes, MAT1A and MAT2A, encode for two homologous MAT catalytic subunits, α1 and α2 (5–7). MAT1A is expressed only in liver, and it encodes the α1 subunit found in two native MAT isoenzymes, which are either a dimer (MAT III) or tetramer (MAT I) of this single subunit (7). MAT2A encodes for a catalytic subunit (α2) found in a native MAT isoenzyme (MAT II), which is widely distributed (5–7). MAT2A also predominates in the fetal liver and is progressively replaced by MAT1A during development (8, 9). MAT isoforms differ in their kinetic and regulatory properties. MAT I and MAT II have low Km values for methionine, whereas MAT III has a high Km for methionine (10). MAT I and MAT III are reversibly inactivated by nitric oxide and hydroxyl radicals (11–13), whereas MAT II is inhibited by physiological concentrations of AdoMet (14, 15). MAT III is thought to be responsible for clearing methionine after a protein-rich meal, whereas MAT I, like MAT II outside of the liver, maintains the basal AdoMet content required by the liver under fasting conditions (16).
The existence of a gene, MAT1A, specifically expressed in the adult liver and the observation that mutations in this gene lead to isolated hypermethioninemia further supported the view of the liver as the principal methionine-metabolizing tissue of the body (1, 17). Isolated hypermethioninemia has been considered a benign inborn error of metabolism, because patients with mutations in the MAT1A gene are generally asymptomatic (17). However, the observation that MAT I/III activity is impaired in patients with liver cirrhosis (18, 19) and the demonstration that AdoMet treatment increases survival in patients with alcoholic liver cirrhosis (3, 10, 20) suggest that impaired synthesis of AdoMet may be a necessary but not sufficient component of the pathogenesis of cirrhosis in some patients with this syndrome.
In adult liver, increased expression of MAT2A is associated with rapid growth or dedifferentiation of the liver. We showed a switch in gene expression from MAT1A to MAT2A in human liver cancer (21), from 12 to 24 h after partial hepatectomy in the rat (22) and after treatment with thioacetamide (23). Using a cell line model that differs only in the type of MAT expressed, we showed that cells expressing MAT1A exhibited the slowest rate of growth, whereas the opposite was true for cells expressing MAT2A (24). Because of the differences in kinetic and regulatory properties, a switch in MAT expression is likely to affect the steady-state AdoMet level, methylation reactions, and consequently gene expression. Consistent with this, we found that cells that express MAT1A have much higher levels of AdoMet than cells that express MAT2A (24). Thus, the relative expression of MAT isoenzymes in liver is likely to influence the rate of liver growth and possibly facilitates the development of liver damage and hepatocarcinogenesis. To address many of these possibilities, we have developed a MAT1A knockout mouse model by using the targeted gene disruption approach.
Methods
Materials.
[32P]dCTP (3,000 Ci/mmol) and methyl-l-[3H]methionine (214 mCi/mmol) were purchased from DuPont/NEN or Amersham Pharmacia. The Total RNA isolation kit was obtained from Promega. All restriction enzymes were obtained from either Promega or GIBCO. All other reagents were of analytical grade and were obtained from commercial sources.
Generation of the MAT1A Knockout Mice.
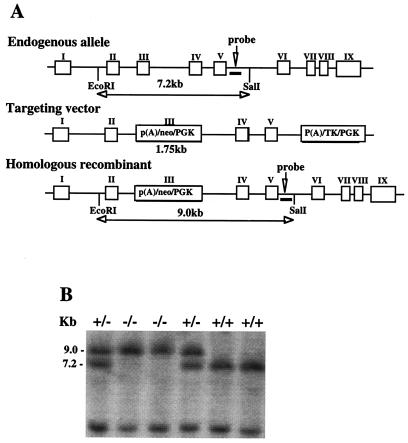
A genomic library from a 129 SvJ mouse strain (Stratagene) was screened with an EcoRI fragment of the rat MAT1A cDNA clone pSSRL (25). Of 500,000 independent recombinants, we obtained three overlapping clones containing a total of 21 kb and covering the entire MAT1A transcription unit. The structural organization of the gene (Fig. 1A) is identical to that reported for the C57BL/6 mouse strain (26). The targeting vector was derived from a 7.2-kb EcoRI-SalI DNA fragment containing exons 2–5, subcloned into the plasmid pUC18 (Fig. 1A). The MAT1A gene was interrupted by the neomycin selection marker in exon 3. For this purpose, the phosphoglycerate kinase (PGK) neocassette was subcloned into the blunt-ended NcoI site of this exon. The PGK promoter driving the herpes simplex virus thymidine kinase gene (PGK-tk cassette) was inserted into the blunt-ended SalI site.
Figure 1.
(A) Structure of the mouse MAT1A gene, targeting construct, and after homologous recombination. (B) Southern blot analysis of tail genomic DNA from offspring of a heterozygous mating.
The targeting vector (30 μg) was linearized with EcoRI and introduced into the embryonic stem cells (129/Sv-derived) by electroporation (performed by Genome Systems, St. Louis). Genomic DNA of resistant clones were screened for recombination events by Southern blot analysis by using a 0.85-kb NcoI fragment probe (Fig. 1A). A targeted clone was used for injection into the blastocysts of C57BL/6J mice and transferred into pseudopregnant female mice as described (27). Chimeric male offspring were bred to C57BL/6J females, and agouti F1 offspring were tested for transmission of the disrupted allele by Southern blot analysis of EcoRI-SalI-digested genomic DNA by using the NcoI fragment probe. Heterozygous matings of the F1 mice were carried out to produce homozygous F2 mutant mice.
Animal Experiments.
Mice were fed a standard diet (Harlan Teklad irradiated mouse diet 7912), unless indicated otherwise. Three- and eight-month-old male homozygous and wild-type littermates were killed for routine histologic examination of the liver. In addition, liver specimens were snap frozen for subsequent Northern and Western blot analyses, determination of MAT activity as described under saturating concentrations of the substrates, 5 mM methionine, and 5 mM ATP (28); levels of AdoMet, S-adenosylhomocysteine (AdoHcy), and methylthioadenosine (MTA), as determined by HPLC after the procedure described by Fell et al. (29) and modified by Miller et al. (30); and glutathione (GSH) and oxidized GSH, as described by Reed et al. (31). MAT activity was also determined in the kidney homogenates of these animals. At time of death, serum samples were collected and deproteinized by ultrafiltration, and methionine levels were analyzed by using a Beckman 6300-aa analyzer, following the procedure recommended by the manufacturer for physiological sample analysis.
In one series of experiments, 3-month-old male knockout mice and wild-type littermates were fed the amino acid-defined low methionine (0.17%) choline-free diet, Lombardi choline-deficient (CD) diet (Dyets, Bethlehem, PA), or control diet (0.4% methionine, 0.2% choline, Harlan Teklad) for 6 days. Four groups of animals were established (five mice per group): wild-type and MAT1A knockout mice on the control diet and wild-type and knockout mice on the CD diet. At the end of the treatment, animals were killed, and livers were removed. Liver samples were formalin fixed and embedded in paraffin for general histology.
Liver Histology.
Sections from formalin-fixed liver tissue from 3- and 8-month old MAT1A knockout and wild-type littermates, fed either a CD diet or a control diet for 6 days, or a normal diet for 3 or 8 months, were examined by two pathologists blinded to the animals' identity and treatment. All sections were stained with Masson's trichrome or hematoxylin and eosin. Liver tissues from four knockout and four wild-type animals were examined and compared.
Nucleic Acids Extraction and Northern Hybridization Analysis.
RNA was isolated from frozen liver specimens according to the method of Chomczynski and Sacchi (32). RNA concentration was determined spectrophotometrically before use, and integrity was checked by electrophoresis with subsequent ethidium bromide staining. Electrophoresis of RNA and gel blotting were carried out as described (33).
MAT1A and MAT2A cDNA probes were as described (21–23). The probes (cDNA fragments) for betaine homocysteine methyltransfease (BHMT), AdoHcy hydrolase (SAHH), and phosphatidylethanolamine N-methyltransferase 2 (PEMT2) were cloned by reverse transcriptase–PCR (34–36). The probes for glycine N-methyltransferase (GNMT), cystathionine β-synthase (CBS), and α fetoprotein (AFP) were the generous gifts of C. Wagner (Vanderbilt University, Nashville, TN), J. T. Brosnan (Memorial University, Newfoundland, Canada), and J. L. Danan, (Centre de Recherche sur l'Endocrinologie Moleculaire, Meudon, France), respectively.
Northern hybridization analysis was performed on total RNA by using standard procedures, as described (21–23, 33), with specific cDNA probes for MAT1A, MAT2A, GNMT, SAHH, PEMT2, CBS, BHMT, and AFP. All probes were labeled with [32P]dCTP by using a random-primer kit (Primer-It II Kit, Stratagene) or the Megaprime DNA labeling system (Amersham Pharmacia). To ensure equal loading of RNA samples and transfer in each of the lanes, before hybridization, membranes were rinsed with ethidium bromide and photographed, and the same membranes were also rehybridized with 32P-labeled 18S rRNA cDNA probe (22). Autoradiography and densitometry (Gel Documentation System, Scientific Technologies, Carlsbad, CA and NIH image 1.60 software program) were used to quantitate relative RNA. Results of the Northern blot analysis were normalized to 18S rRNA.
Western Blot Analysis.
The steady-state levels of liver-specific MAT and proliferating cell nuclear antigen (PCNA) were measured as described (23). Anti-PCNA antibodies were obtained from Zymed.
DNA Methylation Status.
Genomic DNA was isolated from livers of MAT1A knockout and wild-type mice, as described (37). DNA methylation at the CpG site assay was carried out essentially as described (38).
Affymetrix genechip Analysis.
The RNA samples were processed as recommended by Affymetrix (Santa Clara, CA). Briefly, after column cleanup of the RNA [Qiagen RNeasy (Chatsworth, CA)], cDNA was synthesized by using an oligo-dT primer attached to a sequence of the T7 promoter region. This cDNA was then used to perform in vitro transcription by using a bacteriophage T7 RNA polymerase promoter and incorporating biotin-labeled nucleotides [Enzo BioArray kit (Enzo Biochem)]. The in vitro transcription reaction product was purified and analyzed via agarose gel electrophoresis to confirm a size distribution ranging from 500 to 1,200 bp. cRNA was fragmented and hybridized at 45°C overnight to an Affymetrix murine U74A array. The arrays were washed, stained, and scanned by using a Fluidics Station 400 and Hewlett–Packard scanner, components of the Affymetrix genechip Instrument System. Finally, the results were analyzed by using genechip analysis suite software (Ver. 3.3, produced by Affymetrix) and the fold change between the hybridization intensity of the MAT1A homozygous deletion and wild-type samples obtained.
Statistical Analysis.
Data are given as mean ± SD. For changes in metabolites, methylation, MAT activity, and liver weight, statistical analysis was performed by using the Kruskall–Wallis test (STATWORKS). For changes in mRNA and protein levels, ratios of the respective mRNA or protein to 18S or Coomassie blue staining densitometric values were compared between knockouts and wild-type littermates by Student's t test. Significance was defined as P < 0.05.
Results
Generation of MAT1A Knockout Mice.
A sequence-replacement gene-targeting vector (Fig. 1A), designed to interrupt the coding sequences of the murine MAT1A gene in exon 3, was used for electroporation of embryonic stem cells. After homologous recombination, the EcoRI-SalI fragment increased from 7.2 to 9.0 kb, which can be easily differentiated from the wild-type allele on Southern blot analysis (Fig. 1B). One targeted clone of about 300 was identified by Southern blot analysis, used for injection into the blastocysts of C57BL/6J mice, and transferred into pseudopregnant female mice for generation of chimeric mice. Chimeric males were bred with C57BL/6J females, and the agouti F1 offspring were tested for transmission of the disrupted allele by Southern blot analysis (Fig. 1B). Heterozygous matings of the F1 mice were carried out to produce homozygous F2 mutant mice. From 271 offspring, 86 wild-type mice, 111 heterozygotes, and 74 knockout homozygotes were identified.
MAT1A Is Not Expressed in MAT1A Knockouts, but MAT2A and AFP Are Induced.
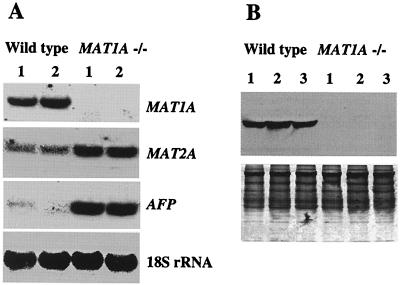
Absence of MAT1A expression is confirmed on both Northern (Fig. 2A) and Western blot analyses (Fig. 2B). In heterozygotes, the MAT1A mRNA and protein levels are reduced by 50% (results not shown). In the MAT1A knockouts, the steady-state mRNA level of MAT2A is induced by 3-fold (P < 0.05 vs. wild type). Because MAT2A expression is associated with rapid growth and dedifferentiation of the liver, we examined the expression of AFP. In the knockouts, AFP mRNA levels are also markedly increased (Fig. 2A).
Figure 2.
(A) Northern blot analysis of MAT1A, MAT2A, and AFP in MAT1A knockout and wild-type littermates. Liver RNA (30 μg each lane) samples obtained from two wild-type and two MAT1A knockout mice were analyzed by Northern blot hybridization with a 32P-labeled MAT1A cDNA probe, as described in Methods. The same membrane was then sequentially rehybridized with 32P-labeled MAT2A, AFP, and 18S rRNA cDNA probes. A representative Northern blot is shown. (B) Hepatic steady-state liver-specific MAT protein level in MAT1A knockout and wild-type littermates. Liver cytosol (50 μg/lane) obtained from three wild-type and three MAT1A knockout mice was analyzed by Western blot analysis by using anti-rat liver-specific MAT antibodies, as described in Methods. Equivalent protein loading was assured by Coomassie-blue staining of gels after transblotting (Lower).
Expression of Genes Involved in Methionine Metabolism.
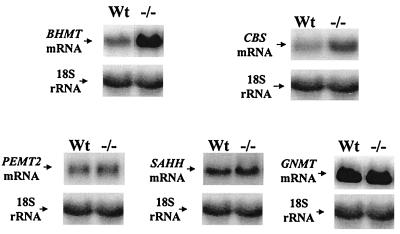
We next examined the expression of genes involved in methionine metabolism, which included SAHH, CBS, BHMT, PEMT2, and GNMT. As shown in Fig. 3, the mRNA levels of CBS and BHMT are increased by 3- to 4-fold, whereas the mRNA levels of SAHH, GNMT, and PEMT2 remained unchanged.
Figure 3.
Expression of BHMT, CBS, PEMT2, SAHH, and GNMT in the MAT1A knockout mouse. Liver RNA (30 μg each lane) samples obtained from four wild-type and four MAT1A knockout mice were analyzed by Northern blot hybridization with specific cDNA probes, as described in Methods. The same membranes were then rehybridized with 32P-labeled 18S rRNA cDNA probe to ensure equal loading. Representative Northern blots are shown.
Effect of MAT1A Disruption on Plasma Methionine Level, MAT Activities, and Hepatic Levels of Methionine Cycle Metabolites and DNA Methylation.
Plasma methionine level increased 776% in the knockout animals (wild type = 48.2 ± 0.2 μM, knockouts = 422 ± 4.5 μM; results represent mean ± SD from n = 5 animals, P < 0.001). Table 1 summarizes hepatic MAT activity and levels of methionine cycle metabolites in 3-month-old knockouts as compared with 3-month-old wild-type littermates. Hepatic MAT activity measured under saturating substrate concentrations was decreased by nearly 70% in the knockouts. By comparison, MAT activity in the kidneys of the knockout animals was unchanged (wild type = 1.1 ± 0.02, knockouts = 1.37 ± 0.1 pmol/min/mg; results represent mean ± SD from n = 5 animals each). Hepatic AdoMet levels fell by 74% in the knockouts, whereas AdoHcy and MTA levels were unchanged. Hepatic GSH and oxidized GSH (GSSG) levels both fell by 40–45%, so that the ratio of GSH to GSSG was unaffected. Overall, DNA methylation in the liver was not affected.
Table 1.
Hepatic MAT activity, levels of methionine cycle metabolites, and DNA methylation
| Parameter | MAT1A+/+ | MAT1A−/− |
|---|---|---|
| MAT activity, pmol/min/mg | 31.89 ± 5.58 | 10.25 ± 1.63* |
| AdoMet, nmol/mg | 0.19 ± 0.04 | 0.05 ± 0.02* |
| AdoHcy, nmol/mg | 0.06 ± 0.02 | 0.07 ± 0.03 |
| MTA, nmol/mg | 0.10 ± 0.02 | 0.10 ± 0.03 |
| GSH, nmol/mg | 46.13 ± 5.97 | 27.82 ± 4.28* |
| GSSG, nmol/mg | 4.23 ± 0.85 | 2.32 ± 0.3* |
| Overall DNA methylation† | 1.00 ± 0.16 | 1.18 ± 0.16 |
Results represent mean ± SD from five or more animals each. Liver specimens were obtained from 3-month-old knockouts and wild-type littermates for determination of MAT activity, methionine cycle metabolites, and DNA methylation, as described in Methods.
, P < 0.05 vs. wild-type littermates by the Kruskall–Wallis test.
Overall DNA methylation measurements were normalized with respect to wild-type values. [3H]-dCTP incorporation in wild-type animals was 9,600 dpm/μg DNA.
Phenotype of the MAT1A Knockout Mice and Response to CD Diet.
At 3 months of age, MAT1A knockout mice had body weights similar to wild-type littermates. However, their liver weights were significantly increased (wild type = 1.04 ± 0.06 gm, knockout = 1.42 ± 0.02 gm; results represent mean ± SD from three wild-type and four knockout animals; P < 0.05). At this age, the liver is histologically normal in the MAT1A knockout mice fed a normal diet (results not shown).
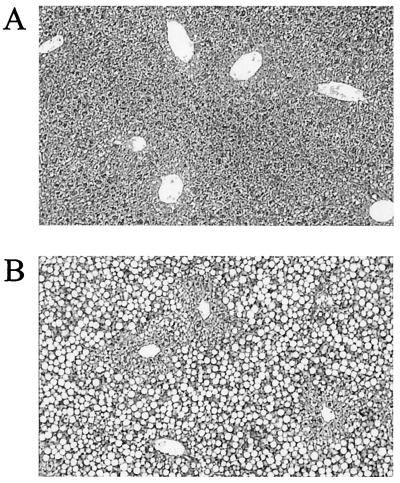
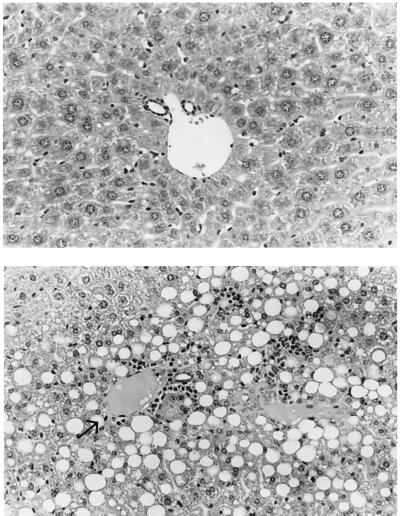
CD diet feeding for 6 days in wild-type animals caused little histologic change or only mild steatosis, whereas it induced severe macrovesicular steatosis in the livers of the knockout animals (Fig. 4). The livers of the 8-month-old wild-type littermates remained normal histologically, but the livers of 8-month-old knockout animals fed a normal diet exhibited macrovesicular steatosis involving 25–50% of hepatocytes and mononuclear cell infiltration, mainly in the periportal areas (Fig. 5).
Figure 4.
Effect of CD diet on liver histology in the MAT1A knockout mouse. Trichrome-stained sections of representative liver samples of wild-type (A) and MAT1A knockout mice (B) fed a CD diet for 6 days. Minimal steatosis or no change was seen in the livers of the wild-type littermates (A), whereas severe macrovesicular steatosis was observed in more than 75% of hepatocytes in the knockout mice (B) (×100). Representative histologic changes are shown from four animals.
Figure 5.
Liver histology from 8-month-old MAT1A knockout mice fed a normal diet. (Upper) Wild type. The portal tract demonstrates a portal vein and two small duct structures. No portal or lobular inflammation or lobular fatty change is seen. Hematoxylin and eosin × 148. (Lower) MAT1A knockout mouse. The portal tract at the left of the field (arrow) shows mild portal lymphocytic infiltrates with an unremarkable bile duct. The hepatocytes adjacent to the portal tract show macrovesicular fatty change. In addition, focal areas of inflammation are also seen involving the liver cells that contain fat (steatohepatitis); the inflammatory component is chiefly lymphocytic. Hematoxylin and eosin × 148. Representative histologic changes are shown from three knockout mice.
Differential Expression of Genes in Knockout Mice.
To better understand the phenotype of the MAT1A knockouts, the Affymetrix genechip system was used to analyze differential expression of genes. Table 2 summarizes genes that are induced in the knockout mouse, whereas Table 3 summarizes genes that are down-regulated in the knockout mouse. Although there is no clear pattern in the observed changes, to facilitate the discussion, we have divided the affected genes into three general categories: (i) those involved in immune response, (ii) those involved in cell proliferation, differentiation and signaling, and (iii) miscellaneous. Induction of BHMT was confirmed by using Northern blot analysis (Fig. 3), and induction of orosomucoid and proliferating cell nuclear antigen was confirmed by using Northern and Western blot analysis, respectively (results not shown).
Table 2.
Genes induced in the MAT1A knockout mouse
| Genes | Fold induction |
|---|---|
| Immune response genes | |
| Orosomucoid/α 1-acid glycoprotein (M12566) | 52.7 |
| Serum amyloid A1 (M17790) | 40.5 |
| Metallothionein 1 (V00835) | 20.5 |
| Histocompatibility 2, class II antigen A, α (X52643) | 19.9 |
| Major histocompatibility complex class II H2-I-A-β gene (k haplotype) (M21932) | 8.7 |
| Intracellular calcium-binding protein (MRP8) (M83218) | 8.6 |
| Metallothionein 2 (K02236) | 6.5 |
| Myeloperoxidase (X15313) | 4.0 |
| Lipopolysaccharide-binding protein (X99347) | 3.1 |
| Interleukin 10 (M37897) | 2.8 |
| IgE-binding factor (M10062) | 2.5 |
| Fas antigen (M83649) | 2.4 |
| Arachidonate 12-lipoxygenase (Y14334) | 2.3 |
| CD14 antigen (X13333) | 2.2 |
| Genes involved in cell proliferation, differentiation, and signaling | |
| Early growth response 1 (EGR01 Krox-24) (M28845) | 14.3 |
| Chromatin assembly factor-I p150 subunit (AJ132771) | 8.3 |
| Insulin-like growth factor-binding protein 5 (L12447) | 3.9 |
| Peroxisome proliferator activator receptor γ (U10374) | 3.8 |
| Guanylate nucleotide-binding protein 1 (M55544) | 3.6 |
| Heat-shock protein 86 (J04633) | 3.6 |
| Ras-related YPT-1 protein (Y00094) | 3.1 |
| TYRO protein tyrosine kinase-binding protein (AF024637) | 2.7 |
| Guanine nucleotide-binding protein, β 1 (U29055) | 2.6 |
| 10.0 | |
| DNA polymerase Z catalytic subunit (AF083464) | 2.6 |
| Protein tyrosine phosphatase (PRL-1) (U84411) | 2.5 |
| Protein phosphatase 1, catalytic subunit, β isoform (M27073) | 2.4 |
| cGMP phosphodiesterase (PDE9A*1) (AF031147) | 2.3 |
| Proliferating cell nuclear antigen (X57800) | 2.1 |
| Miscellaneous genes | |
| Cytochrome P450 17 (M64863) | 11.3 |
| Glycerol kinase (U48403) | 7.6 |
| Nicotinamide N-methyltransferase (U86108) | 7.3 |
| Peptidylglycine α-amidating monooxygenase (U79523) | 6.1 |
| Asparagine synthetase (U38940) | 5.7 |
| Lysozyme P (X51547) | 4.5 |
| Carbonic anhydrase II (M25944) | 4.5 |
| 6-phosphofructo-2-kinase/fructose-2,6- biphosphatase (X98848) | 4.1 |
| Aldehyde dehydrogenase (U96401) | 3.5 |
| Glucose-6-phosphate dehydrogenase (Z84471) | 3.3 |
| Cathepsin C (U74683) | 3.2 |
| BHMT (AF033381) | 2.8 |
| Phospholipid transfer protein (U28960) | 2.8 |
Table 3.
Genes down-regulated in the MAT1A knockout mouse
| Genes | Fold decrease |
|---|---|
| Immune response genes | |
| P6-5 gene (M27347) | 5.1 |
| Cytokine inducible SH2-containing protein (D89613) | 4.3 |
| Interferon α family gene 5 (X01971) | 3.6 |
| Interferon regulatory factor 5 (AF028725) | 2.5 |
| ζ-Chain-associated protein kinase (70kD) (U04379) | 2.6 |
| Genes involved in cell proliferation, differentiation, and signaling | |
| Hepatocyte nuclear factor-3/Forkhead homolog 1 like (AF010405) | 7.8 |
| Vascular endothelial growth factor-related factor (U43836) | 6.3 |
| Growth arrest-specific 2 (M21828) | 5.7 |
| Hepatocyte nuclear factor 3 γ (X74938) | 4.2 |
| Mitogen-activated protein kinase 5/extracellular regulated kinase (AB019374) | 4.2 |
| Fibroblast growth factor receptor 4 (X59927) | 3.6 |
| β actin (M12481) | 3.1 |
| Growth factor-inducible immediate early gene (X61940) | 2.9 |
| Insulin-like growth factor-binding complex (U66900) | 2.8 |
| Cyclin F (Z47766) | 2.2 |
| Miscellaneous genes | |
| Cytochrome P450 4A10 (AB018421) | 5.0 |
| 3-ketosteroid reductase (L41519) | 4.4 |
| Membrane glycoprotein (U97107) | 3.6 |
| Cytochrome P450 4a14 (Y11638) | 3.3 |
| Mitochondrial ribosomal protein S12 (Y11682) | 3.2 |
| Selenium-binding protein 1 (M32032) | 2.9 |
| Peroxismal/mitochondrial dienoyl-CoA isomerase (AF030343) | 2.6 |
Discussion
As indicated in the introduction, decreased activity of liver-specific MAT is well known in various liver diseases and may contribute to the pathogenesis of liver injury (3, 10). In addition to posttranslational regulation of liver-specific MAT by nitric oxide and hydroxyl radicals (11–13), we have recently shown that the expression of MAT1A is reduced in the livers of patients with various causes of liver cirrhosis (39). Furthermore, the often-observed reduced hepatic AdoMet levels in liver diseases can set up a vicious cycle, because AdoMet level modulates the expression of MAT1A and MAT2A. We showed recently that in primary cultures of rat hepatocytes, AdoMet level and MAT1A expression fell, whereas MAT2A expression was induced (40). The changes in MAT expression were prevented by AdoMet treatment (40). AdoMet transcriptionally activates MAT1A (40), whereas it suppresses MAT2A (41). Thus, a normal hepatic AdoMet level may be important in maintaining the “differentiated” state of the organ. As a corollary, expression of MAT1A may be important in maintaining the normal hepatic AdoMet level, and its absence may predispose the liver to injury and a “dedifferentiated” state.
Disruption of MAT1A, as expected, causes hypermethioninemia and a marked reduction of liver AdoMet content, confirming the central role of this organ in methionine metabolism. The reduced AdoMet content occurred despite induction of MAT2A, which is consistent with known differences in the kinetic and regulatory properties of the different isoenzymes and our previous works. Hepatic glutathione levels also fell, which agrees with numerous reports showing that reduction of liver MAT activity leads to a reduction of glutathione (reviewed in refs. 3 and 10). The hepatic content of two other metabolites of methionine metabolism, AdoHcy and MTA, were normal in the knockout mice. The expression of other genes involved in methionine metabolism was either increased, such as nicotinamide N-methyltransferase, CBS, and BHMT, or constant, such as GNMT, PEMT2, and SAHH. The fall in hepatic AdoMet level is likely responsible for the induction in MAT2A expression, as we have shown previously (40), and BHMT (T. Garrow and C. Castro, personal communication), but the mechanism leading to an increase in the mRNA content of nicotinamide N-methyltransferase and CBS in the knockout mouse is unknown. The present results suggest that in liver, the AdoMet level regulates the expression of a number of genes involved in methionine metabolism, a subject that will require further study.
Although AdoMet content was markedly reduced in 3-month-old knockout mice, global hepatic DNA methylation was not affected. In previous work, when the AdoMet level fell to the level we observed in the knockouts, there was global DNA hypomethylation (22, 23, 42, 43). However, in these experiments, AdoMet levels fell as a consequence of administration of potent hepatotoxic agents, such as thioacetamide, ethanol, carbon tetrachloride, or a CD diet. These results suggest that in these models, global DNA hypomethylation may be the result of a combination of reduced hepatic AdoMet content and liver injury followed by regeneration. The observation that global DNA methylation is unaffected in the MAT1A knockout mouse, however, does not preclude the possibility that specific genes may be hypomethylated.
Results obtained from the MAT1A knockout mouse model are consistent with our hypothesis that lack of MAT1A predisposes the liver to injury and a “dedifferentiated” state. First, the expression of many growth-related genes such as MAT2A, AFP, early growth response, and proliferating cell nuclear antigen, is induced in knockout mice. Accordingly, knockout mice exhibit increased liver weights as early as 3 months of age. Second, 3-month-old MAT1A knockout mice developed severe fatty liver after being fed a CD diet for only 6 days, and 8-month-old MAT1A knockout mice developed fatty liver and periportal inflammation on a normal diet. Third, genechip analysis revealed the expression of many acute phase markers, and immune related genes are markedly induced in the knockout mouse. How absence of MAT1A leads to these changes is unknown at the present and will require further study. Nevertheless, the observation that so many genes are altered indicates that maintaining normal hepatic AdoMet levels and/or AdoMet/AdoHcy ratio, accomplished by normal expression of MAT1A and activities of MAT I/III, is very important for liver development and function.
In contrast to findings in MAT1A knockout mice, human MAT I/III deficiency has not been associated with hepatic pathology (44). However, the number of individuals identified with mutations in the MAT1A gene is small (11 patients and 3 pedigrees) and, with the exception of a 43-year-old individual, they are young (under 14 years) (reviewed in ref. 44). Therefore, the possibility that these patients may be at risk for developing liver pathology at a later age cannot be excluded. It is also possible that other factors, such as the relative absence of choline dehydrogenase, makes humans less susceptible to choline-deficiency-related liver pathology. Thus, the choline content of the human diet might also be an important consideration in determining the susceptibility of MAT I/III-deficient patients to liver injury.
Although hepatic pathology has not been described, demyelination and other central nervous system diseases have been reported in three individuals with severe loss of MAT I/III activity. However, two other patients have normal brain myelination, despite the fact that both are homozygous for a mutation predicted to encode a severely truncated MAT I/III. The relationship between hepatic MAT I/III activity and the presence of central nervous system pathology therefore remains unclear (reviewed in ref. 44). The availability of the MAT1A knockout mouse model should facilitate studies to address this question.
In summary, we have developed a MAT1A knockout mouse model by a targeted disruption of the MAT1A gene. The absence of MAT1A resulted in marked depletion of hepatic AdoMet level, hypermethioninemia, and marked changes in the expression of many enzymes of methionine metabolism, as well as increased expression of many acute phase markers and growth-related genes. In the absence of MAT1A, mice developed nonalcoholic steatohepatitis and increased liver proliferation. Because nonalcoholic steatohepatitis is considered a progressive disease implicated in many cases of cirrhosis, these results suggest that AdoMet deficiency might play a key role in the initiation of this liver disease in some patients. This model should be instrumental in delineating the role of MAT1A in liver growth and injury and central nervous system pathology. Finally, this work supports the concept that patients with mutations in MAT1A, although generally asymptomatic, may be more susceptible to liver injury.
Acknowledgments
This work was supported by National Institutes of Health Grants DK-51719 (S.C.L.), AA-12677 (S.C.L. and J.M.M.), Plan Nacional de I + D SAF 98/0132 (J.M.M., F.J.C., and M.A.A.), Salud y Farmacia 99/0038 (J.M.M.), Ortiz de Landazuri Grant, Gobierno de Navarra (J.M.M.), Europharma (J.M.M.), Knoll (J.M.M.), and Professional Staff Association Grant no. 6–268-0–0, University of Southern California School of Medicine (S.C.L.). We thank C. Miqueo, E. Fernández, E. Mirpuri, and Dr. L. Montuenga for technical assistance, and Dr. L. Poirier for helpful comments on the CD diet.
Abbreviations
- AdoMet
S-adenosylmethionine
- AFP
α fetoprotein
- BHMT
betaine homocysteine methyltransferase
- CBS
cystathionine β-synthase
- CD
choline deficient
- GSH
glutathione
- GNMT
glycine N-methyltransferase
- MAT
methionine adenosyltransferase
- PEMT
phosphatidylethanolamine N-methyltransferase
- AdoHcy
S-adenosylhomocysteine
- SAHH
AdoHcy hydrolase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kinsell L W, Harper H A, Marton H C, Michael G D, Weiss H A. Science. 1947;106:589–594. [Google Scholar]
- 2.Cantoni G L. J Biol Chem. 1953;204:403–416. [PubMed] [Google Scholar]
- 3.Mato J M, Alvarez L, Ortiz P, Pajares M A. Pharmacol Ther. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein J D. J Nutr Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 5.Horikawa S, Tsukada K. FEBS Lett. 1992;312:37–41. doi: 10.1016/0014-5793(92)81405-b. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez L, Corrales F, Martín-Duce A, Mato J M. Biochem J. 1993;293:481–486. doi: 10.1042/bj2930481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotb M, Mudd S H, Mato J M, Geller A M, Kredich N M, Chou J Y, Cantoni G L. Trends Genet. 1997;13:51–52. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 8.Horikawa S, Ozasa H, Ota K, Tsukada K. FEBS Lett. 1993;330:307–311. doi: 10.1016/0014-5793(93)80894-z. [DOI] [PubMed] [Google Scholar]
- 9.Gil B, Casado M, Pajares M, Boscá L, Mato J M, Martín-Sanz P, Alvarez L. Hepatology. 1996;24:876–881. doi: 10.1002/hep.510240420. [DOI] [PubMed] [Google Scholar]
- 10.Lu S C. Int J Biochem Cell Biol. 2000;32:391–395. doi: 10.1016/s1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- 11.Avila M A, Mingorance J, Martínez-Chantar M L, Casado M, Martín-Sanz P, Boscá L, Mato J M. Hepatology. 1997;25:391–396. doi: 10.1002/hep.510250222. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Góngora E, Ruiz F, Mingorance J, An W, Corrales F J, Mato J M. FASEB J. 1997;11:1013–1019. doi: 10.1096/fasebj.11.12.9337154. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Mato I, Castro I, Ruiz F A, Corrales F J, Mato J M. J Biol Chem. 1999;274:17075–17080. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan D M, Hoffman J. Biochemistry. 1983;22:1636–1641. doi: 10.1021/bi00276a017. [DOI] [PubMed] [Google Scholar]
- 15.Halim A B, LeGros L, Geller A, Kotb M. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 16.Sánchez del Pino M, Corrales F J, Mato J M. J Biol Chem. 2000;275:23476–23482. doi: 10.1074/jbc.M002730200. [DOI] [PubMed] [Google Scholar]
- 17.Mudd S H, Levy H L, Skovby F. In: The Metabolic Basis of Inherited Diseases. Scriver C R, Beauder R, Charles W S, Valle D, editors. New York: McGraw–Hill; 1994. pp. 1279–1327. [Google Scholar]
- 18.Martin-Duce A, Ortiz P, Cabrero C, Mato J M. Hepatology. 1988;8:65–68. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 19.Cabrero C, Duce A M, Ortiz P, Alemany A, Mato J M. Hepatology. 1988;8:1530–1534. doi: 10.1002/hep.1840080610. [DOI] [PubMed] [Google Scholar]
- 20.Mato J M, Cámara J, Ortiz P, Rodés J Spanish Collaborative Group for the Study of Alcoholic Liver Cirrhosis. J Hepatol. 1999;30:1081–1089. doi: 10.1016/s0168-8278(99)80263-3. [DOI] [PubMed] [Google Scholar]
- 21.Cai J, Sun W M, Hwang J J, Stain S, Lu S C. Hepatology. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z Z, Mao Z, Cai J, Lu S C. Am J Physiol. 1998;275:G14–G21. doi: 10.1152/ajpgi.1998.275.1.G14. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z Z, Mato J M, Kanel G, Lu S C. Hepatology. 1999;29:1471–1478. doi: 10.1002/hep.510290525. [DOI] [PubMed] [Google Scholar]
- 24.Cai J, Mao Z, Hwang J J, Lu S C. Cancer Res. 1998;58:1444–1450. [PubMed] [Google Scholar]
- 25.Alvarez L, Asunción M, Corrales F, Pajares M A, Mato J M. FEBS Lett. 1991;290:142–146. doi: 10.1016/0014-5793(91)81245-4. [DOI] [PubMed] [Google Scholar]
- 26.Sakata S F, Shelly L L, Ruppert S, Schutz G, Chou J Y. J Biol Chem. 1993;268:13978–13986. [PubMed] [Google Scholar]
- 27.Chou X Y, Morreau H, Rottier R, Davis D, Bonten E, Gillemans N, Wenger D, Grosveld E G, Doherty R, Suzuki K, et al. Genes Dev. 1995;9:2623–2634. doi: 10.1101/gad.9.21.2623. [DOI] [PubMed] [Google Scholar]
- 28.Cabrero C, Puerta J, Alemany S. Eur J Biochem. 1987;170:299–304. doi: 10.1111/j.1432-1033.1987.tb13699.x. [DOI] [PubMed] [Google Scholar]
- 29.Fell D, Benjamin L E, Steele R D. J Chromatogr. 1985;345:150–156. doi: 10.1016/0378-4347(85)80146-8. [DOI] [PubMed] [Google Scholar]
- 30.Miller J W, Nadeau M R, Smith J, Smith D, Selhub J. Biochem J. 1994;298:415–419. doi: 10.1042/bj2980415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed D J, Babson J R, Beatty P W, Brodie A E, Ellis W W, Potter D W. Anal Biochem. 1980;106:55–62. doi: 10.1016/0003-2697(80)90118-9. [DOI] [PubMed] [Google Scholar]
- 32.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 33.Avila M A, Carretero M V, Rodriguez E N, Mato J M. Gastroenterology. 1998;114:364–371. doi: 10.1016/s0016-5085(98)70489-5. [DOI] [PubMed] [Google Scholar]
- 34.Neece D J, Griffiths M A, Garrow T A. Gene. 2000;250:31–40. doi: 10.1016/s0378-1119(00)00191-8. [DOI] [PubMed] [Google Scholar]
- 35.Bethin K E, Cimato T, Ettinger M J. J Biol Chem. 1995;270:20703–20711. doi: 10.1074/jbc.270.35.20703. [DOI] [PubMed] [Google Scholar]
- 36.Walkey C J, Donohue L R, Bronson R, Agellon L B, Vance D E. Proc Natl Acad Sci USA. 1997;94:12880–12885. doi: 10.1073/pnas.94.24.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres L, Avila M A, Carretero M V, Latasa M U, Caballería J, López-Rodas G, Boukaba A, Lu S C, Franco L, Mato J M. FASEB J. 2000;14:95–102. doi: 10.1096/fasebj.14.1.95. [DOI] [PubMed] [Google Scholar]
- 38.Pogribny I, Yi P, James S J. Biochem Biophys Res Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 39.Avila M A, Berasain C, Torres L, Martin-Duce A, Yang H P, Prieto J, Lu S C, Caballería J, Rodés J, Mato J M. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Trevijano E R, Latasa M U, Carretero M V, Berasain C, Mato J M, Avila M, A. FASEB J. 2000;14:2511–2518. doi: 10.1096/fj.00-0121com. [DOI] [PubMed] [Google Scholar]
- 41.Yang H P, Huang Z Z, Zeng Z H, Lu S C. Am J Physiol. 2001;280:184–190. doi: 10.1152/ajpgi.2001.280.2.G184. [DOI] [PubMed] [Google Scholar]
- 42.Lu S C, Huang Z Z, Yang H P, Mato J M, Avila M A, Tsukamoto H. Am J Physiol. 2000;279:G178–G185. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- 43.Varela-Moreiras G, Alonso-Aperte E, Rubio M, Gassó M, Deulofeu R, Alvarez L, Caballería J, Rodés J, Mato J M. Hepatology. 1995;22:1310–1315. [PubMed] [Google Scholar]
- 44.Chou J Y. Pharmacol Ther. 2000;85:1–9. doi: 10.1016/s0163-7258(99)00047-9. [DOI] [PubMed] [Google Scholar]