Abstract
Background
The current spread of the gene encoding the metallo-ß-lactamase NDM-1 in Enterobacteriaceae is linked to a variety of surrounding genetic structures and plasmid scaffolds.
Methodology
The whole sequence of plasmid pGUE-NDM carrying the bla NDM-1 gene was determined by high-density pyrosequencing and a genomic comparative analysis with other bla NDM-1-negative IncFII was performed.
Principal Findings
Plasmid pGUE-NDM replicating in Escherichia coli confers resistance to many antibiotic molecules including β-lactams, aminoglycosides, trimethoprim, and sulfonamides. It is 87,022 bp in-size and carries the two β-lactamase genes bla NDM-1 and bla OXA-1, together with three aminoglycoside resistance genes aacA4, aadA2, and aacC2. Comparative analysis of the multidrug resistance locus contained a module encompassing the bla NDM-1 gene that is actually conserved among different structures identified in other enterobacterial isolates. This module was constituted by the bla NDM-1 gene, a fragment of insertion sequence ISAba125 and a bleomycin resistance encoding gene.
Significance
This is the first characterized bla NDM-1-carrying IncFII-type plasmid. Such association between the bla NDM-1 gene and an IncFII-type plasmid backbone is extremely worrisome considering that this plasmid type is known to spread efficiently, as examplified with the worldwide dissemination of bla CTX-M-15-borne IncFII plasmids.
Introduction
Metallo-ß-lactamase (MBL) NDM-1 (New Dehli Metallo-ß-lactamase) corresponds to one of the latest and most important resistance trait identified in Gram-negative rods [1]. The bla NDM-1 gene has been first identified in the UK, India, and Pakistan, either in various enterobacterial isolates, but also in Acinetobacter sp., Pseudomonas sp., or Vibrio sp. [2]–[6]. Occurrence of NDM-1 producers in hospitalized patients in the UK was related in many cases with previous hospitalizations in the Indian subcontinent [2]. Additionally, there have been numerous reports of NDM-1-producing Enterobacteriaceae worldwide (infections or colonizations), most often recovered from patients who presented a link with the Indian subcontinent, and in some cases with the Balkans and Middle-East region [6]–[11].
Five bla NDM-1-bearing plasmids have been fully sequenced, being pHK-NDM (Genbank n°HQ451074), p271A (JF785549), pNDM-1_Dok01 (AP012208), pNDM10505 (JF503991) and pNDM-KN (JN157804), respectively corresponding to IncL/M-, IncN2-, and IncA/C-type plasmid scaffolds [12]–[16]. pHK-NDM is an IncL/M 88,803-bp in-size plasmid harboring the bla NDM-1 gene together with the 16S rRNA methylase armA gene, thus conferring high level of resistance to β-lactams and aminoglycosides, and is highly related to plasmids carrying the bla CTX-M-3 gene [12]. Plasmid p271A is 35,947-bp in size and harbors the bla NDM-1 gene as a single resistance gene within an IncN2-type plasmid scaffold on which a new replicase gene was identified [13]. Plasmids pNDM-1_Dok01, pNDM10505 and pNDM-KN belong to the IncA/C broad-host range plasmid family. In addition to the bla NDM-1 gene, those plasmids carry additional resistance genes including a bla CMY-2-like gene, together with an rmtC or armA 16S RNA methylase gene. Their scaffolds are very similar to those of other IncA/C but bla NDM-1-negative plasmids, known to be responsible for the spread of bla CMY-2-like genes in Enterobacteriaceae in the USA, Canada, and Europe [14], [15].
Overall, the bla NDM-1 gene has been more frequently reported onto broad host-range IncA/C-type plasmids, either in clinical or in environmental isolates recovered from the New Delhi area [6]. In addition, some other plasmid scaffolds have been associated with the bla NDM-1 gene including IncF, IncL/M, together with untypeable plasmids [6], [8]. In particular, IncF-type plasmids were involved in bla NDM-1 acquisition in isolates recovered in France, India, and Switzerland [8].
IncFII-type plasmids are narrow-host range plasmids that are frequently identified among E. coli strains [16]. Those plasmids are known to be involved in the worldwide dissemination of the bla CTX-M-15 gene in the epidemic ST131 E. coli clone [17]. They are characterized by several toxin-antitoxin addiction systems confering stability during bacterial cell division. The IncFII-type pC15-1a (GenBank n° NC_005327) and pEK516 (EU935738) plasmids carrying the bla CTX-M-15 gene have been fully sequenced [17], [18]. In particular, plasmid pEK516 obtained from the UK epidemic CTX-M-15-producing strain D represents the prototypic IncFII-plasmid from the MLST-defined ST131 E. coli lineage [18]. Unlike plasmids belonging to other incompatibilty groups, the backbones of IncF plasmids exhibit a significant heterogeneity in term of size and number of replicons [19].
The aim of this study was to characterize in detail an IncFII-type plasmid harboring the bla NDM-1 gene, in order to possibly identify genetic features explaining the successful spread of that gene. That plasmid had been recovered from an E. coli isolate belonging to ST131 [20] that corresponds to the main genetic background involved in the worldwide distribution of CTX-M-15 producers [21].
Results
General features
Our study was initiated by the isolation of a multidrug-resistant E. coli strain GUE that had been community-acquired in India [20]. E. coli isolate GUE was resistant to most β-lactams (remaining susceptible to aztreonam) and showed reduced susceptibility to carbapenems, minimum inhibitory concentrations (MIC) of imipenem, ertapenem, and meropenem being at 3, 3 and 2 µg/ml, respectively (Table 1). It was also resistant to gentamicin, kanamycin, tobramycin, sulfonamides, tetracycline, and fluoroquinolones, but remained susceptible to amikacin, chloramphenicol, rifampicin, and colistin.
Table 1. MICs of ß-lactams for E. coli clinical isolate Gue, E. coli J53 pGUE-NDM and E. coli J53 reference strains.
| β-lactams | MIC (µg/ml) | ||
| E. coli isolate GUE | E. coli J53 pGUE-NDM | E. coli J53 | |
| Amoxicillin | >256 | >256 | 4 |
| Amoxicillin + CLAa | 256 | >256 | 4 |
| Ticarcillin | >256 | >256 | 2 |
| Ticarcillin + CLAa | 256 | 256 | 2 |
| Cephalothin | >256 | >256 | 4 |
| Cefotaxime | 128 | 128 | 0.06 |
| Ceftazidime | >256 | >256 | 0.06 |
| Cefepime | 32 | 32 | 0.03 |
| Aztreonam | 0.125 | 0.06 | 0.06 |
| Meropenem | 2 | 0.75 | 0.03 |
| Ertapenem | 3 | 0.5 | 0.03 |
| Doripenem | 1.5 | 0.5 | 0.03 |
| Imipenem | 3 | 1.5 | 0.12 |
CLA; clavulanic acid (4 µg/ml) TZB.
Plasmid features
Conjugation assays allowed to transfer the bla NDM-1 gene and identified a ca. 87-kb plasmid named pGUE-NDM. The E. coli transconjugant showed resistance to penicillin/inhibitor combinations and broad-spectrum cephalosporins, and reduced susceptibilty to carbapenems (Table 1). It was also resistant to kanamycin, gentamicin, tobramycin, trimethoprim, and sulfonamides. Plasmid pGUE-NDM was assigned to the IncFII incompatibility group using the PCR-Based Replicon Typing method. Conjugation frequencies were observed at high rates (2×10−4 transconjugants per donor cell).
Sequence analysis of plasmid pGUE-NDM
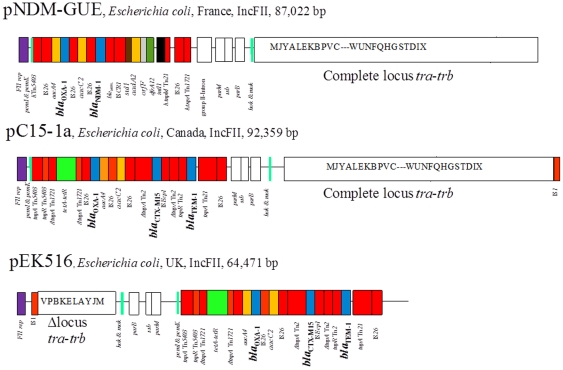
Whole plasmid sequencing identified plasmid pGUE-NDM to be 87,022 bp in-size with a GC content of 53% and 47 open reading frames (ORF) (Figure 1 and Table S1). BLAST analysis of the complete nucleotide sequence was performed in comparison with the reference IncFII plasmid pC15-1a (Genbank n°AY458016) and with plasmid pEK516 UK (Genbank n° EU935738). Comparative DNA sequence analysis confirmed that pGUE-NDM possessed an IncFII-type backbone and exhibited a significant synteny within the two scaffolds, with the exception of regions containing accessory genes (Table S1).
Figure 1. Major structural features of pGUE-NDM, encoding NDM-1 MBL, in comparison with incFII plasmids pC15-1a (genbank n°AY458016) and pEK516 (Genbank n°EU935738) CTX-M-15 encoding plasmids.
Resistance genes are indicated by colored boxes as follows: green, tetracycline resistance (tetA, tetR); azure, β-lactams resistance genes (bla NDM-1, bla OXA-1, bla CTX-M-15, bla TEM-1); orange, aminoglycosides resistance genes (aacA4, aacC2, aadA2); dark green, trimethoprime resistance gene (dfrA12). Transposon-related genes and insertion sequences are indicated in red boxes. Class 1 integrase gene is indicated by a black box. Partitioning-associated genes (parB, parM, ssb) are indicated by black-squared white boxes. The locus tra-trb is indicated by a squared white box with capital letters indicating the respective genes. The toxin-antitoxin genes (pemI/pemK, hok/mok)are indicated by light blue vertical lines. Replicons are indicated by purple boxes.
The backbone of pGUE-NDM included a complete array of genes involved in replication, conjugation and partition. The replication operon was composed by four ORFs exhibiting high identity with IncFII-specific replicase genes. The toxin/antitoxin addiction systems system pemI/pemK and hok/mok were identified as previously described for other FII-plasmids (Figure 1), together with the parB, parM genes encoding proteins involved in plasmid stability (Figure 1 and Table S1). A complete transfer operon (locus tra-trb) was identified being involved in the plasmid dissemination.
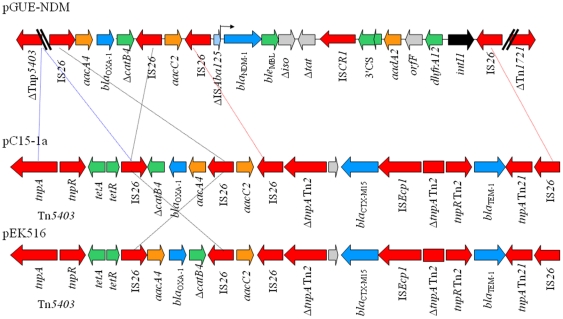
The bla NDM-1 gene was localized in a multidrug resistance (MDR) region of 20,181 bp. This region was bracketed by two copies of insertion sequence IS26 in opposite orientations creating an IS26-made compound transposon that was not bracketed by a target site duplication. Immediately upstream of the bla NDM-1 gene, a remnant of ISAba125 insertion sequence was identified (Figure 2). This truncated ISAba125 contained the −35 promoter sequence leading to the bla NDM-1 gene expression [13]. Upstream of the ΔISAba125, two IS26 were identified bracketing the aacC2 aminoglycoside resistance gene. Then, three gene cassettes being part of a remnant of a class 1 integron, namely aacC4, bla OXA-1 and a truncated catB4 genes were identified, that latter gene being truncated by another IS26 element (Figure 2). Downstream of the bla NDM-1 gene, the ble MBL gene encoding resistance to bleomycin was identified followed by a truncated phosphorybosilanthranilate isomerase gene (Δiso), and then by a truncated twin-arginine translocation pathway signal protein gene (Δtat), as previously found on other plasmid scaffolds (Figure 3). Then, the ISCR1 insertion sequence was identified [22], followed by a class 1 integron structure containing three gene cassettes, namely aadA2 encoding resistance to streptomycin and spectinomycin, orfF of unknown function, and dfrA12 encoding resistance to trimethoprim, and then the class 1 integrase gene. Finally, an additional copy of IS26 truncating the Tn1721 transposase gene was identified (Figure 2). Analysis of surrounding sequences of each IS26 elements revealed that none of them was bracketed by direct repeat (DR) sequences. Similarly, no DR was identified that would suggest that some of these elements may form an IS26-made transposons. This result reinforced the hypothesis of successive homologous recombination events at the origin of the global structure of this MDR region.
Figure 2. Schematic representation of multi-drug resistance regions of pGUE-NDM, pC15-1a and pEK516 incFII plasmids.
The genes and their corresponding transcriptional orientations are indicated by horizontal arrows. Function of each genes are indicated by colored arrows as follows: green, tetracycline resistance; azure, β-lactams resistance genes; orange, aminoglycosides resistance genes; dark green, trimethoprim resistance gene. Transposon-related genes and insertion sequences are indicated in red arrows. Class 1 integrase gene is indicated by black arrow. Deletion is indicated by blue dashed lines. Homologous recombination event leading to inversion is indicated by black dashed lines. Homologous recombination event leading to allelic exchange of bla CTX-M-15 locus by bla NDM-1 locus is indicated by red dashed lines. The accession numbers were: pC15-1a (genbank n°AY458016) and pEK516 (genbank n°EU935738).
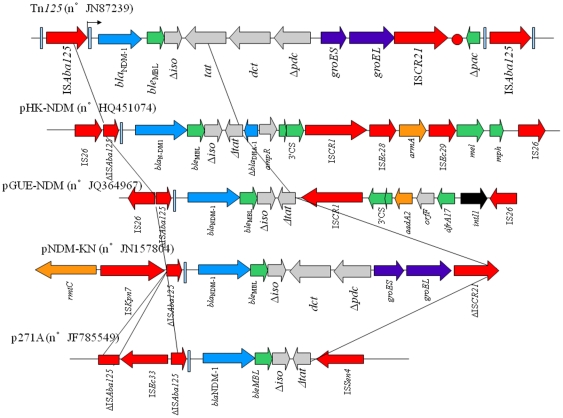
Figure 3. Schematic representation of the DNA sequences surrounding the bla NDM-1 genes in Acinetobacter baumannii strain ML, E. coli HK-NDM NDM-1 encoding plasmid pHK-NDM, E. coli 271 p271a NDM-1 harboring plasmid, K. pneumoniae pNDM-KN harboring plasmid, and E. coli GUE NDM-1 harboring pGUE-NDM plasmid.
Gene names are abbreviated according to their corresponding proteins: Δiso for phosphoribosylanthranilate isomerase; tat for twin-arginine translocation pathway signal sequence protein; dvt for divalent cation tolerance protein; Δpac for truncated phospholipid acetyltransferase. The oriIS of ISCR21 is indicated by a circle. Function of each genes are indicated by colored arrows as follows: green, non-aminoglycosides non-β-lactams resistance genes; azure, β-lactams resistance genes; orange, aminoglycosides resistance genes. Transposon-related genes and insertion sequences are indicated in red arrows. Class 1 integrase gene is indicated by black arrow. GroES, GroEL are indicated in dark blue. ORF of unknown function are indicated by grey arrows. Conserved bla NDM-1 locus was indicated by vertical black lines. The Genbank accession numbers were: pHK-NDM n°HQ451074, p271A n°JF785549, pNDM-KN pending, pGUE-NDM pending and Tn125 pending.
Evolution of the bla NDM-1 genetic context
In order to get further insights into the bla NDM-1 gene acquisition, a comparison of the genetic structures previously identified with that identified on plasmid pGUE-NDM was performed (Figure 3). Interestingly, a common module that we termed NDM module was systematically identified. This module was composed of the ISAba125 fragment containing the −35 promoter region, the bla NDM-1 gene, the bleomycin resistance gene ble MBL, and a truncated phosphoribosylanthranilate isomerase. Recently, composite transposon Tn125 has been identified in several A. baumannii isolates [23], [24]. This 10,099-bp in-size transposon of was made of two copies of ISAba125 bracketing a 7,925-bp fragment and was inserted into the chromosome of different strains (Figure 3). Transposon Tn125 also contained the NDM module, together with the groES and groEL genes and the ISCR21 insertion sequence (Figure 3). It seems that the NDM module has integrated the pGUE-NDM backbone by a recombination event mediated by IS26 elements. Overall, a series of IS26-mediated recombination events has likely been at the origin of the formation of the large resistance gene array identified on plasmid pGUE-NDM.
Discussion
The bla NDM-1 gene is now identified worldwide and it has been speculated that particular genetic features could be at the origin of that wide diffusion. After the characterization of IncA/C, IncN2, and IncL/M plasmids bearing the bla NDM-1 gene, our study characterized an IncFII plasmid which backbone is known to be a major vehicle for dissemination of the bla CTX-M-15 gene [17]. It might be hypothetized that the endemicity of the bla CTX-M-15-positive and IncFII plasmid in the Indian subcontinent has created a favorable context for hosting and therefore disseminating the bla NDM-1 gene. That finding is particularly threatening when considering the explosive diffusion of the bla CTX-M-15 gene that has been witnessed worldwide during the last decade.
Analysis of the genetic features of plasmid pGUE-NDM did not identify any particular element that would positively or negatively interfere into its spreading potency such as addiction systems, partitioning systems or virulence factors in comparison to others IncFII-type plasmids. Indeed, the backbone part of the plasmid (being replication system, maintenance systems and transfer systems) corresponded to one previously identified. However, it appeared that acquisition of the bla NDM-1-containing module resulted from a series of IS26-related recombination events. Our study further underlines that the current dissemination of the bla NDM-1 gene is associated to a variety of genetic backgrounds.
Materials and Methods
Antimicrobial agents and MIC determinations
Susceptibility testing was performed by disk diffusion assay (Sanofi-Diagnostic Pasteur, Marnes-la-Coquette, France) and interpreted aaccording to CLSI [25]. The MICs for carbapenems were determined by Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar plates at 37°C. The production of MBLs was evaluated using Etest, combining imipenem and EDTA as recommended by the manufacturer (AB bioMérieux).
Plasmid identification and preparation
Plasmid pGUE-NDM was assigned to the IncFII incompatibility group using the PCR-Based Replicon Typing (PBRT)method [26]. Conjugation experiments were performed as previously described using azide-resistant E. coli strain J53 [27]. Transconjugants were selected on Trypticase-Soy media containing 100 µg/ml of ticarcillin and 100 µg/ml of sodium azide.
High-density pyrosequencing and sequence assembly
Plasmid DNA was isolated from the E. coli tranconjugant using Qiagen Maxiprep kit (Qiagen, Courtaboeuf, France). The complete sequencing work flow of the Illumina Genome Analyzer IIx system (Illumina Inc., San Diego, CA) was performed by the DNAVision company (Gosselles, Belgium) and is described at www.dnavision.com.
Genome assembly and annotation
Reads from each sample were trimmed to remove poor quality sequence using the following procedure: keeping only reads for which the quality of all the fifty first bases is greater or equal to ten in both reads; for each read trimming the tail of the sequence from the first position for which the quality is smaller than ten; trimming poly-A sequence artifacts; and trimming adapter sequences. Then, the assemblies were carried out using Velvet2 assembler [28] in order to produce contigs from Illumina GAIIx reads. A total of ca. 12,000 high-quality reads were derived from the pGUE-NDM library, of which 482 contig were obtained using Velvet2 assembler including 14 contigs de novo. Contigs were considered as DNA contamination if showing at least 90% similarity with E. coli K12 substrain MG1655 genomic DNA sequences (NCBI accession number NC_000913.2) using the BLASTn algorithm. Among the 482 contigs, 430 were considered as DNA contaminations. A single contiguous sequence was obtained from the 52 contigs obtained using PCR-based gap closure et PCR combinations to close gaps and verify the position of each contig.
Genome comparison
The BLASTp algorithm was used to search for protein similarities by using as a reference the E. coli K12 chromosome sequence. The criterion used to evaluate the deduced amino acid sequence homology was >50% similarity at the amino acid level and >50% coverage of protein length.
Nucleotide sequence accession numbers
The pGUE-NDM plasmid sequence was submitted to the GenBank database and can be found under accession number JQ364967.
Transparency declaration
None to declare.
Supporting Information
Orfs identified in pGUE-NDM.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded mostly by a grant from the INSERM (Institut National de la Santé et de la Recherche Médicale France) (U914), by a grant-in-aid of the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and by a grant of the European Community (TEMPOtest-QC, HEALTH-2009-241742). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of bla OXA-23 with bla NDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65:2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66:1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 5.Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, et al. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011;55:3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 7.Livermore DM, Walsh TR, Toleman M, Woodford N. Balkan NDM-1: escape or transplant? Lancet Infect Dis. 2011;11:164. doi: 10.1016/S1473-3099(11)70048-2. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of bla NDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Fortineau N, Nordmann P. International transfer of NDM-1-producing Klebsiella pneumoniae from Iraq to France. Antimicrob Agents Chemother. 2011;55:1821–1822. doi: 10.1128/AAC.01761-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Al Maskari Z, Al Rashdi F, Bernabeu S, Nordmann P. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J Antimicrob Chemother. 2011;66:304–306. doi: 10.1093/jac/dkq428. [DOI] [PubMed] [Google Scholar]
- 11.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, et al. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One. 2011;6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, et al. Complete sequencing of the bla NDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One. 2011;6:e25334. doi: 10.1371/journal.pone.0025334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. Evolution of IncA/C bla CMY-2 carrying plasmids by acquisition of the bla NDM-1 carbapenemase gene. Antimicrob Agents Chemother. 2012;56:783–786. doi: 10.1128/AAC.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, et al. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum ß-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004;48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, et al. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborn AM, Da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146:2267–2275. doi: 10.1099/00221287-146-9-2267. [DOI] [PubMed] [Google Scholar]
- 20.Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis. 2010;10:832. doi: 10.1016/S1473-3099(10)70279-6. [DOI] [PubMed] [Google Scholar]
- 21.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 22.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Göttig S, et al. Molecular characterization of bla NDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother. 2011;66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, et al. Tn125-related acquisition of bla NDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 2012. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA.
- 26.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Lartigue MF, Poirel L, Aubert D, Nordmann P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring ß-lactamase gene bla CTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother. 2006;50:1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orfs identified in pGUE-NDM.
(DOC)