Abstract
Mitochondria supply energy for physiological function and they participate in the regulation of other cellular events including apoptosis, calcium homeostasis and production of reactive oxygen species. Thus, mitochondria play a critical role in the cells. However, dysfunction of mitochondria is related to a variety of pathological processes and diseases. MicroRNAs (miRNAs) are a class of small noncoding RNAs about 22 nucleotides long, and they can bind to the 3′ un-translated region (3′UTR) of mRNAs, thereby inhibiting mRNA translation or promoting mRNA degradation. We summarize the molecular regulation of mitochondrial metabolism, structure and function by miRNAs. Modulation of miRNAs levels may provide a new therapeutic approach for the treatment of mitochondria-related diseases.
Mitochondria
Mitochondria are main oxidative phosphorylation reaction and energy production organelles, and they supply energy for cellular physiological functions. Besides supplying energy, mitochondria participate in other cellular events. One of the well known cellular roles of mitochondria is their involvement in the complex apoptotic signaling pathways [Lee et al., 2004; Suen et al., 2008]. Additionally, mitochondria regulate calcium homeostasis [Chen et al., 2005; O’ Rourke, 2004; Vandecasteele et al., 2001] and produce reactive oxygen species (ROS) [Chen et al., 2003; Miyamoto et al., 2005]. Thus, mitochondria play a critical role in the cells.
However, the dysfunction of mitochondria is related to a variety of pathological processes and diseases. For example, changes in mitochondrial function and ultrastructure have been observed in mental disorders [Gong et al., 2011]. Mitochondrial DNA (mtDNA) mutations and deletions [Kujoth et al., 2005], and ROS over production [Lemieux et al., 2010] have been shown to contribute to the aging process. The decreased expression of mitochondrial genes [Hsieh et al., 2004] and disrupted mitochondrial structure have been associated with abnormal human development [Au et al., 2005]. Mitochondria form dynamic networks, and constantly undergo fission and fusion to maintain their integrity and quantity [Detmer and Chan, 2007]. The impaired balance of mitochondrial fission and fusion [Wang et al., 2009] and altered morphology of the cristae [Baloyannis, 2006] are related to Alzheimer’s disease. Furthermore, a deficiency in electron transport chain has been noted in diabetes [Ritov et al., 2010].
Given the important role of mitochondria in controlling the cellular physiology and the potential of deregulated mitochondria to cause pathology, it is necessary to understand the molecular regulation of mitochondrial metabolism, structure and function.
MicroRNAs (miRNAs)
miRNAs are a class of small noncoding RNAs about 22 nucleotides long, and they can bind to the 3′ un-translated region (3′UTR) of mRNAs, thereby inhibiting mRNA translation or promoting mRNA degradation [Lee et al., 2003]. This miRNA mediated regulation is sequence-specific and occurs at post-transcriptional level [He and Hannon, 2004]. Recently, miRNA has been also shown to target 5′ UTR or the open reading frames of targeted mRNA [Moretti et al., 2010]. miRNAs have been found in mitochondria [Bian et al., 2010; Kren et al., 2009], and they can contribute to mitochondrial dysfunction [Li et al., 2011]. Growing evidence has demonstrated that miRNA can play a significant role in the regulation of development, differentiation, proliferation, apoptosis as well as tumorigenesis [Cai et al., 2009]. Therefore, there is considerable interest in exploring regulation of mitochondria by miRNAs. This review is focused on recent findings concerning the role of miRNAs in controlling mitochondrial function.
miRNAs can affect mitochondrial metabolism
A fundamental function of mitochondria is to produce ATP through oxidative phosphorylation, thereby providing energy for the cellular activities. Mitochondria have their own genome, transcription and translation systems, but they also require proteins encoded by the nucleus [Cannino et al., 2007]. Cytochrome c oxidase IV (COXIV) is a key nuclear-encoded protein within the electron transfer chain in mitochondria, and participates in ATP production. The alteration of COXIV protein levels is able to affect mitochondrial function. miR-338 is a brain-specific miRNA expressed in neuronal tissue [Kim et al., 2004; Wienholds et al., 2005]. The miR-338 has been shown to regulate the expression of COXIV. COXIV 3′UTR contains miR-338 target And enforced expression of miR-338 reduces COXIV mRNA as well as protein levels. On the contrary, inhibition of endogenous miR-338 by its specific antagomir results in an increase in COXIV mRNA and protein levels. Functional study revealed that the over expression of miR-338 can significantly reduce mitochondrial oxygen consumption, mitochondrial metabolic activity, and ATP production [Aschrafi et al., 2008].
Glutaminase is important for mitochondrial metabolism and it converts glutamine to glutamate, which is further catabolized through the tricarboxylic acid cycle for the production of ATP or serves as the substrate for glutathione synthesis in mitochondria. The miR-23a and miR-23b (miR-23a/b) have been demonstrated to participate in targeting glutaminase [Gao et al., 2009] and can directly repress glutaminase expression. Modulation of miR-23a/b through their antagomirs can affect glutaminase expression levels.
The miR-210, a miRNA significantly upregulated during hypoxic stress in many cell types [Corn, 2008; Kulshreshtha et al., 2007], is reported to be involved in repressing mitochondrial respiration and the associated downstream functions [Chan et al., 2009; Favaro et al., 2010]. For example, mitochondrial iron sulfur cluster homologue (ISCU) has been identified as a target of miR-210. Iron sulfur clusters are assembled in mitochondria by a complex series of chaperones and enzymes [Mühlenhoff and Lill, 2000], which are then exported to the cytoplasm for assembling into the relevant functional protein [Rouault and Tong, 2008; Tong and Rouault, 2000]. A decrease in ISCU can influence the activity of enzymes requiring iron sulfur clusters. The miR-210 can repress the expression of ISCU by directly binding to ISCU 3′UTR. Both gain-and loss-of-function assays have demonstrated that miR-210 is necessary and sufficient for the downregulation of ISCU during hypoxia. Consequently, miR-210 is able to affect aconitase and the activity of mitochondrial complex.
Certain miRNAs are able to regulate insulin gene expression, biosynthesis and the secretion [Herrera et al., 2010; Poy et al., 2004; Xia et al., 2011; Zampetaki et al., 2010]. For example, miR-15a promotes insulin biosynthesis by inhibiting the expression of endogenous uncoupling-protein 2 (UCP-2) in mouse β-cells. The UCP-2 is a member of the mitochondrial inner membrane carrier family of proteins and it facilitates uncoupling of oxygen consumption [Bordone et al., 2006; Bouillaud et al., 1985]. The miR-15a regulates oxygen consumption and ATP generation through targeting UCP-2. Mitochondrial dysfunction is related to insulin resistance [Cheng et al., 2009; Kim et al., 2008; Lowell and Shulman, 2005], but the underlying mechanism is not fully understood. A recent report shows that miR-126 is actively involved in insulin resistance by targeting insulin receptor substrate-1 (IRS-1) [Ryu et al., 2011]. Thus, miRNAs can be considered as a target for developing effective treatment for diabetes.
Interestingly, miR-696 regulates fatty acid oxidation capacity and mitochondrial biogenesis through targeting peroxisome proliferator-activated receptor gamma co activator 1-alpha (PGC-1α) [Aoi et al., 2010]. The PGC-1α promotes aerobic metabolism and mitochondrial biogenesis in skeletal muscle [Puigserver et al., 1998; Wu et al., 2002; Wu et al., 1999]. Both the fatty acid oxidation and mtDNA content are reduced by miR-696 over expression but increased upon transfection of its inhibitor.
miRNAs participate in the regulation of mitochondria-mediated apoptosis
Apoptosis can be initiated though the extrinsic and/or intrinsic pathways. The extrinsic pathway is initiated by the binding of death ligands such as Fas ligand (FasL) or tumor necrosis factor α (TNF-α) to their corresponding death receptors, Fas or TNF receptor-1 (TNFR-1), respectively. Upon ligand binding, the death receptors undergo trimerization and recruit Fas-associated death domain protein (FADD) that then associates with procaspase-8 to form death-inducing signaling complex (DISC) [Li et al., 2010b]. However, the intrinsic pathway is initiated through mitochondria. A death signal induces the release of mitochondrial pro-apoptotic proteins such as cytochrome c [Li et al., 1997], mitochondrial apoptosis-inducing factor [Susin et al., 1999] and Smac/Diablo [Du et al., 2000; Verhagen et al., 2000]. Cytochrome c forms a complex with Apaf-1 and procaspase-9 resulting in the activation of caspase-9. Smac/Diablo can associate with inhibitor of apoptosis proteins thereby counteracting their inhibitory effects on caspases. The intrinsic pathway is regulated by the Bcl-2 family members. For example, in response to proapoptotic stimuli, the cytosolic Bax and Bad translocate to mitochondria and permeabilize the outer mitochondrial membrane leading to the release of proteins in the mitochondrial intermembrane space into the cytosol. In contrast, Bcl-2 and Bcl-xL are able to associate with Bax and Bad thereby quenching their death inducing potential [Antignani and Youle, 2006; Cleland et al., 2011; Karbowski et al., 2006].
The miR-15a and the miR-16-1 (miR-15a/16-1) induce apoptosis through the regulation of mitochondrial function. They reside as a cluster at the chromosomal region 13q14 and are frequently deleted or down-regulated in chronic lymphocytic leukemia [Calin et al., 2005]. miR-15a and 16-1 regulate multiple oncogenic activities including Bcl-2 and Mcl1. Further, miR-15a promotes mitochondrial dysfunction indicated by cytochrome c release into the cytosol and the disruption of mitochondrial membrane potential [Gao et al., 2010]. The miR-143 is specifically expressed in normal colon cells, and its expression is decreased in human colon cancer tumors. ERK5 stimulates cell proliferation [Nishimoto and Nishida, 2006] and it has been identified as a target of miR-143 [Akao et al., 2006]. Induction of apoptosis by interfering with mitochondrial function can be mediated through miR-143 targeting of ERK5 pathway [Nakagawa et al., 2007]. The miR-1 is a muscle specific miRNA [Chen et al., 2006]. Upon apoptotic stimulation, the miR-1 expression is increased with a concomitant release of cytochrome c from mitochondria and a decrease in membrane potential [Yu et al., 2008].
miRNAs control mitochondrial morphology
Mitochondria constantly undergo biogenesis, fusion and fission. They form dynamic networks that are necessary for the maintenance of organelle fidelity [Berman et al., 2008; Cassidy-Stone et al., 2008; Edwards et al., 2010; Tatsuta and Langer, 2008; Yang et al., 2008]. The morphological integrity of mitochondria is vital for their function. Mitochondrial biogenesis involves mitochondrial DNA replication and mass increase. In contrast, mitochondrial DNA replication does not occur during mitochondrial fission. Instead, the existing mitochondrial DNA divides into the newly fission mitochondria [Berman et al., 2008; Tatsuta and Langer, 2008]. Upon fission mitochondrial numbers are increased while their sizes are decreased. The mitochondrial fusion and fission participate in the regulation of apoptosis. Mitochondrial fusion is able to inhibit apoptosis, whereas mitochondrial fission is involved in the initiation of apoptosis [Cassidy-Stone et al., 2008]. Excessive mitochondrial fission is involved in the pathogenesis of many diseases such as diabetic neuropathy [Edwards et al., 2010] and brain and skeletal muscle disorders [Yang et al., 2006; Yang et al., 2008].
miRNAs have been recently shown to be involved in controlling mitochondrial dynamics. The miR-30 family includes 5 members, miR-30a, miR-30b, miR-30c, miR-30d and miR-30e, and they are abundantly expressed in the heart. miR-30a, miR-30b and miR-30d levels were decreased substantially upon hydrogen peroxide treatment, whereas miR-30c and miR-30e levels remained unaltered. In searching for the downstream targets, p53 was identified as the direct target of miR-30 family members. Mitochondrial fission requires the activity of a dynamin-related protein-1 (DRP1) [Frank et al., 2001], which is a GTPase that causes scission of the mitochondrial outer membrane, resulting in fission of mitochondrial tubules into fragments. The p53 activates Drp1 transcriptionally and consequently leads to apoptosis. Thus the miR-30 family members can regulate mitochondria fission and apoptosis through p53 and Drp-1 axis [Li et al., 2010a].
The miR-499 encoded by intron of myosin gene is a cardiac-abundant miRNA under physiological conditions [Kim et al., 2006; van Rooij et al., 2009], but is down-regulated during apoptosis. The miR-499 can prevent apoptosis by targeting calcinurin, which dephosphorylates and activates Drp1, resulting in fission of mitochondria and apoptosis. Interestingly, the expression of miR-499 is also transcriptionally regulated by p53 [Wang et al., 2011].
Despite the fact that several miRNAs are involved in the mitochondrial fission machinery, the maintenance of mitochondrial equilibrium is very complex. Other factors such as mitofusion and Fis-1 are vital in regulating mitochondrial fusion and fission. The detailed molecular mechanism of miRNAs in regulating the mitochondrial network remains largely unknown.
miRNAs are present in mitochondria
miRNAs are known to be encoded by nuclear genome and to modulate gene expression mainly in the cytosol. Intriguingly, recent studies using a miRNA microarray system to survey miRNA expression at a genome-wide scale in the rigorously purified mitochondria found that unique miRNAs are enriched in mitochondria independent of total cellular abundance [Bandiera et al., 2011; Barrey et al., 2011; Bian et al., 2010; Kren et al., 2009]. However, miRNAs retained in mitochondria are cell type-dependent. 15 nuclear encoded miRNAs are uniquely and reproducibly identified in mitochondria isolated from adult rat livers [Kren et al., 2009]. Twenty miRNAs are reported in mouse liver mitochondria including miR-122, miR-805 and miR-609 [Bian et al., 2010]. In the mitochondria isolated from the human myotubes there are more than twenty miRNAs [Barrey et al., 2011]. Thirteen miRNAs such as miR-1973, miR-1275 and miR-494 are significantly enriched in the mitochondria from HeLa cells [Bandiera et al., 2011]. These findings, and the miRNAs present in mitochondria are summarized in Table 1.
Table 1.
miRNAs identified in mitochondria from different tissue or cell types
| Adult rat liver | Mouse liver | Human myotubes | HeLa cell line |
|---|---|---|---|
| miR-130a | miR-122 | miR-720 | miR-1973 |
| miR-130b | miR-805 | miR-133b | miR-1275 |
| miR-140* | miR-690 | miR-1974 | miR-494 |
| miR-320 | miR-689 | miR-24 | miR-513a-5p |
| miR-494 | miR-494 | miR-133a | miR-1246 |
| miR-671 | miR-705 | miR-125a-5p | miR-328-5p |
| miR-202 | miR-721 | miR-1979 | miR-1908 |
| miR-705 | miR-720 | miR-103 | miR-1972 |
| miR-709 | miR-188-5p | miR-125b | miR-1977 |
| miR-721 | miR-101 | miR-103 | miR-638 |
| miR-761 | let-7f | miR-221 | miR-1974 |
| miR-763 | miR-711 | miR-23a | miR-1978 |
| miR-198 | miR-432 | let-7b | miR-1201 |
| miR-765 | miR-181b | miR-423-3p | |
| miR-361-5p | miR-106a | ||
| miR-680 | miR-23b | ||
| miR-181d | miR-92a | ||
| miR-29c | miR-193b | ||
| miR-29a | miR-365 | ||
| miR-762 | miR-93 |
Not only miRNAs are present in mitochondria, other components of miRNA machinery are also detectable in mitochondria. For example, miRNAs require binding to the RNA-induced silencing complex (RISC) before they can modulate the expression of their target genes [Parker and Sheth, 2007]. The argonaute Ago2 is the main and a key active protein of the RISC complex. Recently, it is shown that Ago2 is present in mitochondria [Bandiera et al., 2011; Bian et al., 2010].
The physiology and pathology of miRNAs in mitochondria remain unknown. Streptozotocin (STZ) is known to induce type 1 diabetes and mitochondrial dysfunction [Ghosh et al., 2004]. Mitochondria-associated miRNAs are significantly altered upon STZ treatment, with miR-494, miR-202-5p, miR-134 and miR-155 increased, while miR-705 and miR-122 decreased [Bian et al., 2010]. Elucidating the roles of miRNAs in mitochondria will provide the basic framework to investigate their functions in mitochondria and to unravel their potential in designing new therapeutic strategies for mitochondrial diseases.
miRNA and mitophagy
Autophagy is a process of catabolism of cellular components, and necessary for the maintenance of cell homeostasis [Mortensen et al., 2010; Yang and Klionsky, 2010]. It is induced by nutrient deprivation and a variety of physiological and pathological conditions. The morphological character of autophagy is autophagosome formation that encompasses the cellular components, and fuses with lysosome to degrade it. Mitophagy is a type of autophagy, which selectively degrades mitochondria [Kim et al., 2007]. Mitophagy regulates mitochondria number to match metabolic or developmental demand [Kundu et al., 2008], and also is a form of quality control to remove damaged mitochondria [Kundu et al., 2008; Okamoto K, 2009; Tolkovsky et al., 2002]. In mammalian cells, upon mitochondrial membrane depolarization PINK1 triggers Parkin translocation from cytosol to mitochondria where Parkin ubiquitylates mitochondrial proteins [Geisler et al., 2010], thereby inducing damaged mitochondria to form autophagosomes (Figure 1).
Figure 1. A schematic model of mitophagy.
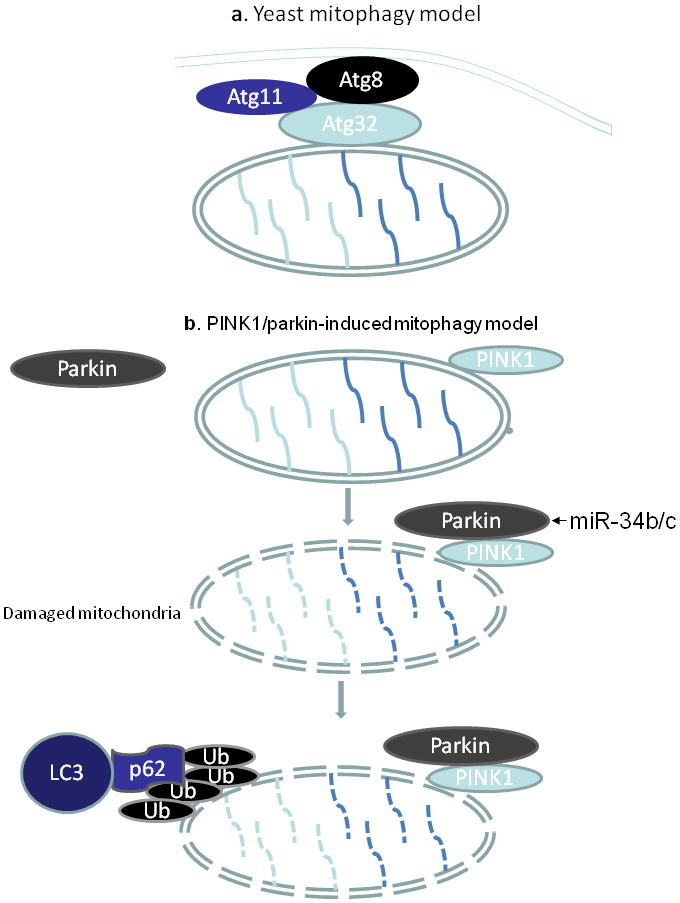
a. In yeast, Atg32 interacts with Atg 8 and Atg11 to recruit mitochondria into autophagosomes. b. PINK1/parkin-induced mitophagy in mammals. In the absence of mitochondrial injury, Parkin is located in cytosol. Upon mitochondrial damage, PINK1 triggers Parkin translocation to mitochondria where Parkin ubiquitylates mitochondrial proteins. This leads to the association of p62 with LC3, resulting in autophagosome formation.
Recent work has suggested a role for miRNAs in autophagy, including miR-101 [Frankel et al., 2011], miR-204 [Xiao et al., 2011], miR-30a [Zhu et al., 2009]. miRNAs target transcripts of autophagy-related proteins thereby influencing their function in autophagy. For example, Parkin functions in a pathway that links ubiquitylation with mitophagy [Geisler et al., 2010]. In Parkinson’s disease miR-34b/c downregulation is an early event [Miñones-Moyano et al., 2011]. It remains to be elucidated as to whether the early deregulation of miR-34b/c is responsible for the downstream transcriptome alterations underlying mitochondrial dysfunction. In some human tumors, miR-21 levels are significantly increased, and it down regulates the expression of PTEN [Meng et al., 2007; Zhang et al., 2010]. The PTEN regulates PINK1 that is involved in mitophagy pathway (Fig. 1). Further investigations are required to clearly elucidate the exact mechanism by which miRNAs control autophagy and mitophagy.
Perspective
It is obvious that miRNAs play a critical role in regulating mitochondrial function under physiological and pathological conditions. The relationship between the aberrant distribution of miRNAs in mitochondria and mitochondrial dysfunction needs to be fully established. Mitochondrial dysfunction is related to a variety of diseases. miRNAs have been used as diagnostic biomarkers in some of the diseases [Cogswell et al., 2008]. However, specific miRNAs that can be employed as diagnostic markers for mitochondria-related diseases have to be identified and validated. Further understanding of the specific functional consequence of modulating miRNA levels may eventually lead to miRNAs-based therapy for the treatment of mitochondria-related diseases. Overall, the elucidation of miRNAs in regulating mitochondrial activities may fill some of the gap in the knowledge on the aspects of cell biology and the pathogenesis of diseases, and may eventually lead to the development of pharmacological interventions for mitochondrial diseases.
Acknowledgments
We thank Suling Ding and Qian Li for their assistance in preparing this review. This work was supported by grants 5R01CA107506, 1R01HL102202 and 5R21HL092315 from the National Institutes of Health, Bethesda, MD.
References
- Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Aoi W, Naito Y, Mizushima K, Takanami Y, Kawai Y, Ichikawa H, Yoshikawa T. The microRNA miR-696 regulates PGC-1α in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab. 2010;298:E799–806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au HK, Yeh TS, Kao SH, Tzeng CR, Hsieh RH. Abnormal mitochondrial structure in human unfertilized oocytes and arrested embryos. Ann N Y Acad Sci. 2005;1042:177–185. doi: 10.1196/annals.1338.020. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chrétien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear Outsourcing of RNA Interference Components to Human Mitochondria. PloS one. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and Mature microRNA in Human Mitochondria. PloS one. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman S, Pineda F, Hardwick J. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ. 2008;15:1147–1152. doi: 10.1038/cdd.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, Zen K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaud F, Ricquier D, Thibault J, Weissenbach J. Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proc Natl Acad Sci USA. 1985;82:445–448. doi: 10.1073/pnas.82.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Cannino G, Di Liegro C, Rinaldi A. Nuclear-mitochondrial interaction. Mitochondrion. 2007;7:359–366. doi: 10.1016/j.mito.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang X, Kubo H, Harris DM, Mills GD, Moyer J, Berretta R, Potts ST, Marsh JD, Houser SR. Ca2+ influx-induced sarcoplasmic reticulum Ca2+ overload causes mitochondrial-dependent apoptosis in ventricular myocytes. Circ Res. 2005;97:1009–1017. doi: 10.1161/01.RES.0000189270.72915.D1. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Guo S, Copps K, Dong X, Kollipara R, Rodgers JT, Depinho RA, Puigserver P, White MF. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15:1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland MM, Norris KL, Karbowski M, Wang C, Suen DF, Jiao S, George NM, Luo X, Li Z, Youle RJ. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ. 2011;18:235–247. doi: 10.1038/cdd.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putativebiomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Corn PG. Hypoxic regulation of miR-210: Shrinking targets expand HIF-1’ s Influence. Cancer Biol Ther. 2008;7:265–267. doi: 10.4161/cbt.7.2.5745. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Edwards J, Quattrini A, Lentz S, Figueroa-Romero C, Cerri F, Backus C, Hong Y, Feldman E. Diabetes regulates mitochondrial biogenesis and fission in mouse neurons. Diabetologia. 2010;53:160–169. doi: 10.1007/s00125-009-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, Vojnovic B, Piresdas Neves R, Glazer P, Iborra F, Ivan M, Ragoussis J, ALH MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PloS one. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, Jäättelä M, Lund AH. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011 doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SM, Chen C, Wu J, Tan Y, Yu K, Xing CY, Ye A, Yin L, Jiang L. Synergistic apoptosis induction in leukemic cells by miR-15a/16–1 and arsenic trioxide. Biochem Biophys Res Commun. 2010;403:203–208. doi: 10.1016/j.bbrc.2010.10.137. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Qi D, An D, Pulinilkunnil T, Abrahani A, Kuo KH, Wambolt RB, Allard M, Innis SM, Rodrigues B. Brief episode of STZ-induced hyperglycemia produces cardiac abnormalities in rats fed a diet rich in n-6 PUFA. Am J Physiol Heart Circ Physiol. 2004;287:H2518–2527. doi: 10.1152/ajpheart.00480.2004. [DOI] [PubMed] [Google Scholar]
- Gong Y, Chai Y, Ding JH, Sun XL, Hu G. Chronic mild stress damages mitochondrial ultrastructure and function in mouse brain. Neurosci Lett. 2011;488:76–80. doi: 10.1016/j.neulet.2010.11.006. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Herrera B, Lockstone H, Taylor J, Ria M, Barrett A, Collins S, Kaisaki P, Argoud K, Fernandez C, Travers M, Grew JP, Randall JC, Gloyn AL, Gauguier D, McCarthy MI, Lindgren CM. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–1109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh RH, Au HK, Yeh TS, Chang SJ, Cheng YF, Tzeng CR. Decreased expression of mitochondrial genes in human unfertilized oocytes and arrested embryos. Fertil Steril. 2004;81:912–918. doi: 10.1016/j.fertnstert.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Cui XS, Kim EJ, KIM WJ, Kim NH. New porcine microRNA genes found by homology search. Genome. 2006;49:1283–1286. doi: 10.1139/g06-120. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren BT, Wong P, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Galin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–11. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux H, Vazquez EJ, Fujioka H, Hoppel CL. Decrease in Mitochondrial Function in Rat Cardiac Permeabilized Fibers Correlates With the Aging Phenotype. J Gerontol A Biol Sci Med Sci. 2010;65:1157–1164. doi: 10.1093/gerona/glq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010a;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Bates DJ, An J, Terry DA, Wang E. Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol Aging. 2011;32:944–955. doi: 10.1016/j.neurobiolaging.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Li P, Jayarama S, Ganesh L, Mordi D, Carr R, Kanteti P, Hay N, Prabhakar BS. Akt-phosphorylated mitogen-activated kinase-activating death domain protein (MADD) inhibits TRAIL-induced apoptosis by blocking Fas-associated death domain (FADD) association with death receptor 4. J Biol Chem. 2010b;285:22713–22722. doi: 10.1074/jbc.M110.105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim Biophys Acta. 2000;1459:370–382. doi: 10.1016/s0005-2728(00)00174-2. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Martí E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Howes AL, Adams JW, Dorn GW, 2nd, Brown JH. Ca2+ dysregulation induces mitochondrial depolarization and apoptosis: role of Na+/Ca2+ exchanger and AKT. J Biol Chem. 2005;280:38505–38512. doi: 10.1074/jbc.M505223200. [DOI] [PubMed] [Google Scholar]
- Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Iinuma M, Naoe T, Nozawa Y, Akao Y. Characterized mechanism of α-mangostin-induced cell death: Caspase-independent apoptosis with release of endonuclease-G from mitochondria and increased miR-143 expression in human colorectal cancer DLD-1 cells. Bioorg Med Chem. 2007;15:5620–5628. doi: 10.1016/j.bmc.2007.04.071. [DOI] [PubMed] [Google Scholar]
- Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke B. Evidence for mitochondrial K+ channels and their role in cardioprotection. Circ Res. 2004;94:420–432. doi: 10.1161/01.RES.0000117583.66950.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto KK-ON, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, MacDonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Ritov VB, Menshikova EV, Azuma K, Wood R, Toledo FG, Goodpaster BH, Ruderman NB, Kelley DE. Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Am J Physiol Endocrinol Metab. 2010;298:E49–58. doi: 10.1152/ajpendo.00317.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PloS one. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- Tong WH, Rouault T. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–5700. doi: 10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJJ, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecasteele G, Szabadkai G, Rizzuto R. Mitochondrial calcium homeostasis: mechanisms and molecules. IUBMB life. 2001;52:213–219. doi: 10.1080/15216540152846028. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, BMS Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xia HQ, Pan Y, Peng J, Lu GX. Over-expression of miR375 reduces glucose-induced insulin secretion in Nit-1 cells. Mol Biol Rep. 2011;38:3061–3065. doi: 10.1007/s11033-010-9973-9. [DOI] [PubMed] [Google Scholar]
- Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, Ni X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35. doi: 10.1186/1423-0127-18-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, Li Y. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun. 2008;376:548–552. doi: 10.1016/j.bbrc.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]


