Abstract
MicroRNAs (miRNAs) are recognized as being central players in many biological processes and cellular pathways. Their roles in disease have been highlighted first by observation of their aberrant expression profiles in human tumors, and then by in vitro and in vivo functional studies in transformed cells and model organisms. One of the most commonly observed features of miRNAs in malignancies is a defect in their production. Although several causes may be associated with this phenomenon, such as upstream oncogenic/tumor-suppressor defects and alterations in the miRNA-processing machinery, epigenetic inactivation is the prime suspect. The number of miRNAs with putative growth-inhibitory functions undergoing promoter CpG island hypermethylation in human cancer is growing fast and more detailed biological studies are necessary. The recognition of miR-124a and miR-34b/c as bona fide tumor-suppressor miRNAs undergoing DNA methylation-associated silencing in a wide spectrum of human neoplasms is a good starting point to be followed by other candidate miRNAs. Most importantly, even at this early stage, the transcriptional repression of miRNAs by hypermethylation of their corresponding promoter loci seems to be a common feature of all human tumors. This will have translational consequences for the management of the disease.
Keywords: DNA methylation, microRNAs, tumor-suppressor genes
miRNAs: Processing, targets and transport
Non-coding RNAs (ncRNAs) were formerly considered to be transcriptional noise, a byproduct of RNA polymerase during the synthesis of functional RNA. However, more than 90% of the human genome is transcribed in a network of overlapping transcripts without protein-coding capacity (Birney et al., 2007). Moreover, ncRNA sequences and their secondary structures are known to be subject to purifying selection; the evolution of ncRNAs is not consistent with neutralist explanations, thereby indicating that there is selective pressure on them (Ponjavic et al., 2007). ncRNAs are involved in different processes, including not only those involved in protein synthesis (transfer and ribosomal RNA), but also gene regulation, X-chromosome inactivation (Xu et al., 2007) and telomerase maintenance (Azzalin et al., 2007).
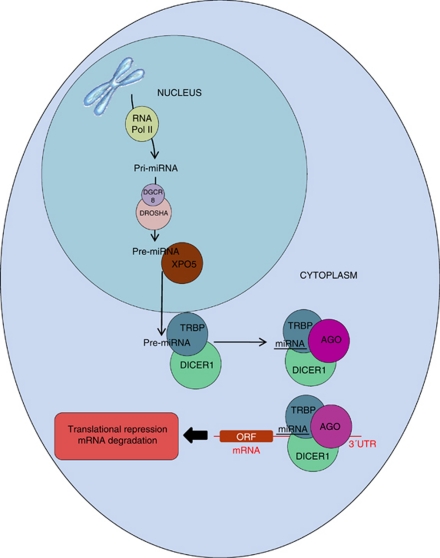
The most widely studied class of ncRNAs are microRNAs (miRNAs), which are small ncRNAs of ∼22-nt and are involved in post-translational gene silencing by controlling mRNA translation into proteins. It has been predicted that miRNAs regulate the translation rate of more than 60% of protein-coding genes (Friedman et al., 2009), and so participate in the regulation of all cellular processes. Like mRNAs, miRNAs are mainly transcribed by RNA polymerase-II (Lee et al., 2004), although miRNA synthesis is known to occur by RNA polymerase-III in those miRNAs that reside near tRNA, Alu and mammalian-wide interspersed sequences (Borchert et al., 2006). RNA polymerase-II synthesizes a primary transcript called pri-miRNA. The primary transcripts are long, poly-(A)-tailed and capped species measuring several kilobases. This transcript is processed by Drosha, a ribonuclease-III protein (Lee et al., 2003), in collaboration with DGCR8 (DiGeorge Syndrome Critical Region Gene-8), the microprocessor protein (Han et al., 2006) responsible for anchoring the pri-miRNA in the complex. The processing of the pri-miRNA produces a 70-nt-long molecule (Gregory et al., 2004), named pre-miRNA, which migrates to the cytoplasm by using the nuclear export protein Exportin-5 (XPO5) (Yi et al., 2003). In the cytoplasm, the pre-miRNA is released in a GTP-dependent manner and processed by Dicer (a ribonuclease-III protein) in a complex with TRBP, a double-stranded RNA-binding protein, to produce the functional 22-nt-long miRNA (Hutvágner et al., 2001), which targets the mRNA. The ∼22-nt molecules are loaded by the Dicer–TRPB complex into a member of the Argonaute (Ago) Protein Subfamily to form the RNA-induced silencing complex (RISC). These Ago proteins are the catalytic components of the RISC (Liu et al., 2004). The miRNA biosynthesis pathway is summarized in the Figure 1.
Figure 1.
The miRNA biosynthesis pathway from the nucleus to the cytosol. Ago, Argonaute protein; DGCR8, DiGeorge Syndrome Critical Region Gene-8; miRNA, microRNA; ORF, open reading frame; TAR-binding protein; UTR, untranslated region; XPO5, exportin-5; TRBP.
The genes targeted by the miRNAs are defined by complementary base pairs between the miRNA loaded into the complex and, more often, the 3′-untranslated (UTR) region of the mRNA. The complementarity between the miRNA and the targeted mRNA is imperfect, the seed region of the miRNA (nucleotides 2–8) being essential to associate the miRNA with its target mRNA (Lai, 2002; Lewis et al., 2003). Usually, the 3′UTR region of the mRNA is bound to multiple miRNAs, improving the repressive effect of the miRNA machinery (Grimson et al., 2007). mRNA cleavage is one of the mechanisms by which miRNA regulates gene expression. In this mechanism, the miRNAs act as small interference RNAs (siRNAs), another type of ncRNA produced by DICER from double-stranded RNA sequences, mostly of exogenous origin. siRNAs direct RNA cleavage through the RNA interference pathway in extensive base pairing that is complementary between the siRNA and the targeted mRNA (Plasterk, 2002). The entrance of the miRNA in the RNA interference pathway depends on the degree of complementarity between the miRNA and the targeted mRNA (Hutvágner and Zamore, 2002). However, almost none of the miRNA described was highly complementary to its targeted sequence in the mRNA, preventing its entrance into the RNA interference pathway (Elbashir et al., 2001). In fact, the disruption of protein synthesis in the ribosomes through inhibition of translational initiation is the predominant mechanism of gene silencing mediated by miRNAs. The RISC-bound mRNAs localize in the processing bodies (P bodies) in the cytoplasm, where they accumulate or are degraded (Pillai et al., 2005). AU-rich elements (AREs) in the 3′UTR of unstable mRNAs dictate their degradation (Shaw and Kamen, 1986). miRNAs and Dicer are involved in this mechanism of mRNA turnover. Many miRNAs present a complementary sequence to the ARE sequence characteristic of unstable mRNAs, the cooperation between miRNA, Dicer and Ago proteins, and ARE-binding proteins being essential for mRNA decay (Jing et al., 2005).
Although gene silencing is the most thoroughly studied role of miRNAs, the capability to upregulate gene transcription during cell-cycle arrest has been described; miRNA369-3 and let-7 oscillate between the ability to repress and activate, depending on the cell cycle. In proliferating cells, they repress the translation of their target genes, whereas in cell arrest, which is often the step preceding cell differentiation, they promote activation (Vasudevan et al., 2007). Another example of miRNA-mediated activation of mRNA translation is the case of miR-10a (Ørom et al., 2008). This miRNA enhances ribosomal protein synthesis by binding to the 5′UTR of the target mRNAs. The major tumor-suppressor genes, p53 and RB, negatively regulate ribosome synthesis (Ruggero and Pandolfi, 2003), and a very well known oncogene, MYC, positively regulates protein production (Boon et al., 2001). As for the described genes, miR-10a is involved in tumor progression (Ørom et al., 2008). miRNAs are known to target proteins directly. miRNAs can act as direct inhibitors of ribonucleoprotein activity in an miRNA-mediated silencing process called decoy. miR-328 interacts directly, independently of the Ago protein, with the heterogeneous ribonucleoprotein (hnRNP) E2. This interaction prevents the translational repression by hnRNP E2 of CEBPA mRNA, a key regulator of myeloid differentiation to granulocytes. Downregulation of miR-328 allows the hnRNP E2 inhibition of myeloid differentiation and thus, tumor progression (Eiring et al., 2010). siRNAs have the ability to direct gene silencing by heterochromatin formation. RNA-induced transcription silencing is a mechanism described in yeast (Verdel et al., 2004). The RNA-induced transcription silencing complex recruits histone methyltransferases after siRNA loading, directing heterochromatin formation in the DNA locus.
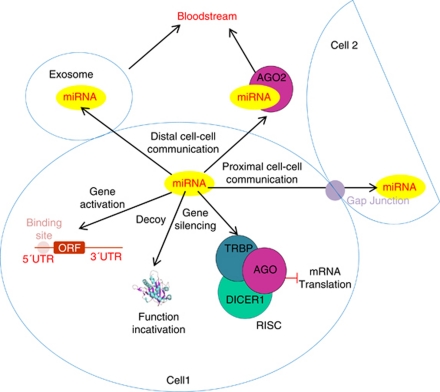
miRNAs circulate in the bloodstream in a very stable form; different mechanisms have been proposed to explain the high stability of the circulating RNA in an RNase-rich environment. Under normal conditions, many cell types, such as B- and T-cells, mast cells and reticulocytes, have the capacity to produce exosomes, which are small membrane vesicles that are released into the extracellular matrix with the plasma membrane. Tumoral cells in melanoma are also known to produce exosomes (Mears et al., 2004). The exosomes carry proteins, mRNA and miRNA. The mRNA present in the exosomes can be translated in the recipient cells, suggesting a functional role for the miRNA. This mechanism of RNA delivery is called ‘exosomal shuttle RNA' and may constitute a way by which the donor cell regulates protein production in the recipient cell (Valadi et al., 2007). In this paper, the authors found 121 miRNAs in the exosomes produced by mast cells. Taking into account that every miRNA has the potential to interfere with up to 200 mRNAs (Krek et al., 2005), the regulatory capacity of the miRNAs present in the exosomes may modulate up to 24 000 mRNAs. More recently, it has been found that most of the circulating miRNA in plasma is confined to protein complexes rather than membrane vesicles. These proteins, like exosomes, protect miRNA from degradation, there being at least two different populations of circulating RNA. The ribonucleoprotein complex contains Ago-2, the protein that mediates RNA silencing in the RISC, suggesting that cells release functional silencing complexes into the circulation to modulate gene expression in the recipient cells (Arroyo et al., 2011). Cell–cell communication can be mediated by direct contact between the donor and the recipient cells by gap junction intercellular connections. miRNAs can pass from one cell to the other by these junctions and modulate the gene expression of the targeted genes in the receptor cell (Lim et al., 2011). These authors showed the exchange of miRNAs between bone marrow stromal cells and metastatic breast cancer cells by this mechanism; specific miRNAs target CXCL12, a chemokine involved in cell communication and signal transduction, cell proliferation is reduced and cells enter quiescence, suggesting a role for miRNAs in the dormancy of breast cancer cells in the bone marrow and explaining the recurrence of metastatic breast cancer from the bone marrow many years after primary tumor elimination (Willis et al., 2010). Figure 2 summarizes the different mechanisms by which miRNAs regulate gene expression.
Figure 2.
Mechanisms by which miRNAs regulate gene expression within a cell and in neighboring cells. Ago, Argonaute protein; ORF, open reading frame; RISC, RNA-induced silencing complex; TRBP, TAR-binding protein; UTR, untranslated region.
The regulation of gene expression by miRNAs, their role in targeting different genes and their capacity to move between cells illustrate the complexity of transcriptome regulation in the cells. In addition, RNA editing increases this complexity by generating new molecules from existing ones (reviewed by Bass, 2002). RNA editing occurs by the deamination of adenosines to inosines, performed by adenosine deaminase acting on RNA enzymes (ADAR), mostly within the non-coding regions of the transcripts. The newly generated nucleotide base pairs have cytosine instead of thymidine, promoting the synthesis of new proteins from a single open reading frame (Burns et al., 1997) and altering miRNA targeting by editing of the miRNAs or the mRNAs (Borchert et al., 2009). Moreover, the role of pseudogenes in miRNA homeostasis has been described. Pseudogenes are commonly considered to be non-functional genomic loci and to have no biological relevance, although preservation of the nucleotide sequences suggests that they are under selective pressure. Pseudogenes can be transcribed or not, and those that are transcribed show a high complementarity with the protein-coding gene from the 5′ to the 3′UTR. Taking into account that miRNAs commonly regulate gene silencing by 3′UTR sequence complementarity, pseudogenes may act to regulate miRNA activity on the coding gene by competing for the miRNA molecules present in the cell. This mechanism of miRNA regulation has been modeled by studying PTEN (tumor-suppressor gene)/PTENP1 (PTEN pseudogene) expression levels in tumoral cell lines (Poliseno et al., 2010). The authors demonstrated the role of PTEN transcripts in modulating miRNA targeting by examining PTENP1 levels. This study provides a new perspective on transcriptional regulation. Considering the genomic rearrangements, copy-number alterations and translocations that many cancer cells suffer and that give rise to altered levels of the transcripts, changes in the tumoral cell phenotype can be due not only to the protein alteration itself, but also to global RNA deregulation by altered competition for miRNAs (Poliseno et al., 2010).
miRNAs: Cancer involvement and deregulation mechanisms
Cancer development involves genetic and epigenetic alterations that give rise to an uncontrolled overgrowth of cells (Jones and Baylin, 2007; Berdasco and Esteller, 2010). Research has tended to focus on coding genes and the proteins they produce, classifying them as oncogenes, if they promote cell growth, or tumor-suppressor genes, if they prevent overgrowth. With the discovery of miRNAs in Caenorhabditis elegans in the late 1990s (Fire et al., 1998) and their regulatory role in gene expression, a new front in cancer research was initiated.
As with classical genes, the non-coding loci in the DNA (ncRNAs) can be subjected to genetic and epigenetic alterations. These alterations modify the expression profile of the ncRNAs and thus alter the mechanism they regulate. Chromosome abnormalities (deletions, translocations, copy-number alterations), DNA mutations and epigenetic deregulation of the ncRNAs or the genes involved in their biogenesis have been described in tumor progression. The first association between miRNA and cancer development was described in chronic lymphocytic leukemia with chromosome 13q14 deletion. This deletion deregulates miR-15 and miR-16 (Calin et al., 2002). Most of the targets of these two miRNAs are involved in cell growth and cell cycle (for example, Anxa2 and Cdc2), apoptosis (Grp78 and Bcl2), and oncogenes and tumor-suppressor genes (Wt1 and Rab8B) (Calin et al., 2008). Surprisingly, miRNAs are frequently located in fragile regions of the chromosomes as has been shown in ovarian and breast carcinomas and melanomas (Calin et al., 2004; Zhang et al., 2006). miR-16 is also involved in the mRNA decay of tumor necrosis factor-α (Jing et al., 2005) by ARE sequence binding in the 3′UTR of the mRNA. This suggests that a single miRNA involved in the regulation of multiple mRNAs can use different mechanisms to achieve gene silencing: not only basepair complementary to specific sequences in the targeted genes, but also recognition of the ARE instability signal. Mutations of the miRNA sequences can also disrupt their regulatory role. Mutations in the primary transcripts of miR-15a and miR-16-1 are responsible for their reduced expression levels in chronic lymphocytic leukemias (Calin et al., 2005). Polymorphisms have been described in a set of miRNAs in non-small cell lung cancer and a single single-nucleotide polymorphism in has-mir-196a2-3p is associated with survival in patients with non-small cell lung cancer, probably owing to a defect in binding to the target gene (Hu et al., 2008).
let-7 is one of the most widely studied miRNA families in cancer. Alterations of let-7 function have been described in several human cancer types, such as lung cancer (Takamizawa et al., 2004), colorectal cancer (Akao et al., 2006), head and neck cancer (Yu et al., 2011), and ovarian cancer (Wendler et al., 2011). It acts mainly as a tumor-suppressor miRNA. On the one hand, the members of this family negatively regulate cell-cycle-related genes such as RAS or cyclins (Johnson et al., 2005). Mutations or alterations of the expression profile of these oncogenes are involved in cancer development. On the other hand, the let-7 miRNA family influences apoptosis owing to its negative regulation of the Fas gene (Wang et al., 2011). Fas is a death receptor in peripheral T-cells, which is involved in activation-induced cell death (AICD) in the presence of the Fas ligand, a member of the tumor necrosis factor cytokine family (Krueger et al., 2003). In cervical carcinomas as compared with normal tissue, let-7 is overexpressed and, as a consequence, the level of Fas gene expression is reduced (Das et al., 2000; Lee et al., 2008). In this case, the let-7 miRNA family acts as an oncomiRNA (tumor-promoting miRNA).
miRNA biosynthesis is also subjected to extracellular signaling. Under mitogenic signals, the mitogen-activated protein kinase (MAPK) Erk mediates the phosphorylation of TRBP, one of the components of the DICER1 complex. TRBP phosphorylation enhances miRNA expression by increasing the stability of the biosynthesis complex (Paroo et al., 2009). Surprisingly, under tumorigenic signals, only miRNAs involved in cell growth promotion are upregulated when the MAPK/Erk pathway is activated, and let-7 miRNA expression levels, which are mainly involved in cell growth suppression, are reduced. let-7 downregulation has been shown under type-I collagen activation of the transforming growth factor-β1-mediated expression of MT1–MMP (membrane type-1-matrix metalloproteinase) and Erk pathway activation in pancreatic ductal adenocarcinoma (Dangi-Garimella et al., 2011). This mechanism modulates the metastatic capacity of the tumoral cells by enhancing the expression of metalloproteinases, which are proteins involved in collagen degradation, a key process for promoting cell invasion through the extracellular matrix (Ottaviano et al., 2006), and let-7 downregulation (Dangi-Garimella et al., 2011).
Alterations in the genes involved in miRNA expression can disrupt the correct function of these miRNAs. Overexpression of the oncogenic miRNA miR-17-92 cluster has been reported in human cancer (Volinia et al., 2006). This miRNA family is regulated by c-Myc, a transcriptional factor overexpressed in multiple human cancers (Chang et al., 2008). Overexpression of c-Myc activates miR-17-92 cluster expression. This family regulates the gene silencing of many genes involved in cell cycle and proliferation. The miR-17-92 cluster confers resistance to cell senescence and apoptosis by silencing several tumor-suppressor genes such as PTEN (Olive et al., 2009) and p21 (Hong et al., 2010). Angiogenesis is a key step in tumor development and metastasis, providing nutrients and oxygen to overgrowing cells. The miR-17-92 cluster confers a metastatic capacity on tumoral cells by inhibiting Tsp1 (anti-angiogenic thrombospondin-1) and CTGF (connective tissue growth factor), which are genes involved in angiogenesis (Dews et al., 2006).
Once transcribed, miRNAs are processed and exported from the nucleus to the cytoplasm. Alterations in the processing machinery can also lead to deregulation of functional miRNAs. Reduction of the expression levels of Drosha and Dicer has been described in ovarian cancer patients, leading to a downregulation of mature miRNAs (Merritt et al., 2008). Germline mutations in the DICER1 gene have been described in familial ovarian and pulmonary neoplasm (Rio Frio et al., 2011), and kidney neoplasm (Bahubeshi et al., 2010). An miRNA family, miR-103/107, targets Dicer to downregulate its translation into the protein and attenuate global miRNA synthesis. This mechanism creates a feedback relationship taking into account that miR-103/107 are both generated by and regulators of Dicer, producing a downscaling of Dicer levels but not their complete depletion, maintaining a basal level to regulate the miRNA pathway (Martello et al., 2010). Overexpression of the miR-130/107 family in breast cancer is associated with epithelial–mesenchymal transition and metastasis due to the downregulation of miR-200 levels. The miR-200 family regulates the silencing of ZEB1/ZEB2, which are genes involved in the epithelial–mesenchymal transition (Burk et al., 2008).
Latent infection by Epstein–Barr virus is associated with the development of human cancers such as Burkitt lymphoma, Hodgkin lymphoma and pharyngeal carcinomas (Pagano, 2002). The viral genome encodes its own miRNAs, identified in human B-cells infected by Epstein–Barr virus (Pfeffer et al., 2004). miR-BART6-5p, one of the miRNAs encoded by the virus, targets Dicer and suppresses its expression, altering global miRNA patterns in the infected cells (Iizasa et al., 2010).
TARBP2, one of the components of the DICER1 complex, presents frameshift mutations that cause diminished protein expression levels in sporadic and hereditary colorectal carcinomas. This reduced expression causes a defect in pre-miRNA processing and promotes tumor progression (Melo et al., 2009). XPO5 is the protein in charge of miRNA export from the nucleus to the cytoplasm. Inactivating mutations in the XPO5 gene appear in human carcinomas with satellite instability; these mutations trap pre-miRNA in the nucleus and deregulate the mature miRNA pattern in cancer cells (Melo et al., 2010).
miRNAs: The impact of aberrant DNA methylation profiles
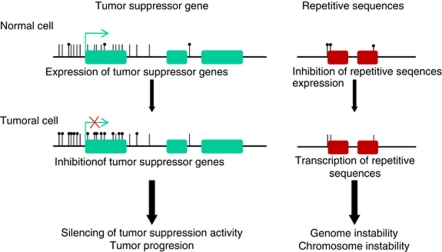
One of the most common causes of the loss of tumor-suppressor miRNAs in human cancer is the silencing of their primary transcripts by CpG island promoter hypermethylation (Saito et al., 2006; Lujambio et al., 2007, 2008; Toyota et al., 2008; Huang et al., 2009). In cancer, cells undergo global hypomethylation of the DNA (Feinberg and Vogelstein, 1983). This contributes to genome instability and the transcription of silenced transposable sequences, promoting chromosomal rearrangements and genome disruption, a characteristic feature of tumoral cells. However, the CpG islands of the promoter regions of the tumor-suppressor genes undergo DNA hypermethylation, leading to gene silencing and promotion of cancer development (Jones and Baylin, 2007; Berdasco and Esteller, 2010). The DNA methylation profile of tumors can be used as a signature to define tumor type, clinical prognosis and treatment response (Esteller, 2008; Rodríguez-Paredes and Esteller, 2011). miRNAs transcribed from CpG islands undergo DNA methylation-associated repression with a similar chromatin context to coding genes, such as binding of the transcriptional repressor methyl-CpG-binding domain proteins (Lujambio et al., 2007; Urdinguio et al., 2010), and a histone modification profile associated with silencing, such as in the case of loss of acetylation of histones H3 and H4 (Lujambio et al., 2007; Toyota et al., 2008). A similar chromatin-repressive environment has been described for other ncRNAs, such as transcribed-ultraconserved regions, which also undergo promoter CpG island hypermethylation in cancer cells (Lujambio et al., 2010). Figure 3 illustrates aberrant DNA methylation patterns that occur in tumoral cells.
Figure 3.
Aberrant DNA methylation patterns in human cancer. Heavily methylated repetitive sequences undergo hypomethylation events in transformed cells that might cause genome instability (right panel), whereas unmethylated promoter CpG islands of coding genes and miRNAs with tumor-suppressor features become hypermethylated and silenced in cancer cells (left panel).
Epigenetic silencing of the tumor-suppressor miR-124a by aberrant DNA hypermethylation of the colorectal cancer cell line HCT-116 leads to cyclin-D kinase-6 (CDK6) overexpression, its target. Transfection of the tumoral cell line with miR-124a diminishes CDK6 protein levels and retinoblastoma (Rb) phosphorylation, one of the targets of the kinase (Lujambio et al., 2007). Both proteins are involved in cell-cycle progression. Rb is an antiproliferative tumor-suppressor gene that, when phosphorylated by CDK6, diminishes its antiproliferative activity (Grossel and Hinds, 2006). Hypermethylation of this miRNA has also been observed in glioblastoma multiforme (Silber et al., 2008), gastric cancer (Ando et al., 2009), hematopoietic malignancies (Agirre et al., 2009; Roman-Gomez et al., 2009), cervical cancer (Wilting et al., 2010) and hepatocellular carcinoma (Furuta et al., 2010).
miR-145 is a well-known tumor-suppressor miRNA that is downregulated in many human cancers owing to aberrant DNA methylation of its promoter and/or p53 mutations (Suh et al., 2011). This miRNA is a pluripotency repressor; it regulates OCT, SOX2 and KLF4 gene silencing in human embryonic stem cells, genes required for cell self-renewal and pluripotency maintenance (Xu et al., 2009). In fact, they are three of the four factors known to acquire de novo pluripotency (Takahashi and Yamanaka, 2006). Reprogramming to pluripotency resembles the de-differentiation process that cancer cells undergo during tumor formation. In acute myeloid leukemia, the oncogene c-kit is overexpressed in 60–80% of patients (Kindler et al., 2004). miR-193a has been described as a key tumor-suppressor miRNA that targets c-kit and, thus, prevents cell overgrowth and promotes cell differentiation. This miRNA is regulated by DNA methylation and its aberrant silencing by DNA hypermethylation has been described in acute myeloid leukemia cell lines (Gao et al., 2011).
miR-335 was identified in a set of miRNAs involved in the suppression of breast cancer metastasis to the lungs and bone (Tavazoie et al., 2008). It was found to inhibit metastasis by targeting the genes involved in cell invasion and migration such as the transcription factor SOX4 and the extracellular matrix protein tenascin-C. This miRNA undergoes genetic deletion and promoter hypermethylation silencing in metastatic cell populations derived from primary breast tumors. Cells that undergo miR-335 silencing in the population have a selective advantage over the others, acquiring metastatic capacity and colonizing new tissues (Png et al., 2011). The genetic and epigenetic silencing of this miRNA have been observed in ovarian cancer samples, and its deletion is correlated with the malignant progression of breast and ovarian tumors. miR-129-2 is another miRNA involved in SOX4 silencing and it is also regulated by promoter methylation. Aberrant miR-192-2 DNA hypermethylation has been found in endometrial (Huang et al., 2009), colorectal (Bandres et al., 2009) and gastric cancer (Shen et al., 2010).
miR-199a expression is regulated by DNA methylation of the locus from which it is transcribed (Cheung et al., 2011). Downregulation of this miRNA owing to aberrant DNA hypermethylation of its promoter and its association with invasive properties of the tumoral cells have been described in gastric (Ueda et al., 2010), ovarian (Yin et al., 2010) and testicular (Cheung et al., 2011) tumors. This miRNA regulates the silencing of many genes involved in tumor progression. In testicular cancer, it targets PODXL, an anti-adhesive protein that inhibits cell–cell interaction, a key feature of the cells that acquire an invasive phenotype in the primary tumor (Cheung et al., 2011).
Nuclear factor-κB is a transcription factor that regulates the proinflammatory response. Its activation is linked to cancer development by its role in the regulation of the secretion of cytokines and chemokines by the immune cells in the tumoral microenvironment. Its activation is regulated by the IκB kinases (IKKs) by phosphorylation and further degradation of the nuclear factor-κB inhibitor, IκBα. This cascade promotes the translocation of nuclear factor-κB to the nucleus, where it promotes the gene expression of the inflammatory cytokines involved in cell proliferation. In most cancer cells, the IKKs and, for instance, nuclear factor-κB are constitutively active (Chen et al., 2007). miR-199a regulates the post-transcriptional silencing of IKKβ, one of the members of the IKK complex. Aberrant downregulation of the miRNA activates the IKK pathway by aberrant translation of IKKβ. This promotes the secretion of proinflammatory molecules characteristic of tumor development (Chen et al., 2008).
miR-34 is a tumor-suppressor miRNA family regulated by the transcriptional factor p53 (Tarasov et al., 2007) and DNA methylation (Lujambio et al., 2008; Toyota et al., 2008; Tsai et al., 2011). p53 is one of the best studied tumor-suppressor genes; it is called the ‘guardian of the genome' because it promotes apoptosis under DNA damage. It regulates the expression of many oncogenes such as c-Myc, Bcl2, and cell cytokines and miRNAs involved in cell proliferation. Inactivation of miR-34 members is a common feature of many tumor types. Downregulation of miR-34 owing to p53 inactivation or DNA hypermethylation of its promoter alters the Notch pathway (Pang et al., 2010), which is involved in the promotion of cell invasion (Bin Hafeez et al., 2009) and apoptosis of cancer cells. One of the targets of miR-34 is SIRT1, an oncogene that decreases the ability of p53 to promote cell-cycle arrest by deacetylating the protein. In this way, cells create a positive feedback loop between p53 and miR-34 for tumor prevention mediated by SIRT1 silencing (Yamakuchi et al., 2008). Altered expression profile of this miRNA family has been described in gastric (Tsai et al., 2011), ovarian (Corney et al., 2010), colorectal (Lujambio et al., 2008) and non-small cell lung cancer (Gallardo et al., 2009). Promoter CpG island hypermethylation for both miR-34b/c (Lujambio et al., 2008; Toyota et al., 2008; Roman-Gomez et al., 2009; Tsai et al., 2011) and miR-34a (Lodygin et al., 2008; Chim et al., 2010; Corney et al., 2010) represents a major mechanisms accounting for the loss of these transcripts in human cancer.
Epigenetic silencing of miRNAs is also involved in the acquisition of an invasive phenotype and the development of metastasis (Lujambio and Esteller, 2009). Treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine and further analysis of miRNA expression arrays of metastatic cells from lymph node metastasis of different human cancers showed cancer-specific DNA hypermethylation of the CpG islands in the promoter of three miRNAs, miR-148a, miR-9 family and miR-34b/c. Epigenetic silencing of these miRNAs mediates the activation of metastatic genes such as c-Myc, CDK6 and E2F3 (E2F transcription factor-3, involved in the activation of the expression of cyclins and other proliferative genes), targeted by miR-34b/c downregulation, and TGIF2 (transforming growth factor-β-induced factor-2), targeted by miR-148a (Lujambio et al., 2008). Alterations in these miRNAs have been described in many cancer types such as breast (Lehmann et al., 2008), ovarian (Laios et al., 2008) and pancreatic cancer (Omura et al., 2008). Surprisingly, miR-9 has been described as an oncogene owing to its role in E-cadherin silencing in hepatocellular carcinomas promoting cell invasion (Tan et al., 2010). This suggests the potential dual role of miRNA in promoting or inhibiting tumoral progression, depending on the tissue where they are expressed. Hypermethylation of the miR-9 family has also been reported in hematopoietic malignancies (Roman-Gomez et al., 2009) and renal cell carcinoma (Hildebrandt et al., 2010). Recent reports have also indicated the presence of hypermethylation-associated silencing of some miR-200 family members in cancer cells (Ceppi et al., 2010; Neves et al., 2010; Vrba et al., 2010; Wiklund et al., 2010; Chen et al., 2011). The miR-200 family has an important epithelial–mesenchymal transition-regulatory activity in which it directly targets and inhibits ZEB1 and ZEB2 (Hurteau et al., 2006; Park et al., 2008), transcriptional repressors of E-cadherin, a cell adhesion protein crucial for maintaining epithelial structure (Peinado et al., 2007). Most importantly, the DNA methylation-associated silencing of the miR-200 family is a dynamic process that determines the evolving epithelial–mesenchymal transition phenotypes of colorectal and breast tumors (Davalos et al., 2011).
Other important miRNAs disrupted by promoter CpG island hypermethylation in human cancer include miR-203, which targets ABL1 and is epigenetically inactivated in hematopoietic malignancies (Bueno et al., 2008) and hepatocellular carcinoma (Furuta et al., 2010); miR-181c, which targets the oncogenes NOTCH4 and K-RAS, and undergoes hypermethylation in gastric tumors (Hashimoto et al., 2010); and let-7a-3 in ovarian cancer with an inverse correlation with insulin-like growth factor-II expression (Lu et al., 2007). The list of miRNAs undergoing promoter CpG island hypermethylation in human cancer is rapidly expanding and a recent epigenomic survey of 450 000 CpG sites revealed that among the 616 miRNAs printed in the microarray, 30–40 miRNAs underwent hypermethylation in colorectal cancer cells (Sandoval et al., 2011):
As mentioned above, cell treatment with the DNA-demethylating agent 5-aza-2′-deoxycytidine and the histone deacetylase inhibitor 4-phenylbutyric acid followed by analysis of transcript expression profiles is a common approach used to identify sequences controlled by the epigenetic marks. Using this approach, the epigenetic silencing of miRNA-127 in prostate and bladder cancer described originally (Saito et al., 2006). This miRNA belongs to an miRNA cluster but only miR-127 presents a CpG island, being the only one regulated by DNA methylation. Silencing of this miRNA by DNA hypermethylation leads to cancer progression owing to overexpression of its target, BCL6, a key regulator of p53 expression.
miRNAs can be transcribed from intronic regions in coding genes. Sometimes, the tumoral phenotype is thought to be the cause of the aberrant hypermethylation of a given gene, but it is also due to the downregulation of the miRNA it hosts. This is the case of miR-342, which is transcribed from the EVL gene (Grady et al., 2008). The EVL promoter is hypermethylated in the early stages of colorectal adenocarcinomas and as a consequence, the gene and the miRNA are both downregulated. EVL is a gene involved in cell polarity and cytoskeleton remodeling, and can affect cell motility. The targets of miR-342 have not been described. Another miRNA aberrantly methylated in the early stages of colorectal cancer development is miR-137 (Balaguer et al., 2010), which is also methylated in oral cancer (Kozaki et al., 2008) and glioblastoma multiforme (Silber et al., 2008). Among the targets of this miRNA, we can highlight LSD-1 (lysine-specific histone demethylase-1), a histone demethylase that has a key role in the epigenetic regulation of gene expression; CDK6 and E2F6, a transcription factor involved in cell-cycle progression. In primary hepatocellular tumors, aberrant hypermethylation of miR-1, a tumor-suppressor miRNA targeting FoxP1, a ubiquitously expressed transcription factor; MET (hepatocyte growth factor receptor) and histone deacetylase-4 has been described. Overexpression of these genes owing to miRNA silencing promotes hepatocellular carcinogenesis (Datta et al., 2008).
Considered from the opposite point of view, aberrant DNA hypermethylation of tumor-suppressor genes can be promoted by an altered miRNA expression profile. In hepatitis B virus-related hepatocellular carcinomas, the levels of miRNA-152 are downregulated relative to those in non-cancerous tissues. miR-152 downregulation promotes the overexpression of DNA methyltransferase-1 (DNMT1), its target, and, as a consequence, aberrant DNA hypermethylation. The relation of the viral infection and the hypermethylation of the DNA is the hepatitis B virus X protein (HBx), the inhibitor of miR-152. This mechanism may also be involved in the methylation of the hepatitis B virus genome, promoting the survival of the virus in the host cells (Huang et al., 2010). The miRNA-29 family is downregulated in lung cancer. This family of miRNAs has sequence complementarity with the 3′UTR of DNMTs 3a and 3b. These two enzymes are frequently upregulated in lung cancer and are associated with poor prognosis in patients. An inverse correlation between DNMT-3a and -3b, and the members of the miRNA-29 family has also been noted. Enforced expression of these miRNAs restores the normal DNA methylation profile by silencing the DNMTs, inducing the expression of tumor-suppressor genes that are aberrantly methylated, and inhibiting tumor growth in lung cancer cell lines and mice (Fabbri et al., 2007).
Table 1 summarizes the best characterized miRNAs dysregulated by DNA hypermethylation and the functional consequences in tumoral cells.
Table 1. Hypermethylated miRNAs in human cancer and their functional consequences.
| miRNA hypermethylated | Chromosome | Target | Functional consequences | Cancer type |
|---|---|---|---|---|
| miR-1 | 20 | FoxP1 | Transcriptional activation | Hepatocarcinoma |
| MET | Cell proliferation, angiogenesis, tumor cell invasion | |||
| HDAC4 | Gene transcription alteration | |||
| miR-9-1 | 1 | FGF family | Cell proliferation | Breast, ovarian, pancreas, hematological, renal |
| Claudin family | Cell communication and growth | |||
| miR-9-3 | 15 | E-cadherin | Cell migration | Colorectal, melanoma, head and neck |
| miR-34a | 1 | CD44 | Cell communication and signal transduction | Hematological, prostate, breast, renal, |
| Notch1 | Transcriptional activation | colorectal, ovarian | ||
| miR-34b/c | 11 | Notch1 | Transcriptional activation | Gastric, ovarian, lung, colon, melanoma, |
| c-Myc | Transcriptional activation | head and neck | ||
| Bcl2 | Cell proliferation | |||
| SIRT1 | Epigenetic gene silencing | |||
| miRNA-124a | 20 | CDK6 | Cell-cycle deregulation | Colon, gastric, hematological, cervical, liver, glioblastoma |
| miR-127 | 14 | BCL6 | p53 downregulation and cell proliferation | Prostate, bladder |
| miR-129-2 | 11 | SOX4 | Cell dedifferentiation and proliferation | Gastric, endometrial, colorectal |
| miR-137 | 1 | CDK6 | Cell-cycle dysregulation | Oral, colorectal, glioblastoma, |
| E2F6 | Cell-cycle dysregulation | |||
| LSD-1 | Gene transcription alteration | |||
| miR-145 | 5 | OCT/SOX2/KLF4 | Cell dedifferentiation | Prostate |
| miR-148a | 7 | TGIF2 | Invasive capacity | Colorectal, melanoma, head and neck, breast |
| miR-129-2 | 11 | SOX4 | Cell dedifferentiation and proliferation | Gastric, endometrial |
| miR-181c | 19 | Notch4 | Transcriptional activation | Gastric |
| K-Ras | Signal transduction | |||
| miRNA-199a | 19 | PODXL | Invasive capacity | Testicular, ovarian |
| IKKβ | Proinflammatory molecules secretion | |||
| miR-200c | 12 | ZEB1/ZEB2 | Epithelial–mesenchymal transition | Colorectal, breast, lung |
| miR-141 | 12 | ZEB1/ZEB2 | Epithelial–mesenchymal transition | Colorectal, breast, lung |
| miR-429 | 1 | ZEB1/ZEB2 | Epithelial–mesenchymal transition | Colorectal, breast, lung |
| miR-203 | 14 | ABL1 | Cell communication and signal transduction | Hematological, liver |
| miR-205 | 1 | ZEB1/ZEB2 | Epithelial–mesenchymal transition | Bladder |
| miRNA-335 | 7 | SOX4/TNC | Cell dedifferentiation and proliferation | Breast |
| miR-342 | 14 | PDGFRA | Cell proliferation | Colorectal |
| RASA1 | Cell proliferation | |||
| let-7a-3 | 22 | IGF-II | Cell communication and signal transduction | Ovarian, breast |
Abbreviation: miRNA, microRNA.
miRNAs: Translational applications
From a clinical point of view, miRNAs have great potential at the diagnostic and therapeutic levels. Microarray technology offers the opportunity to determine the expression of thousands of miRNAs at the same time, generating a very useful miRNA profile to determine the tumor type, patient prognosis and treatment response. Microarray analysis highlights a general downregulation of miRNAs in tumors when compared with normal tissues (Lu et al., 2005); furthermore, owing to the remarkable tissue specificity of the miRNAs, they become a very useful tool for defining the origin of the tumor in poorly differentiated cancers (Rosenfeld et al., 2008). In fact, a specific miRNA signature is being used for cancer screening.
The prognosis and survival of patients depend on the stage of the cancer when they are diagnosed. For this reason, one of the most important issues in clinical cancer research is to find early biomarkers of the tumorigenic process. In clear cell renal carcinoma, the miRNA signature distinguishes between metastatic and non-metastatic tumors. The 5-year survival rate of patients with primary metastasis is 10% as compared with 70–90% in non-metastatic patients (Heinzelmann et al., 2011), the miRNA signature being a powerful tool for early diagnosis. Moreover, analysis of circulating tumor-derived miRNAs represents a non-invasive means of diagnosing cancer (Kosaka et al., 2010). The utility of altered miRNA levels in serum has been noted in many cancer types. For example, serum levels of miRNA-141 can distinguish people with prostate cancer from healthy individuals (Mitchell et al., 2008). From an epigenetic point of view, there is a DNA methylation signature for human cancer metastasis (Lujambio et al., 2008). The authors analyzed the methylation profile of lymph node metastatic cancer cells and tumors, and found a relationship between specific miRNA hypermethylation patterns and the metastatic behavior of tumors. The use of miRNA gene hypermethylation to improve cancer diagnosis has been extended recently to many types of biological fluids, such as oral rinses (Langevin et al., 2010), feces (Kalimutho et al., 2011) and colorectal mucosal wash fluids (Kamimae et al., 2011).
miRNAs not only have diagnostic but also therapeutic potential. Inactivation of tumor-promoting miRNAs (Medina et al., 2010; Obad et al., 2011) or restoration of tumor-suppressor miRNAs (Saito et al., 2006; Lujambio et al., 2007, 2008) offers great prospects for cancer treatment. Activation of hypermethylated tumor-suppressor miRNAs can be accomplished by using chromatin-modifying drugs such as DNA methylation inhibitors. To date, two DNMT inhibitors, 5-azacytidine and 5-aza-2′-deoxycytidine, have been approved by the US Food and Drug Administration (FDA) for the treatment of myelodysplastic syndromes and acute myeloid leukemia (Rodríguez-Paredes and Esteller, 2011). Thus, part of the clinical benefit of these drugs might relate to their capacity to hypomethylate and reactivate previously silenced tumor-suppressor miRNAs. Finally, it is worth mentioning the novel area of reactivating miRNAs with low levels of expression in human tumors (Melo et al., 2011). In this approach, a small drug acts as a coactivator of the miRNA-processing machinery, particularly the aforementioned TRBP protein encoded by the TARBP2 gene, to enhance the production of mature miRNAs (Melo et al., 2011). Although it is not known whether this compound, or similar ones, can override the silencing belts imposed on the hypermethylated miRNAs, it is a pharmacological strategy that merits further attention.
Final perspective
From the candidate miRNA approach to whole-epigenome strategies, an increasing number of miRNAs undergoing DNA methylation-associated silencing in cancer cells have been described since the first reported instance in this area (Saito et al., 2006). Although the functional tumor-suppressor roles and a rigorous analysis of the promoter CpG island hypermethylation-related transcriptional repression have been accomplished for a few of them, such as miR-124a and miR-34b/c, from initial observations (Lujambio et al., 2007, 2008; Toyota et al., 2008) to subsequent independent studies (that is, Ando et al., 2009; Furuta et al., 2010; Vázquez et al., 2010; Wilting et al., 2010), much effort lies ahead of us. A good definition of the exact transcription start sites for many of the new candidates and a detailed characterization of their mRNA targets are needed. Most importantly, further cell and mouse models will be required to test their growth-inhibitory function. However, even now, the epigenetic inactivation of miRNAs with tumor-suppressor activities is recognized as a major hallmark of human tumors, probably at the same level as the promoter CpG island hypermethylation of classical coding tumor-suppressor genes (Jones and Baylin, 2007; Berdasco and Esteller, 2010). Their use as biomarkers of the disease and as potential therapy targets will develop and exciting developments in this area are expected in the near future.
Acknowledgments
This work was supported by grants SAF2007-00027-65134, Consolider CSD2006-49, the Lilly Foundation, the Dr Josef Steiner Cancer Research Foundation and the European Research Council Advanced Grant EPINORC.
The authors declare no conflict of interest.
References
- Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, Martin-Subero JI, Cordeu L, Gárate L, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Bahubeshi A, Bal N, Frio TR, Hamel N, Pouchet C, Yilmaz A, et al. Germline DICER1 mutations and familial cystic nephroma. J Med Genet. 2010;47:863–866. doi: 10.1136/jmg.2010.081216. [DOI] [PubMed] [Google Scholar]
- Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, et al. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bin Hafeez B, Adhami VM, Asim M, Siddiqui IA, Bhat KM, Zhong W, et al. Targeted knockdown of Notch1 inhibits invasion of human prostate cancer cells concomitant with inhibition of matrix metalloproteinase-9 and urokinase plasminogen activator. Clin Cancer Res. 2009;15:452–459. doi: 10.1158/1078-0432.CCR-08-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Gilmore BL, Spengler RM, Xing Y, Lanier W, Bhattacharya D, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. miR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKb by miR-199a affects NF-kB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Mor G. Inflammation, cancer and chemoresistance: taking advantage of the Toll-like receptor signaling pathway. Am J Reprod Immunol. 2007;57:93–107. doi: 10.1111/j.1600-0897.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. 2011;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- Cheung HH, Davis AJ, Lee TL, Pang AL, Nagrani S, Rennert OM, et al. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene. 2011;30:3404–3415. doi: 10.1038/onc.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31:745–750. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi-Garimella S, Strouch MJ, Grippo PJ, Bentrem DJ, Munshi HG. Collagen regulation of let-7 in pancreatic cancer involves TGF-β1-mediated membrane type 1-matrix metalloproteinase expression. Oncogene. 2011;30:1002–1008. doi: 10.1038/onc.2010.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H, Koizumi T, Sugimoto T, Chakraborty S, Ichimura T, Hasegawa K, et al. Quantitation of Fas and Fas ligand gene expression in human ovarian, cervical and endometrial carcinomas using real-time quantitative RT-PCR. Br J Cancer. 2000;82:1682–1688. doi: 10.1054/bjoc.2000.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, et al. 2011Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis Oncogenee-pub ahead of print 2 August 2011; doi: 10.1038/onc.2011.383 [DOI] [PMC free article] [PubMed]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- Gao XN, Lin J, Li YH, Gao L, Wang XR, Wang W, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30:3416–3428. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, et al. Epigenetic silencing of the intronic microRNA has-miR.342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossel MJ, Hinds PW. Beyond the cell cycle: a new role for Cdk6 in differentiation. J Cell Biochem. 2006;97:485–493. doi: 10.1002/jcb.20712. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Akiyama Y, Otsubo T, Shimada S, Yuasa Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31:777–784. doi: 10.1093/carcin/bgq013. [DOI] [PubMed] [Google Scholar]
- Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, et al. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, et al. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang Y, Guo Y, Sun S. Downregulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurteau GJ, Spivack SD, Brock GJ. Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT-PCR. Cell Cycle. 2006;5:1951–1956. doi: 10.4161/cc.5.17.3133. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, et al. Editing of Epstein–Barr-virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalimutho M, Di Cecilia S, Del Vecchio Blanco G, Roviello F, Sileri P, Cretella M, et al. Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br J Cancer. 2011;104:1770–1778. doi: 10.1038/bjc.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimae S, Yamamoto E, Yamano HO, Nojima M, Suzuki H, Ashida M, et al. Epigenetic alteration of DNA in mucosal wash fluid predicts invasiveness of colorectal tumors. Cancer Prev Res. 2011;4:674–683. doi: 10.1158/1940-6207.CAPR-10-0214. [DOI] [PubMed] [Google Scholar]
- Kindler T, Breitenbuecher F, Marx A, Beck J, Hess G, Weinkauf B, et al. Efficacy and safety of imatinib in adult patients with c-kit-positive acute myeloid leukemia. Blood. 2004;103:3644–3654. doi: 10.1182/blood-2003-06-2071. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krueger A, Fas SC, Baumann S, Krammer PH. The role of CD95 in the regulation of peripheral T-cell apoptosis. Immunol Rev. 2003;193:58–69. doi: 10.1034/j.1600-065x.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- Lai EC. MicroRNAs are complementary to 3′UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35–49. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31:864–870. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY., et al., , Bae DS. Altered microRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, et al. Epigenetic inactivation of microRNA gene has-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71:1550–1560. doi: 10.1158/0008-5472.CAN-10-2372. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A microRNA targeting Dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–89. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNA in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 22-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, Dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, et al. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviano AJ, Sun L, Ananthanarayanan V, Munshi HG. Extracellular matrix-mediated membrane-type 1 matrix metalloproteinase expression in pancreatic ductal cells is regulated by transforming growth facor-beta1. Cancer Res. 2006;66:7032–7040. doi: 10.1158/0008-5472.CAN-05-4421. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Pagano JS. Viruses and lymphomas. N Engl J Med. 2002;347:79–79. doi: 10.1056/NEJMp020056. [DOI] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli–Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Agirre X, Jiménez-Velasco A, Arqueros V, Vilas-Zornoza A, Rodriguez-Otero P, et al. Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J Clin Oncol. 2009;27:1316–1322. doi: 10.1200/JCO.2008.19.3441. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer. Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn HA, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450 000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Vandenberg SR, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BCM Med. 2008;6:14–31. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32:772–778. doi: 10.1093/carcin/bgr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tan HX, Wang Q, Chen LZ, Huang XH, Chen JS, Fu XH, et al. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol. 2010;27:645–660. doi: 10.1007/s12032-009-9264-2. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, et al. 2011Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer Int J Cancer(in press). [DOI] [PubMed]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Fernandez AF, Lopez-Nieva P, Rossi S, Huertas D, Kulis M, et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics. 2010;5:656–663. doi: 10.4161/epi.5.7.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vázquez I, Maicas M, Marcotegui N, Conchillo A, Guruceaga E, Roman-Gomez J, et al. Silencing of hsa-miR-124 by EVI1 in cell lines and patients with acute myeloid leukemia. Proc Natl Acad Sci USA. 2010;107:167–168. doi: 10.1073/pnas.1011540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression profile signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tang Y, Cui H, Zhao X, Luo X, Pan W, et al. Let-7/miR-98 regulate Fas and Fas-mediated apoptosis. Genes Immun. 2011;12:149–154. doi: 10.1038/gene.2010.53. [DOI] [PubMed] [Google Scholar]
- Wendler A, Keller D, Albrecht C, Peluso JJ, Wehling M. Involvement of let-7/miR-98 microRNAs in the regulation of progesterone receptor membrane component 1 expression in ovarian cancer cells. Oncol Rep. 2011;25:273–279. [PubMed] [Google Scholar]
- Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2010;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- Willis L, Alarcón T, Elia G, Jones JL, Wright NA, Tomlinson IPM, et al. Breast cancer dormancy can be maintained by small numbers of micrometastases. Cancer Res. 2010;70:4310–4317. doi: 10.1158/0008-5472.CAN-09-3144. [DOI] [PubMed] [Google Scholar]
- Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167–181. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2 and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein C. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, et al. TWIST Ting stemness, inflammation and proliferation of epithelial ovarian cell through MIR199A2/214. Oncogene. 2010;29:3545–3553. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CC, Chen YW, Chiou GY, Tsai LL, Huang PI, Chang CY, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. Oral Oncol. 2011;47:202–210. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Glannakakis A, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]