Abstract
N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated ion channels highly permeable to calcium and essential to excitatory neurotransmission. The NMDARs have attracted much attention because of their role in synaptic plasticity and excitotoxicity. Evidence has recently accumulated that NMDARs are negatively regulated by intracellular calcium binding proteins. The calcium-dependent suppression of NMDAR function serves as a feedback mechanism capable of regulating subsequent Ca2+ entry into the postsynaptic cell, and may offer an alternative approach to treating NMDAR-mediated excitotoxic injury. This short review summarizes the recent progress made in understanding the negative modulation of NMDAR function by DREAM/calsenilin/KChIP3, a neuronal calcium sensor (NCS) protein.
Keywords: calcium, excitotoxicity, neuroprotection, NMDA, NR1, NR2B, glutamate, neuronal calcium sensor (NCS) proteins
Introduction
Glutamate functions as the major excitatory neurotransmitter by binding to N-methyl-D-aspartate receptors (NMDARs) that are widespread in the central nervous system. The NMDARs constitute a major class of ionotropic glutamate receptors and play an essential role in synaptic transmission, plasticity, and memory. Activation of NMDARs results in cell membrane depolarization with an equilibrium potential near 0 mV, producing the excitatory postsynaptic potential (EPSP) and leading to an increase of Ca2+ influx into the cell. The intracellular Ca2+ can in turn function as a second messenger, mediating a variety of signaling cascades. Excessive activation of NMDARs by glutamate mediates neuronal damage in many neurological disorders including ischemia and neurodegenerative diseases (Choi et al., 1988; Sattler and Tymianski, 2001).
The NMDARs have long been considered the main target for the treatment of excitotoxicity-related neuronal injury, and a variety of antagonists or blockers of NMDARs have been developed. Unfortunately, the results of clinical trials have been disappointing because of the obvious side effects associated with blocking the physiological roles of NMDARs (Chen and Lipton, 2006). Therefore, a better understanding of the mechanism of how NMDARs can be modulated by regulatory proteins should help in the development of new therapeutic agents to counteract overactive NMDA receptor function, and may represent an alternative to treating NMDAR-mediated excitotoxic injury. This short review focuses on the specific negative modulation of NMDARs by a neuronal calcium sensor (NCS) protein, DREAM/calsenilin/KChIP3.
Structural and functional features of NMDA receptor channels
NMDARs are believed to be heterotetrameric complexes composed of combinations of the obligatory NR1 subunit and NR2 and/or NR3 subunits (Chazot and Stephenson, 1997; Laube et al., 1998; Schorge and Colquhoun, 2003; Furukawa et al., 2005). The NR1 subunit is encoded by a single gene but exists as eight functional splice variants, while the NR2 (NR2A-B) and NR3 (NR3A-B) subunits are encoded by four and two different genes, respectively. The NMDAR subunits form a central ion conductance pathway selective for cations such as Na+, K+, and Ca2+, and share a common membrane topology, with each subunit consisting of four transmembrane (TM) domains (M1–M4). The long extracellular N-terminal regions of NMDAR subunits are organized as a tandem of two domains. The first domain, called the N-terminal domain (NTD) that includes the first 380 amino acids, is involved in tetrameric assembly (Mayer, 2006; Paoletti and Neyton, 2007; Stroebel et al., 2011). The second domain of about 300 amino acids is known as the agonist-binding domain (ABD) that precedes the TM1 domain. The ABD binds glycine (or D-serine) in the NR1 and NR3 subunits, whereas the NR2 ABD binds glutamate (Furukawa et al., 2005; Yao and Mayer, 2006). The pore loop (P loop), or the M2 region, forms the narrowest constriction of the channel ion conductance pathway and determines the permeation properties of NMDARs. The NMDARs feature an intracellular C-terminal tail of about 400–600 residues that has a strong diversity in its amino acid sequence. The C-terminal tails of NMDAR subunits contain a series of short motifs that interact with intracellular factors or binding partners involved in receptor trafficking, anchoring and signaling (Skeberdis et al., 2006; Ryan et al., 2008; Lau et al., 2010).
Activation of NMDARs requires a simultaneous binding of two co-agonists, glutamate, and glycine with different biophysical properties of ion permeation. The typical NMDARs contain NR2 subunits with properties of high permeability to Ca2+ and extracellular Mg2+ block at hyperpolarized membrane potentials (Wrighton et al., 2008; Singh et al., 2012). Different from conventional NR1/NR2 heterotetramers, NR3-containing NMDARs have unique properties with a five to tenfold decrease of Ca2+ permeability, insensitivity to Mg2+ block, and reduced single-channel conductance and open probability, functioning as a negative modulator for NMDA receptor channel function (Das et al., 1998; Sasaki et al., 2002; Nakanishi et al., 2009; Cavara et al., 2010; Henson et al., 2010).
Modulation of NMDAR function by intracellular binding partners
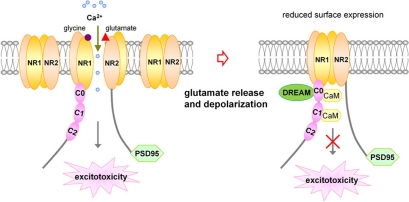
NMDA receptors are also regulated by other intracellular signals and proteins, including calcium, protein kinases, protein phosphatase calcineurin, and calcium-sensitive proteins such as calmodulin (Legendre et al., 1993; Vyklicky, 1993; Lieberman and Mody, 1994; Tong et al., 1995; Ehlers et al., 1996). Calcium-dependent NMDA receptor desensitization and inactivation provides a feedback mechanism capable of regulating subsequent Ca2+ entry into the postsynaptic cell through NMDA channels (Figure 1).
Figure 1.
Schematic representation for inhibitory effect of DREAM/calsenilin/KChIP3 on NMDARs in a Ca2+-sensitive manner. Upon activation of NMDARs by glutamate binding, Ca2+ influx through NMDARs increases the association between DREAM and NR1 subunits, resulting in reduced surface expression of NMDARs, and subsequent inhibition of NMDARs-mediated Ca2+ influx and excitotoxicity. DREAM functions as a Ca2+-sensitive modulator for the negative feedback control of NMDAR function.
So far, a number of NR1 or NR2 subunit binding partners have been identified in the postsynaptic density. The NR1 binding proteins include calmodulin (CaM) (Ehlers et al., 1996; Akyol et al., 2004), Ca2+/CaM-dependent protein kinase II (CaMKII) (Leonard et al., 2002), α-actinin (Wyszynski et al., 1997; Merrill et al., 2007), tubulin (van Rossum et al., 1999), spectrin (Wechsler and Teichberg, 1998), neurofilament (Ehlers et al., 1998), and Yotiao (Lin et al., 1998). Calmodulin binding to the NR1 subunit is Ca2+ dependent and occurs with homomeric NR1 complexes, heteromeric NR1/NR2 subunit complexes from expression systems, and NMDA receptors from the brain. Calmodulin binding to NR1 causes a fourfold reduction in NMDA channel open probability, mediating the negative modulation of NMDAR function (Ehlers et al., 1998).
DREAM/calsenilin/KChIP3, a neuronal calcium sensor and calcium binding EF-hand protein
DREAM/calsenilin/KChIP3 is encoded by the same gene locus. The downstream regulatory element antagonist modulator (DREAM) protein, first identified in the nucleus as a Ca2+-regulated transcriptional repressor through its binding to DNA at specific regulatory elements, contains four Ca2+-binding EF-hand domains and belongs to the NCS family (Carrion et al., 1999; Burgoyne, 2007). DREAM was named for its ability to block gene expression in its Ca2+-free form via direct binding with the downstream regulatory element (DRE) sequence in target genes such as preprodynorphin (PPD), c-fos, Hrk, Na+, and Ca2+ exchanger NCX3 (Carrion et al., 1999; Sanz et al., 2001; Gomez-Villafuertes et al., 2005). DREAM was also named calsenilin or Kv channel interacting protein 3 (KChIP3) (Buxbaum et al., 1998; An et al., 2000), indicating that DREAM/calsenilin/KChIP3 has multifunctional properties. In the nucleus the DREAM protein functions as a dimer, whereas outside the nucleus KChIP3 is a monomer and regulates the surface expression and gating kinetics of Kv4 channels (An et al., 2000; Kim and Sheng, 2004; Scannevin et al., 2004; Pioletti et al., 2006; Wang et al., 2007; Wang, 2008).
DREAM/calsenilin/KChIP3 is preferentially expressed in the central nervous system, as well as in non-neuronal tissues (Link et al., 2004; D'Andrea et al., 2005; Savignac et al., 2005). DREAM/calsenilin/KChIP3 knock-out mice display a hypoalgesic phenotype, suggesting a critical role of DREAM/calsenilin/KChIP3 in pain modulation (Cheng et al., 2002). In addition, emerging evidence reveals the role of DREAM/calsenilin/KChIP3 in long-term potentiation (LTP) (Lilliehook et al., 2003) and learning and memory (Alexander et al., 2009; Fontan-Lozano et al., 2009), suggesting a possible connection between DREAM and NMDA function.
KChIP1–3 were initially identified from a rat brain library in yeast two-hybrid (YTH) screens using the cytoplasmic N-terminal domain (amino acids 1–180) of rat Kv4.3 as a bait (An et al., 2000). Similarly, KChIP4 from mouse and human was accidentally cloned using the C-terminal 43 amino acid residues of presenilin 2 (PS2, amino acids 406–448) as a bait in the YTH system (Morohashi et al., 2002). KChIP4, also known as calsenilin-like protein (CALP), binds to PS2 which is known to facilitate intramembranous γ-cleavage of γ-amyloid protein precursor (βAPP) (Morohashi et al., 2002).
KChIP1–4 (216 ∼ 256 amino acids) can co-immunoprecipitate and co-localize with either Kv4 from co-transfected cells or Kv4 α-subunits from tissues, and thus constitute integral components of native Kv4 channel complexes (Wang, 2008). KChIP1–4 all share a conserved carboxy-terminal core region that contains four EF-hand-like calcium binding motifs, but have a variable amino-terminal region that causes diverse modulation of Kv4 trafficking and channel function (An et al., 2000; Holmqvist et al., 2002; Scannevin et al., 2004; Cui et al., 2008; Liang et al., 2009, 2010).
Functional interactions between DREAM and NMDA receptors
We and others have investigated mechanisms underlying the functional interactions between DREAM and NMDARs. Findings from co-immunoprecipitation experiments show that DREAM antibody can immunoprecipitate endogenous NR1 subunit and DREAM protein from rat hippocampal tissue (Zhang et al., 2010). In the reciprocal co-IP studies in HEK 293 cells expressing DREAM and NR1-1a (NR1a) proteins, NR1 antibody can also immunoprecipitate DREAM along with the NR1 subunit. GST pull-down assays reveal that the N-terminus of DREAM directly interacts with the NR1a C-terminus, and that the DREAM-NR1 interaction is sensitive to Ca2+ and depends on the EF hand domains of DREAM (Zhang et al., 2010).
PSD-95 is a major scaffolding protein in the postsynaptic density, tethering NMDARs to signaling proteins, and is critical for NMDA receptor function (Kim and Sheng, 2004). Wu et al. generated a line of transgenic mice (TgDREAM) overexpressing a dominant active DREAM mutant, and compared NMDA receptor-mediated EPSCs in TgDREAM and wild-type mice under conditions of various stimulation intensities (Wu et al., 2010). They found that the amplitude of NMDA receptor-mediated EPSCs in TgDREAM mice is significantly reduced compared to that in wild-type mice (Wu et al., 2010). In addition, LTD is significantly reduced in TgDREAM mice whereas LTP is not affected by DREAM, demonstrating that DREAM interacts with PSD-95, and that the interaction is negatively regulated by calcium (Wu et al., 2010). In Xenopus oocytes expressing NR2B-containing NMDARs alone or together with DREAM, two-electrode voltage clamp recordings show that, in the absence of DREAM, the peak currents of NMDA channels activated by glutamate (plus glycine) are suppressed by DREAM, and the current decrease is caused by a reduction in the density of NMDARs at the cell surface (Figure 1; Zhang et al., 2010).
Fontan-Lozano et al. recently provided another piece of evidence that DREAM negatively regulates the function of NMDA receptors (Fontan-Lozano et al., 2011). By taking advantage of mice lacking the DREAM protein, they demonstrated that the facilitated learning induced by decreased expression of Kv4.2 in dream−/− mice requires the activation of NMDA receptors containing the NR2B subunit (Fontan-Lozano et al., 2011). This study not only indicates the significance of the balance between Kv4 channel function and NMDAR activity, but also suggests the formation of a functional complex between DREAM/Kv4.2/NMDARs that regulates the synaptic efficacy mediating synaptic plasticity and learning.
Neuroprotective effect of DREAM/calsenilin/KChIP3 over-expression on neuronal excitotoxic injury
Excitotoxicity is caused by overactivation of NMDA receptor function, and inhibition of NMDARs can reverse the neuronal toxicity. The data available so far support both the pro-apoptotic and anti-apoptotic roles of DREAM. In general, the pro-apoptotic role of DREAM closely correlates with its interaction with presenilins, the production of amyloid beta (Aβ) and the modulation in Ca2+ signaling, whereas the anti-apoptotic role of DREAM is conferred by its transcriptional repressor activity on the apoptotic protein Hrk. Jo et al. reported that HeLa cells transiently transfected with DREAM exhibit the morphological and biochemical features of apoptosis and that expression of presenilin potentiates DREAM-induced apoptosis (Jo et al., 2001). Jo et al. also reported that DREAM expression increases in either human neuroblastoma SK-N-BE2(c) cells or rat neuroblastoma B103 cells after exposure to Aβ, but no other apoptotic inducers such as staurosporine, thapsigargin, and calcium ionophore A23187. The pro-apoptotic role of DREAM is selectively induced during Ab toxicity. Because of the involvement of presenilins/γ-secretase in Aβ formation and neuronal death, DREAM coordinates with presenilin activity to play a crucial role in these processes through binding with the C-terminus of presenilins (Jo et al., 2003, 2004). Lilliehook et al. stably expressed DREAM in H4 neuroglioma cells which showed no initiation of apoptosis in the absence of apoptosis triggers, and the apoptosis-associated caspase and calpain activities were not affected by DREAM (Lilliehook et al., 2002). On the other hand, binding of the transcriptional repressor DREAM to the hrk gene avoids inappropriate Hrk expression and apoptosis in hematopoietic progenitor cell lines (Sanz et al., 2001, 2002). Nevertheless, the precise function of DREAM in pro-apoptosis and anti-apoptosis remains to be explored.
From our previous observations, we noticed that cell viability is affected by the amount of exogenous DREAM gene and the method of transfection. However, we could not observe any obvious morphological changes associated with cell death after the transfection of DREAM. To prove the cytoprotective role of DREAM, we utilized cell lines and primary cultured hippocampal neurons with DREAM overexpression or siRNA-mediated knockdown, and evaluated lactate dehydrogenase (LDH) leakage and propidium iodide (PI) uptake both in NMDA and oxygen-glucose deprivation (OGD) -induced excitotoxic injury models (Zhang et al., 2010). Administration of NMDA markedly increases the number of PI-positive cells (dead cells) in NMDAR-transfected CHO cells, whereas co-expression of DREAM greatly reduces PI-positive cells (Zhang et al., 2010). LDH leakage measurement is a sensitive index reflecting the extent of cell damage. Over-expression of DREAM suppresses the NMDA-induced LDH release. OGD is commonly used in vitro to mimic ischemia-reperfusion insult to the brain, and OGD treatment induces a significant increase of LDH release (Dawson et al., 1994). With over-expression of DREAM, however, OGD treatment induces a smaller increase in LDH release. These results indicate that the over-expression of DREAM attenuates NMDAR-mediated excitotoxicity (Zhang et al., 2010).
We have previously tested the effect of DREAM siRNA on NMDA-induced current and excitotoxic injury in hippocampal neurons (Zhang et al., 2010). Knockdown of endogenous DREAM with siRNA results in an increased amplitude of NMDA current recorded by whole-cell patch-clamp assays. DREAM siRNA also significantly exacerbates NMDA-induced cell death in hippocampal neurons. After NMDA exposure, PI-positive cells in the DREAM siRNA group increase compared with control siRNA, indicating the inhibitory effect of DREAM on NMDAR-mediated current and excitotoxic injury (Zhang et al., 2010).
Conclusions
The NCS protein DREAM/calsenilin/KChIP3 acts as an auxiliary subunit and suppresses NMDA receptor channel function. This negative modulation of NMDA receptor function by DREAM likely provides a feedback mechanism by which overactive NMDA receptors are inhibited. Therefore, targeting regulatory proteins of NMDARs may represent an alternative approach to treating NMDAR-mediated excitotoxic damage and providing neuroprotection.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the members of our laboratories for their comments and Greg Vatcher for his careful proof-reading of this manuscript. This work was supported by research grants from the National Science Foundation of China to KeWei Wang (30970919) and to Yun Wang (81161120497, 30830044, and 30925015).
References
- Akyol Z., Bartos J. A., Merrill M. A., Faga L. A., Jaren O. R., Shea M. A., Hell J. W. (2004). Apo-calmodulin binds with its C-terminal domain to the N-methyl- D-aspartate receptor NR1 C0 region. J. Biol. Chem. 279, 2166–2175 10.1074/jbc.M302542200 [DOI] [PubMed] [Google Scholar]
- Alexander J. C., McDermott C. M., Tunur T., Rands V., Stelly C., Karhson D., Bowlby M. R., An W. F., Sweatt J. D., Schrader L. A. (2009). The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn. Mem. 16, 167–177 10.1101/lm.1261709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000). Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 10.1038/35000592 [DOI] [PubMed] [Google Scholar]
- Burgoyne R. D. (2007). Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193 10.1038/nrn2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum J. D., Choi E. K., Luo Y., Lilliehook C., Crowley A. C., Merriam D. E., Wasco W. (1998). Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 4, 1177–1181 10.1038/2673 [DOI] [PubMed] [Google Scholar]
- Carrion A. M., Link W. A., Ledo F., Mellstrom B., Naranjo J. R. (1999). DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80–84 10.1038/18044 [DOI] [PubMed] [Google Scholar]
- Cavara N. A., Orth A., Hicking G., Seebohm G., Hollmann M. (2010). Residues at the tip of the pore loop of NR3B-containing NMDA receptors determine Ca2+ permeability and Mg2+ block. BMC Neurosci. 11, 133 10.1186/1471-2202-11-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazot P. L., Stephenson F. A. (1997). Molecular dissection of native mammalian forebrain NMDA receptors containing the NR1 C2 exon: direct demonstration of NMDA receptors comprising NR1, NR2A, and NR2B subunits within the same complex. J. Neurochem. 69, 2138–2144 [DOI] [PubMed] [Google Scholar]
- Chen H. S., Lipton S. A. (2006). The chemical biology of clinically tolerated NMDA receptor antagonists. J. Neurochem. 97, 1611–1626 10.1111/j.1471-4159.2006.03991.x [DOI] [PubMed] [Google Scholar]
- Cheng H. Y., Pitcher G. M., Laviolette S. R., Whishaw I. Q., Tong K. I., Kockeritz L. K., Wada T., Joza N. A., Crackower M., Goncalves J., Sarosi I., Woodgett J. R., Oliveirados-Santos A. J., Ikura M., van der Kooy D., Salter M. W., Penninger J. M. (2002). DREAM is a critical transcriptional repressor for pain modulation. Cell 108, 31–43 10.1016/S0092-8674(01)00629-8 [DOI] [PubMed] [Google Scholar]
- Choi D. W., Koh J. Y., Peters S. (1988). Pharmacology of glutamate neurotoxicity in cortical cell culture: attenuation by NMDA antagonists. J. Neurosci. 8, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y. Y., Liang P., Wang K. W. (2008). Enhanced trafficking of tetrameric Kv4.3 channels by KChIP1 clamping. Neurochem. Res. 33, 2078–2084 10.1007/s11064-008-9688-7 [DOI] [PubMed] [Google Scholar]
- D'Andrea B., Di Palma T., Mascia A., Motti M. L., Viglietto G., Nitsch L., Zannini M. (2005). The transcriptional repressor DREAM is involved in thyroid gene expression. Exp. Cell Res. 305, 166–178 10.1016/j.yexcr.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Das S., Sasaki Y. F., Rothe T., Premkumar L. S., Takasu M., Crandall J. E., Dikkes P., Conner D. A., Rayudu P. V., Cheung W., Chen H. S., Lipton S. A., Nakanishi N. (1998). Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393, 377–381 10.1038/30748 [DOI] [PubMed] [Google Scholar]
- Dawson T. M., Zhang J., Dawson V. L., Snyder S. H. (1994). Nitric oxide: cellular regulation and neuronal injury. Prog. Brain Res. 103, 365–369 [DOI] [PubMed] [Google Scholar]
- Ehlers M. D., Fung E. T., O'Brien R. J., Huganir R. L. (1998). Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J. Neurosci. 18, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M. D., Zhang S., Bernhadt J. P., Huganir R. L. (1996). Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell 84, 745–755 10.1016/S0092-8674(00)81052-1 [DOI] [PubMed] [Google Scholar]
- Fontan-Lozano A., Romero-Granados R., del-Pozo-Martin Y., Suarez-Pereira I., Delgado-Garcia J. M., Penninger J. M., Carrion A. M. (2009). Lack of DREAM protein enhances learning and memory and slows brain aging. Curr. Biol. 19, 54–60 10.1016/j.cub.2008.11.056 [DOI] [PubMed] [Google Scholar]
- Fontan-Lozano A., Suarez-Pereira I., Gonzalez-Forero D., Carrion A. M. (2011). The A-current modulates learning via NMDA receptors containing the NR2B subunit. PLoS One 6:e24915 10.1371/journal.pone.0024915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H., Singh S. K., Mancusso R., Gouaux E. (2005). Subunit arrangement and function in NMDA receptors. Nature 438, 185–192 10.1038/nature04089 [DOI] [PubMed] [Google Scholar]
- Gomez-Villafuertes R., Torres B., Barrio J., Savignac M., Gabellini N., Rizzato F., Pintado B., Gutierrez-Adan A., Mellstrom B., Carafoli E., Naranjo J. R. (2005). Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J. Neurosci. 25, 10822–10830 10.1523/JNEUROSCI.3912-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson M. A., Roberts A. C., Perez-Otano I., Philpot B. D. (2010). Influence of the NR3A subunit on NMDA receptor functions. Prog. Neurobiol. 91, 23–37 10.1016/j.pneurobio.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist M. H., Cao J., Hernandez-Pineda R., Jacobson M. D., Carroll K. I., Sung M. A., Betty M., Ge P., Gilbride K. J., Brown M. E., Jurman M. E., Lawson D., Silos-Santiago I., Xie Y., Covarrubias M., Rhodes K. J., Distefano P. S., An W. F. (2002). Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. U.S.A. 99, 1035–1040 10.1073/pnas.022509299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo D. G., Chang J. W., Hong H. S., Mook-Jung I., Jung Y. K. (2003). Contribution of presenilin/gamma-secretase to calsenilin-mediated apoptosis. Biochem. Biophys. Res. Commun. 305, 62–66 10.1016/S0006-291X(03)00688-0 [DOI] [PubMed] [Google Scholar]
- Jo D. G., Lee J. Y., Hong Y. M., Song S., Mook-Jung I., Koh J. Y., Jung Y. K. (2004). Induction of pro-apoptotic calsenilin/DREAM/KChIP3 in Alzheimer's disease and cultured neurons after amyloid-beta exposure. J. Neurochem. 88, 604–611 [DOI] [PubMed] [Google Scholar]
- Jo D. G., Kim M. J., Choi Y. H., Kim I. K., Song Y. H., Woo H. N., Chung C. W., Jung Y. K. (2001). Pro-apoptotic function of calsenilin/DREAM/KChIP3. FASEB J. 15, 589–591 10.1096/fj.00-0541fje [DOI] [PubMed] [Google Scholar]
- Kim E., Sheng M. (2004). PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771–781 10.1038/nrn1517 [DOI] [PubMed] [Google Scholar]
- Lau C. G., Takayasu Y., Rodenas-Ruano A., Paternain A. V., Lerma J., Bennett M. V., Zukin R. S. (2010). SNAP-25 is a target of protein kinase C phosphorylation critical to NMDA receptor trafficking. J. Neurosci. 30, 242–254 10.1523/JNEUROSCI.4933-08.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube B., Kuhse J., Betz H. (1998). Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 18, 2954–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Rosenmund C., Westbrook G. L. (1993). Inactivation of NMDA channels in cultured hippocampal neurons by intracellular calcium. J. Neurosci. 13, 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. S., Bayer K. U., Merrill M. A., Lim I. A., Shea M. A., Schulman H., Hell J. W. (2002). Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J. Biol. Chem. 277, 48441–48448 10.1074/jbc.M205164200 [DOI] [PubMed] [Google Scholar]
- Liang P., Chen H., Cui Y., Lei L., Wang K. (2010). Functional rescue of Kv4.3 channel tetramerization mutants by KChIP4a. Biophys. J. 98, 2867–2876 10.1016/j.bpj.2010.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Wang H., Chen H., Cui Y., Gu L., Chai J., Wang K. (2009). Structural Insights into KChIP4a Modulation of Kv4.3 Inactivation. J. Biol. Chem. 284, 4960–4967 10.1074/jbc.M807704200 [DOI] [PubMed] [Google Scholar]
- Lieberman D. N., Mody I. (1994). Regulation of NMDA channel function by endogenous Ca(2+)-dependent phosphatase. Nature 369, 235–239 10.1038/369235a0 [DOI] [PubMed] [Google Scholar]
- Lilliehook C., Bozdagi O., Yao J., Gomez-Ramirez M., Zaidi N. F., Wasco W., Gandy S., Santucci A. C., Haroutunian V., Huntley G. W., Buxbaum J. D. (2003). Altered Abeta formation and long-term potentiation in a calsenilin knock-out. J. Neurosci. 23, 9097–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilliehook C., Chan S., Choi E. K., Zaidi N. F., Wasco W., Mattson M. P., Buxbaum J. D. (2002). Calsenilin enhances apoptosis by altering endoplasmic reticulum calcium signaling. Mol. Cell. Neurosci. 19, 552–559 10.1006/mcne.2001.1096 [DOI] [PubMed] [Google Scholar]
- Lin J. W., Wyszynski M., Madhavan R., Sealock R., Kim J. U., Sheng M. (1998). Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 18, 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W. A., Ledo F., Torres B., Palczewska M., Madsen T. M., Savignac M., Albar J. P., Mellstrom B., Naranjo J. R. (2004). Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J. Neurosci. 24, 5346–5355 10.1523/JNEUROSCI.1460-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. L. (2006). Glutamate receptors at atomic resolution. Nature 440, 456–462 10.1038/nature04709 [DOI] [PubMed] [Google Scholar]
- Merrill M. A., Malik Z., Akyol Z., Bartos J. A., Leonard A. S., Hudmon A., Shea M. A., Hell J. W. (2007). Displacement of alpha-actinin from the NMDA receptor NR1 C0 domain By Ca2+/calmodulin promotes CaMKII binding. Biochemistry 46, 8485–8497 10.1021/bi0623025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi Y., Hatano N., Ohya S., Takikawa R., Watabiki T., Takasugi N., Imaizumi Y., Tomita T., Iwatsubo T. (2002). Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. J. Biol. Chem. 277, 14965–14975 10.1074/jbc.M200897200 [DOI] [PubMed] [Google Scholar]
- Nakanishi N., Tu S., Shin Y., Cui J., Kurokawa T., Zhang D., Chen H. S., Tong G., Lipton S. A. (2009). Neuroprotection by the NR3A subunit of the NMDA receptor. J. Neurosci. 29, 5260–5265 10.1523/JNEUROSCI.1067-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P., Neyton J. (2007). NMDA receptor subunits: function and pharmacology. Curr. Opin. Pharmacol. 7, 39–47 10.1016/j.coph.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Pioletti M., Findeisen F., Hura G. L., Minor D. L. Jr. (2006). Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 13, 987–995 10.1038/nsmb1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan T. J., Emes R. D., Grant S. G., Komiyama N. H. (2008). Evolution of NMDA receptor cytoplasmic interaction domains: implications for organisation of synaptic signalling complexes. BMC Neurosci. 9, 6 10.1186/1471-2202-9-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz C., Horita M., Fernandez-Luna J. L. (2002). Fas signaling and blockade of Bcr-Abl kinase induce apoptotic Hrk protein via DREAM inhibition in human leukemia cells. Haematologica 87, 903–907 [PubMed] [Google Scholar]
- Sanz C., Mellstrom B., Link W. A., Naranjo J. R., Fernandez-Luna J. L. (2001). Interleukin 3-dependent activation of DREAM is involved in transcriptional silencing of the apoptotic Hrk gene in hematopoietic progenitor cells. EMBO J. 20, 2286–2292 10.1093/emboj/20.9.2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y. F., Rothe T., Premkumar L. S., Das S., Cui J., Talantova M. V., Wong H. K., Gong X., Chan S. F., Zhang D., Nakanishi N., Sucher N. J., Lipton S. A. (2002). Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J. Neurophysiol. 87, 2052–2063 10.1152/jn.00531.2001 [DOI] [PubMed] [Google Scholar]
- Sattler R., Tymianski M. (2001). Molecular mechanisms of glutamate receptor-mediated excitotoxic neuronal cell death. Mol. Neurobiol. 24, 107–129 10.1385/MN:24:1-3:107 [DOI] [PubMed] [Google Scholar]
- Savignac M., Pintado B., Gutierrez-Adan A., Palczewska M., Mellstrom B., Naranjo J. R. (2005). Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 24, 3555–3564 10.1038/sj.emboj.7600810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin R. H., Wang K., Jow F., Megules J., Kopsco D. C., Edris W., Carroll K. C., Lü Q., Xu W., Xu Z., Katz A. H., Olland S., Lin L., Taylor M., Stahl M., Malakian K., Somers W., Mosyak L., Bowlby M. R., Chanda P., Rhodes K. J. (2004). Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1. Neuron 41, 587–598 [DOI] [PubMed] [Google Scholar]
- Schorge S., Colquhoun D. (2003). Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J. Neurosci. 23, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Doshi S., Spaethling J. M., Hockenberry A. J., Patel T. P., Geddes-Klein D. M., Lynch D. R., Meaney D. F. (2012). N-methyl-D-aspartate receptor mechanosensitivity is governed by C terminus of NR2B subunit. J. Biol. Chem. 287, 4348–4359 10.1074/jbc.M111.253740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeberdis V. A., Chevaleyre V., Lau C. G., Goldberg J. H., Pettit D. L., Suadicani S. O., Lin Y., Bennett M. V., Yuste R., Castillo P. E., Zukin R. S. (2006). Protein kinase A regulates calcium permeability of NMDA receptors. Nat. Neurosci. 9, 501–510 10.1038/nn1664 [DOI] [PubMed] [Google Scholar]
- Stroebel D., Carvalho S., Paoletti P. (2011). Functional evidence for a twisted conformation of the NMDA receptor GluN2A subunit N-terminal domain. Neuropharmacology 60, 151–158 10.1016/j.neuropharm.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Tong G., Shepherd D., Jahr C. E. (1995). Synaptic desensitization of NMDA receptors by calcineurin. Science 267, 1510–1512 10.1126/science.7878472 [DOI] [PubMed] [Google Scholar]
- van Rossum D., Kuhse J., Betz H. (1999). Dynamic interaction between soluble tubulin and C-terminal domains of N-methyl-D-aspartate receptor subunits. J. Neurochem. 72, 962–973 [DOI] [PubMed] [Google Scholar]
- Vyklicky L. Jr. (1993). Calcium-mediated modulation of N-methylD-aspartate (NMDA) responses in cultured rat hippocampal neurones. J. Physiol. 470, 575–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. (2007). Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat. Neurosci. 10, 32–39 10.1038/nn1822 [DOI] [PubMed] [Google Scholar]
- Wang K. (2008). Modulation by clamping: Kv4 and KChIP interactions. Neurochem. Res. 33, 1964–1969 10.1007/s11064-008-9705-x [DOI] [PubMed] [Google Scholar]
- Wechsler A., Teichberg V. I. (1998). Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. EMBO J. 17, 3931–3939 10.1093/emboj/17.14.3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton D. C., Baker E. J., Chen P. E., Wyllie D. J. (2008). Mg2+ and memantine block of rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J. Physiol. 586, 211–225 10.1113/jphysiol.2007.143164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. J., Mellstrom B., Wang H., Ren M., Domingo S., Kim S. S., Li X. Y., Chen T., Naranjo J. R., Zhuo M. (2010). DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 3, 3 10.1186/1756-6606-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M., Lin J., Rao A., Nigh E., Beggs A. H., Craig A. M., Sheng M. (1997). Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature 385, 439–442 10.1038/385439a0 [DOI] [PubMed] [Google Scholar]
- Yao Y., Mayer M. L. (2006). Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J. Neurosci. 26, 4559–4566 10.1523/JNEUROSCI.0560-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su P., Liang P., Liu T., Liu X., Liu X. Y., Zhang B., Han T., Zhu Y. B., Yin D. M., Li J., Zhou Z., Wang K. W., Wang Y. (2010). The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J. Neurosci. 30, 7575–7586 10.1523/JNEUROSCI.1312-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]